Abstract
Leukocytes are central regulators of inflammation and the target cells of therapies for key diseases, including autoimmune, cardiovascular, and malignant disorders. Efficient in vivo delivery of small interfering RNA (siRNA) to immune cells could thus enable novel treatment strategies with broad applicability. In this report, we develop systemic delivery methods of siRNA encapsulated in lipid nanoparticles (LNP) for durable and potent in vivo RNA interference (RNAi)-mediated silencing in myeloid cells. This work provides the first demonstration of siRNA-mediated silencing in myeloid cell types of nonhuman primates (NHPs) and establishes the feasibility of targeting multiple gene targets in rodent myeloid cells. The therapeutic potential of these formulations was demonstrated using siRNA targeting tumor necrosis factor-α (TNFα) which induced substantial attenuation of disease progression comparable to a potent antibody treatment in a mouse model of rheumatoid arthritis (RA). In summary, we demonstrate a broadly applicable and therapeutically relevant platform for silencing disease genes in immune cells.
Keywords: delivery, immune cell, siRNA
Introduction
The discovery and optimization of RNA interference (RNAi) technologies over the past 10 years has made it possible to routinely develop in vitro picomolar (pM) small interfering RNA (siRNA) inhibitors to any chosen gene within a matter of weeks.1 Significant progress has also been made, through the incorporation of straightforward chemical modifications, in introducing drug-like properties into siRNA duplexes, resulting in improved nuclease stability and reduced immunostimulatory capacity.1,2
While numerous approaches for in vivo siRNA silencing have been reported involving both local and systemic delivery, robust and repeated success with commercially viable formulations has been limited.3 Systemic delivery of siRNA has resulted in silencing of liver-specific genes using several different classes of lipid nanoparticle (LNP)-based formulations. Broadly speaking, LNPs can be classified into those containing cationic or ionizable lipids as their main active component and differ in both their structure and mechanism of uptake.4,5 While both cationic and ionizable lipids can be formulated into LNPs with the same size and encapsulation properties, “ionizable” lipids maintain a nearly neutral charge at physiological pH while “cationic” lipids have an overall slightly positive charge. Recently, in vivo hepatocyte gene silencing has been improved by roughly two orders of magnitude for both classes of LNPs through identification of novel lipids and formulation optimization. Early reports demonstrated efficacious mRNA silencing in hepatocytes at siRNA doses of ~1 mg/kg6,7 and more recent reports show similar efficacy at doses of ~0.01 mg/kg.4,5 Importantly, not only has progress been made in the potency and mechanistic understanding of how these LNP mediate siRNA delivery,8 but also in the translation of this technology: multiple LNP-siRNA therapeutic candidates are currently in clinical testing and late stage preclinical development.9
Despite impressive progress in siRNA delivery to hepatocytes, efficacious systemic siRNA delivery to extra-hepatic cells and tissues remains difficult. One area of particular interest is delivery to immune cells, given their central role in homeostasis and disease. siRNA delivery by an antibody or peptide linked to a cationic entity, such as protamine or poly-arginine has been reported.10,11,12,13 Application of these formulations resulted in the inhibition of viral replication or protection from cytokine induction via delivery of siRNA. Another approach taken by Peer and colleagues employed antibody-targeted lipid particles to deliver siRNA to immune cells via anti-β7 integrin antibody14 or anti-CD11a antibodies,11 resulting in improvement of disease in a dextran sodium sulfate colitis mouse model and HIV resistance in a humanized mouse model, respectively. It has also been reported that siRNA can be delivered to macrophages after oral administration by packaging in yeast particles,15 or via injection of polymer or lipoplex complexes after intraperitoneal administration in chitosan/siRNA particles.16,17 Importantly however, much of the above mentioned work on silencing immune cell genes has used chemically unmodified siRNA duplexes that are known to stimulate innate immune response and could, therefore, result in nonspecific gene modulation.18,19
Innate immune cells, in particular monocytes and macrophages, are critical for inducing and orchestrating global and local immune responses,20 and are centrally involved in disease initiation and progression. Thus, we focused our attention on these cells, which are also professional phagocytes and therefore have the propensity to engulf nanoparticle-based formulations. We developed highly active siRNAs to genes expressed in myeloid cells and optimized the LNP formulations for greater potency, achieving half-maximal silencing doses of 0.2 mg/kg. Using dynamic fluorescence tomography combined with microscopy, we show robust delivery of siRNA to the spleen in mice. These formulations provide efficacious RNAi-mediated silencing of multiple targets in myeloid cells in both healthy rodents and nonhuman primates (NHPs). In addition, we demonstrated that by inhibiting the well-validated target tumor necrosis factor-α (TNFα) in a macrophage-dependent model of rheumatoid arthritis (RA), disease activity was drastically reduced to a level comparable with potent antibody-mediated treatment. Two very recent studies complement and further extend this work. Basha et al. present a detailed analysis establishing the KC2 ionizable lipid as very potent vehicle for siRNA delivery to antigen-presenting cells.21 Leuschner et al. demonstrates the broad therapeutic potential of C12-200-mediated delivery to myeloid cells while focusing on a specific gene target, CCR2.22 Taken together these findings represent a significant advancement in both RNAi technology and drug development as they open possibilities to simultaneously silence gene targets not accessible to small molecules or antibodies in myeloid cells, offering new therapeutic opportunities for diseases with inflammatory etiology.
Results
Significant work has been dedicated toward the development of new materials and formulations for the delivery of siRNA to hepatocytes (see e.g. refs. 4,5). We sought to develop nanoparticles with delivery potential to myeloid cells through the use of lipids and lipid-like materials known as KC2 (an ionizable lipid)4 and C12-200 (a cationic lipid).5 Formulations containing these lipid materials, as well as the excipients: cholesterol, PEG-DMG, and distearoyl phosphatidyl choline were formulated as nanoparticles, encapsulating siRNAs directed to several myeloid-expressed gene targets (including CD45, CD11b, integrin β1, TNFα). The KC2 containing formulations utilized here use a higher molar percent of cationic lipid compared to earlier work,4 further increasing formulation potency. We also optimized the C12-200 containing liposomes increasing the lipid to siRNA ratio in the particle.
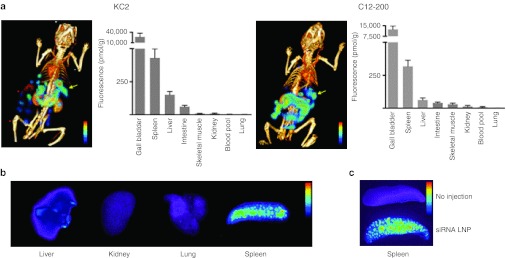
Quantitative in vivo whole body imaging and histological localization of LNP-siRNA distribution. In order to determine LNP-siRNA distribution, we intravenously (i.v.) injected LNP-encapsulated, fluorescently labeled siRNA, and followed whole body fluorescence by fluorescence-mediated tomography/X-ray computed tomography (FMT-CT). FMT-CT is a hybrid imaging approach that quantitates fluorochrome concentration in tissue, while fusion with CT data provides the anatomic localization of the fluorescent signal at high resolution.23 Concentration was sampled 90 minutes after injection of siRNA formulated in KC2 LNP and C12-200 LNP (Figure 1a), and key organs were then analyzed. Fluorescent signal reporting on siRNA concentration was attributed to anatomical structures using hybrid CT data. Interestingly, we found that the spleen is a major distribution site for both LNP preparations, with high fluorescence per gram of tissue (Figure 1a,b). Imaging also provided information about the excretion pathway of LNP siRNA. The signal peaked in the liver, gall bladder and intestine likely reflecting the excretion of the fluorochrome and attached materials. Low signal was observed in the kidneys and the urinary tract (Figure 1 and data not shown). Low siRNA concentration was observed in the lung for both nanoparticles. Ex vivo fluorescence reflectance imaging corroborated these findings with the spleen showing the brightest signal among major organs (Figure 1b,c). Distribution data obtained by imaging are consistent with plasma/tissue siRNA amount derived from a PCR-based method of siRNA quantification,24 which gives us confidence that we are not merely tracking fluorochrome, but fluorescently labeled siRNA. Leuschner et al. have shown that multiple myeloid cell types in the spleen take up C12-200 LNP-formulated siRNA including macrophages, splenic reservoir monocytes, and dendritic cells.22 Similar analysis carried out for the KC2 formulation did not reveal major difference in the cell populations targeted (data not shown).
Figure 1.
In vivo dynamic fluorescence-mediated tomography/X-ray computed tomography (FMT-CT) of lipid nanoparticles (LNP)-small interfering RNA (siRNA) delivery and siRNA localization within reservoir monocyte clusters in the spleen. (a) 3D reconstruction of FMT-CT data of KC2 (left) and C12-200 (right) is shown. The arrow points to the position of splenic signal. Signal in organ systems of interest was quantified, data are shown as mean and SE, n = 4–5 per group. (b). Ex vivo fluorescence reflectance imaging after injection of LNP siRNA side-by-side imaging of respective organs and highlights the signal intensity in the spleen. (c) Fluorescence images of spleens from mice injected or not injected with LNP-siRNA.
Efficient silencing of gene targets in myeloid cells after i.v. injection. Encouraged by the distribution of LNP siRNA to sites of immune cell localization, we initiated experiments to determine whether distribution translates into RNAi silencing activity in leukocytes. Macrophage lineage cells specialize in the removal of foreign material, thus most systemically administered particles are taken up by these cells. In fact, cells of the monocyte/macrophage lineage showed the highest fluorescence signal after i.v. injection of fluorescently labeled siRNA (see below). However, uptake often does not translate into siRNA-induced silencing; macrophages specialize in shuttling cargo into lysosomes for degradation, whereas siRNA needs to reach the cytoplasm to induce cleavage of the target mRNA. Indeed, with earlier-generation LNP formulations,6,7 despite good uptake we have not observed siRNA-mediated gene silencing in macrophages after i.v. administration.
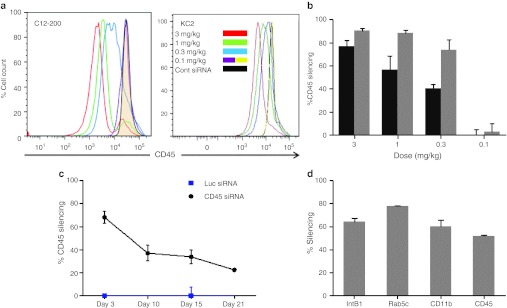
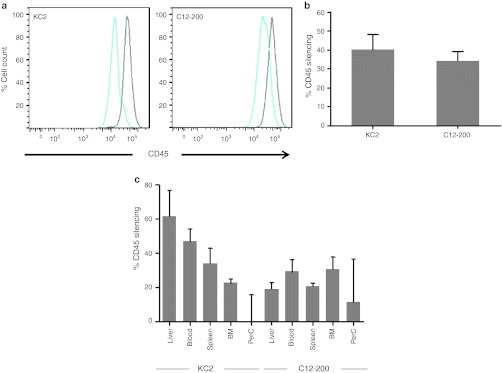
To determine which leukocyte populations are subject to silencing in response to i.v. injected LNP-siRNA, we used a CD45 silencing assay. CD45 is a common, highly expressed leukocyte antigen that occupies up to 10% of the cell surface.25 We assayed CD45 protein knockdown by flow cytometric analysis of leukocytes defined by combinations of specific cell surface markers and then measured the decrease of CD45 expression in each of these cell populations (Figure 2). We compared leukocytes from animals injected with a formulation containing a CD45-specific siRNA versus animals injected with an identical formulation containing a control siRNA targeting luciferase. We surveyed leukocytes isolated from different organs (spleen, liver, peritoneal cavity, bone marrow, and lymph nodes) 3 days after a single i.v. bolus injection (Figure 2a and Supplementary Figure S1b). A 3-day interval between injection and protein level analysis was needed to allow the reduction in mRNA levels to be translated into decreased CD45 protein expression. Using this assay, we found highly effective (~80%) silencing in cells of macrophage lineage (Figure 2a,b), good (~40%) silencing in dendritic cells (Supplementary Figure S1b), some activity (~15%) in B cells (Supplementary Figure S1c, and no silencing in T cells, NK cells, or GR-1+ granulocytes (Supplementary Figure S1c). Silencing in all cell types was examined in multiple tissues including spleen, liver, bone marrow, blood, and peritoneal cavity. These findings imply broad applicability of this technology to diseases that involve innate immune activation, infection, and antigen presentation.
Figure 2.
Efficacious and durable silencing in peritoneal cavity macrophages. (a) Cells were collected at 72 hours after a single intravenous (i.v.) bolus administration of the indicated doses of small interfering RNA (siRNA) in KC2 or C12-200 lipid nanoparticles (LNP) formulation. Leukocytes were stained for CD11b and CD45 and analyzed by flow cytometry. Representative profiles of CD45 staining in CD11b+ population are shown. Groups of three mice were analyzed, a representative experiment out of three independently conducted ones is shown. (b) Quantification of the percent silencing with C12-200 (gray bars) and KC2 (black bars) seen in panel (a). Bars represent average percent silencing within group with standard deviation. (c) Green fluorescent protein (GFP)-expressing peritoneal lavage cells from N5-RAGE GFP mice were transferred into C57BL/6 recipients by intraperitoneal (i.p.) injection. Thirty minutes after transfer mice were treated with 2 mg/kg KC2 formulation encapsulated CD45 or Luc siRNA i.v. At indicated days silencing of CD45 in GFP-expressing peritoneal cavity myeloid cells was monitored. Three mice per group were analyzed. (d) C57BL/6 mice were injected with 0.8 mg/kg of equivalent mixture of four siRNA directed against integrin β1, Rab5c, CD11b, and CD45 or with total amount of Luc siRNA. Twenty four hours post injection peritoneal cavity cells were collected and gene expression was analyzed by quantitative-PCR (Q-PCR). Each gene expression was measured as ratio to GAPDH expression. Knockdown results are expressed as % gene knock down with SEM, relative to the group injected with Luc siRNA. Three mice per group were analyzed.
To address whether ingestion of LNPs might activate leukocytes, we compared mice injected with PBS versus siRNA in LNP. Notably, we used only chemically modified siRNA molecules (2′OMe modified at selected pyrimidine sites) which do not induce cytokine production in the human PBMC assay.18 We investigated leukocyte numbers as well as their activation status in blood, spleen, bone marrow, and peritoneal cavity following an i.v. administration of LNP siRNA. Interestingly, we did not find evidence for overt activation in any of the populations surveyed; there was no increase in costimulatory molecules CD80 and CD86 or in MHCII expression (Supplementary Figure S2). After injection of Luc LNP-siRNA, we observed a minor influx of CD11b− low cells in the peritoneal cavity and slight CD45 upregulation on CD11b+ cells (Supplementary Figure S2).
To corroborate the CD45 silencing data, we conducted silencing experiments with other gene targets, including green fluorescent protein (GFP), CD11b, integrin β1, and TNFα, where we observed similar or greater silencing efficiencies (Supplementary Figure S3 and data not shown). For instance, silencing of integrin β1 in CD11b+ cells from the bone marrow was more pronounced than that of CD45 in the same cells (Supplementary Figure S3). These observations may reflect both intrinsic differences in the potencies of the specific siRNA (half-maximal concentration required for in vitro silencing of 10 pM for integrin β1 versus 90 pM for CD45) and different characteristics in the target mRNA transcripts (e.g., mRNA half-life).
Interestingly, the anatomical location showing the strongest silencing of CD45 after i.v. injection in mice was the peritoneal cavity with up to 90% reduction in CD45 protein expression, followed by significant silencing in the spleen, and only moderate silencing seen in the bone marrow, lymph nodes, and liver (Figure 2 and Supplementary Figure S1, data not shown), using shift in mean fluorescent intensity of the total population as the metric. It is noteworthy that within a population with average silencing even at 20–30%, there are subpopulations of cells with silencing as high as >95% at the lower end of distribution. Given the extreme stability and abundance of CD45, it is likely a gene target that is more refractive to silencing.
Next, in dose response experiments, we assessed the potency of in vivo silencing in peritoneal macrophages and found that C12-200 LNP induced 50% CD45 silencing at doses of ~0.2 mg/kg, and KC2 LNP at doses of ~0.5 mg/kg (Figure 2a,b). This is a significant improvement compared to similarly formulated LNPs using early generation cationic lipids, where no silencing in leukocytes was seen after i.v. injection of at least tenfold higher doses.7 Even compared to LNPs using the KC2 lipid the formulation used here shows a sixfold improvement in potency with 50% silencing at 0.5 mg/kg versus 3 mg/kg21 in peritoneal macrophages. Knowing that peritoneal cells are sessile, we could monitor longevity of silencing in vivo using cells transferred intraperitoneally from the peritoneum of GFP transgenic mice. To this end, we injected cohorts of animals with CD45 or control LNP-siRNA and sacrificed them at different time points. We observed silencing in peritoneal macrophages for up to 3 weeks after a single injection (Figure 2c), which is near identical to the duration of silencing observed in hepatocytes.4,5 We observe a longer silencing duration than that seen in peritoneal macrophages put in culture following liposome treatment.21 This may be a reflection of the differences in the assay systems and the gene targets being assayed (CD45 versus GFP).
With such low doses, it is possible to combine several gene targets for silencing in leukocytes, thus, enabling functional genomics studies in the cells central to inflammatory disorders. To validate this utility, we performed an experiment that mixed siRNA to four independent gene targets formulated in C12-200 LNP, each at the 0.2 mg/kg dose, an estimated IC50 dose for the CD45 siRNA. We included siRNA targeting CD45, CD11b, RAB5c, and integrin β1 into this cocktail. Silencing was monitored in total peritoneal cavity cells 24 hours post injection on the mRNA level by reverse transcription-quantitative PCR. We observed silencing ranging from 50 to 80% for each of the targets as compared to the levels seen in animals dosed with control siRNA (Figure 2d).
Macrophage uptake of LNP-siRNA relies on phagocytosis. Since both KC2 and C12-200 represent potent LNP formulations for myeloid cell silencing in vivo, we chose to investigate the mechanism of cellular uptake for these two formulations. We performed uptake experiments with primary mouse bone marrow-derived macrophages that were treated with fluorescently labeled siRNA in KC2 or C12-200 LNP. C12-200, relative to KC2 LNP, mediated more efficient uptake of the labeled siRNA by primary macrophages in vitro consistent with mRNA-silencing results (Figure 3a). To characterize the mechanism of cellular uptake, we co-exposed primary macrophages to markers of different endocytic pathways. We saw ~60–70% colocalization of labeled siRNA particles with fluorescent latex beads that due to their large 1 µm size enter these cells by phagocytosis (Figure 3b,c).26 We saw substantially less colocalization with markers of other pathways: dextran, a marker of macropinocytosis (~40%), and transferrin, a marker of clathrin-mediated endocytosis (10%) (Supplementary Figure S4a–c). Latex beads and siRNA-containing vesicles colocalized most prominently in the perinuclear region; these likely represent vesicles that have already undergone lysosome fusion (Figure 3b). Since phagosomes are known to contain the EEA1 marker27 the compartments in which siRNA signal is observed likely correspond to the EEA1 positive structures reported by Basha et al.21 In addition, LNP-siRNA uptake was inhibited by Cytochalasian D and by Dynasore (inhibitors of actin rearrangement and dynamin, respectively, both previously shown to inhibit different steps of phagocytosis (Supplementary Figure S4d,e)). We therefore concluded that the primary mechanism of LNP siRNA internalization in macrophages was phagocytosis.
Figure 3.
Uptake of KC2 and C12-200 formulations in primary macrophages. (a) Quantitation of cellular uptake of Alexa Fluor-647 labeled small interfering RNA (siRNA) (AF647-si) in KC2 and C12-200 formulations in primary macrophages in vitro. RFU, relative fluorescence units. Data are average of 20 fields from three independent wells. (b) Colocalization of AF647-si in C12-200 or KC2 formulations with a marker of phagocytosis Alexa-488 labeled 1 µmol/l latex beads in primary macrophages, 1 hour time point C12-200, 4-hour time point KC2, arrows indicate signal overlap. (c) Quantitation of percent colocalization of AF488 beads with A647-si in KC2 or C12-200 formulations (4 hour time point). Data are the average of 20 fields from three different independent replicate wells for a total of 60 fields ± SEM.
Silencing of myeloid genes in vivo occurs in both tissue-resident and splenic reservoir cells of monocyte/macrophage lineage. A key question for therapeutic gene silencing in leukocytes is whether delivery can be achieved to circulating and splenic monocyte/macrophages, including splenic reservoir monocytes that migrate in high numbers to inflammatory sites such as acute myocardial infarcts.28,29,30 One technical challenge in assessing in vivo gene silencing in leukocytes is their migratory nature. The site of initial LNP-siRNA uptake and the ultimate cell destination when protein downregulation is detectable may not coincide. To address this issue, we injected mice i.v. with LNP-siRNA and isolated monocytes/macrophages from bone marrow, blood, spleen, and peritoneal cavity at 15, 60, and 120 minutes post injection. The cells were then seeded onto plastic to allow time for downregulation of CD45 protein expression. This strategy interrupted the migratory path of the cells, and determined the site of effective uptake independent of subsequent cell relocation. Interestingly, we did not observe any silencing in the bone marrow in this assay, whereas blood monocytes reached maximum silencing at 15 minutes, splenic cells at 1 hour, and peritoneal macrophages at 2 hours after injection with KC2 LNP-formulated CD45 siRNA (Figure 4a). The maximal levels of CD45 silencing seen in this in vivo/ in vitro assay (>50%) were comparable to the levels reached 3 days post injection in vivo (Figure 4a). Silencing kinetics were faster following injection of C12-200 LNP-formulated siRNA, with maximal blood silencing as early as 5 minutes post injection (data not shown). These data indicate that LNP siRNA effectively reached the central pool of circulating and splenic monocytes, as well as resident tissue macrophages. Due to the fast migratory kinetics of myeloid cells, many circulating and splenic monocytes relocate to target tissue after ingesting LNP siRNA. Some of these transfected cells appear to migrate to the peritoneal cavity.
Figure 4.
Fast and efficient silencing in central monocytes/macrophages coupled with delayed but durable silencing in resident peritoneal cavity myeloid cells. (a) Cells were collected at 15 minutes, 1 hour, or 2 hours after a single intravenous (i.v.) bolus administration of 3 mg/kg of small interfering RNA (siRNA) in KC2 formulation. Leukocytes were seeded on plastic tissue culture plates and adherent cells were cultured for 3 days. Then harvested cells were stained for CD11b and CD45 and analyzed by flow cytometry. Mean fluorescent intensity shifts are shown when comparing CD45 siRNA to Luc siRNA in KC2 lipid nanoparticles (LNP). (b) Green fluorescent protein (GFP)-expressing peritoneal lavage cells from N5RAGE GFP mice were transferred into C57BL/6 recipients by i.p. injection. Thirty minutes after transfer mice were treated with 2 mg/kg KC2 formulation encapsulated CD45 or Luc-specific siRNA i.v. 72 hours later peritoneal cells were collected and the level of CD45 in GFP positive and negative peritoneal macrophage was quantified by flow cytometry. (c, d) C12-200 (c) or KC2 (d) encapsulated Alexa647 labeled siRNAs was injected at 1 mg/kg i.v. Blood, spleen, and peritoneal lavage were collected at indicated time points. Macrophages were identified by surface marker staining (see Materials and Methods) and the Alexa647 signal ±SEM is plotted.
The efficient silencing seen in mouse peritoneal macrophages 3 days after i.v. injection was surprising, and may be (i) a functional consequence of gene silencing, (ii) due to migration of the cells that had been targeted while circulating in blood or residing in the spleen, and/or (iii) a result of local accumulation of LNP siRNA in the peritoneal cavity and uptake by resident macrophages. Notably, the kinetics of LNP accumulation in peritoneal cavity macrophages was significantly slower than that observed in blood or spleen (Figure 4c,d). Using GFP-expressing transgenic mice, we found that equally efficient silencing of GFP can also be measured in the peritoneal cavity macrophages, therefore excluding a role for endogenous gene knockdown in localization of these cells to the peritoneal cavity (Supplementary Figure S3a).
To determine whether LNP internalization first occurred in circulation or locally in the peritoneal cavity, we transferred resident GFP+ peritoneal cells into the peritoneum of wild-type mice and injected recipient mice i.v. with LNP-siRNA targeting CD45 30 minutes after cell transfer. CD45 in both, GFP+ and well as GFP− cells, was silenced to a nearly identical degree in vivo (Figure 4b). This experiment established that gene targets in sessile resident peritoneal cells31 are also silenced by i.v. injected LNP-siRNA.
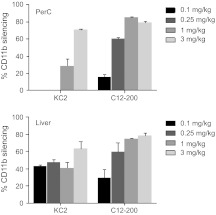
Silencing mRNA specific for the monocyte/macrophage lineage. While the CD45 assay allowed us to follow many leukocyte cell types in parallel, it necessitated isolation of leukocytes from different tissues to identify cell types by surface staining. Previous work demonstrated that some tissue resident leukocytes cannot be isolated,32 thereby making it impossible to assess the degree of silencing in such cells by flow cytometry. We therefore decided to focus on all cells of the monocyte/macrophage lineage by devising siRNA against CD11b (Mac-1) and thus enabling total tissue mRNA analysis of silencing in the cells of interest. This siRNA was very active in vitro with an IC50 of about 4 pM. We identified a 24-hour time point when mRNA for CD11b was significantly silenced, but cell surface protein was still unaffected, to avoid potential effects specific to CD11b function. We performed in vivo titration of LNP encapsulated CD11b siRNA and found it required ~0.3 mg/kg siRNA in C12-200 and ~1 mg/kg siRNA in KC2 LNP to achieve 50% silencing at the mRNA level in the peritoneal cavity cells (Figure 5). These in vivo potencies were very similar to those found for the CD45 protein and they demonstrate the robustness and reproducibility of silencing across several targets, using mRNA and protein as readouts.
Figure 5.
Macrophage-specific CD11b silencing. mRNA ratio of CD11b to GAPDH was measured in peritoneal cavity or total perfused liver lobes 24 hours after bolus intravenous (i.v.) injection of small interfering RNA (siRNA) in lipid nanoparticles (LNP). Bars represent the mean level from three animals per group, three biological replicas per sample, with SEM in animals treated with CD11b siRNA relative to Luc siRNA-treated animals.
We proceeded to test CD11b silencing in different organs, measuring the ratio of CD11b mRNA level to GAPDH. In spleen and peritoneal cavity, CD11b mRNA was reduced to similar levels when compared with CD45 silencing (Figures 2 and 5 and Supplementary Figure S1), namely, ~30% in the spleen and 70–90% in peritoneal cavity. Interestingly, we also found efficient CD11b knockdown in the liver (Figure 5). We believe that the near absence of CD45 silencing in the macrophages isolated from liver reflects the fact that a true resident population of Kupffer cells is not isolated by the liver digestion protocols employed and was therefore not accessible to flow cytometric analysis.32 CD11b knockdown was also normalized to another macrophage-specific marker, F4/80, and was shown to be very similar to the values obtained by normalization to GAPDH (data not shown): This indicates that observed CD11b knockdown is not due to the loss of macrophages.
CD45 and CD11b silencing is RNAi mediated. To confirm that the knockdown of myeloid gene mRNA observed in rodents was mediated by an RNAi silencing mechanism, we isolated CD11b+ cells from mice treated with either CD45 or CD11b LNP-siRNA. mRNA from these cells was then subjected to rapid amplification of cDNA ends (5′-RACE), a method previously used to demonstrate siRNA-mediated cleavage.6,33 5′-RACE analysis of peritoneal cavity macrophage-derived mRNA from animals treated with LNP-siRNA revealed products of the expected sizes for both CD45 and CD11b amplicons in their respective cohorts (Supplementary Figure S5). Sequence analysis of cloned PCR products demonstrated that 46 out of 48 and 24 out of 24 PCR products were derived from the predicted cleavage event at position (CTGGCTGAA/TTTCAGAGCA) for CD45 siRNA in KC2 and C12-200 LNP, respectively; for CD11b, 29 out of 48 and 20 out of 24 PCR products were cleaved at position (TTGTCTCAA/CTGTGATGGA) in KC2 and C12-200 LNP, correspondingly. No specific cleavage site PCR products were derived from the 5′-RACE samples treated with LNP-encapsulated control siRNA (out of 140 sequenced products) except for one likely contaminant clone with CD45 cleavage product derived from CD11b siRNA in C12-200 LNP treated cells. These results clearly demonstrate that the effect of siRNA in LNP treatment on CD45 and CD11b expression levels observed is due to cleavage of the mRNA transcript via an RNAi mechanism.
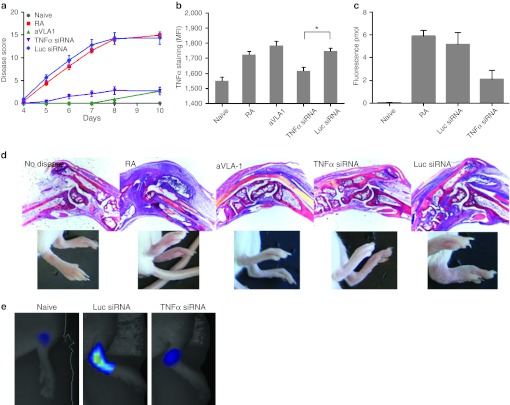
siRNA-mediated silencing in immune cells substantially inhibits disease progression in a mouse model of RA. To determine whether the effective silencing in macrophages translated into disease modifying activity, we tested TNFα-specific siRNA in an antibody-induced arthritis mouse model in which systemic inhibition of soluble TNFα has been previously shown by several groups to be highly effective.34 Using siRNA targeting TNFα, we found inhibition of paw swelling in two independent experiments with C12-200 LNP (Figure 6a). In fact, the anti-inflammatory activity was comparable to anti-VLA1 i.v. antibody treatment (Figure 6a,d), which has been previously demonstrated to be at least as effective as systemic TNFα inhibition.35 In agreement with the near complete absence of redness and swelling in the joints and digits, histological analyses of paw sections showed significantly decreased edema, synovial inflammation, and inflammatory cell infiltration in TNFα siRNA-treated animals (Figure 6d). To quantitate the degree of inflammation in siRNA-treated animals, we used in vivo fluorescence tomography of arthritic joints after injection of a pan-cathepsin sensor, which reports on protease activity.36 Using this method, we found that TNFα-specific LNP siRNA reduced joint inflammation by more than half (Figure 6c,e). In splenic macrophages from arthritic mice treated with TNFα targeting but not with control siRNA, we found a decrease of intracellular TNFα staining indicating effective silencing (Figure 6b).Overall, the effect of treatment on mean fluorescent intensity value is highly significant according to an ANOVA (F = 17.061, P < 10−5). Tukey's post-hoc tests indicate that the pairwise difference between mean MFI in the TNFα siRNA group and the Luc siRNA group is significant (P = 0.0197; see star in Figure 6b).
Figure 6.
Inhibition of collagen mAb-induced arthritis by small interfering RNA (siRNA) against tumor necrosis factor-α (TNFα). (a) Preventative treatment of mice with siRNA against TNFα (purple curve) but not against Luc (blue curve) in C12-200 formulation decreases arthritis score. Anti-VLA1 mAb given several hours before arthitogenic antibody mix was used as a positive control (green curve) as previously shown.35 Mice were treated with anticollagen mAb's at day 0, followed by lipopolysaccharide (LPS) on day 3. Mice receiving only arthitogenic antibodies are graphed in red. 0.5 mg/kg siRNA in C12-200 formulation was administered on days −3, 0, 4, and 7. All mice were scored on days 4–8 and 10. Each limb was evaluated and scored on a 0–4 scale. Results are expressed as the mean arthritic score (±SEM) of all four limbs (maximum score of 16). Groups of 10 mice per condition were used; one out of two experiments is shown, both showed similar protection. (b) Intensity of intracellular TNFα staining in CD11b+ splenic macrophages from diseased mice at day 10. Groups of three mice were analyzed; bars represent average of the mean fluorescent intensity in the group with SEM, statistical analysis as described in the Results section. (c) Detection of inflammatory macrophage activity by fluorescence-mediated tomography (FMT) imaging. Arthritic mice treated with siRNAs targeted to TNFα or Luc were injected with Prosense-750, a pan-specific cathepsin substrate, and imaged by FMT on day 7. Fluorescence concentration reports on protease activity in the joint and was quantified as described in Supplementary Materials and Methods, n = 5, 2 paws per mouse were analyzed. Data are mean ± SEM. (d) Joint histology at 5x magnification in H&E staining correlated with images of the paws. (e) Representative images of macrophage activity measurement by FMT quantified in panel c. Naive and siRNA-treated animals are shown.
In vivo silencing in circulating monocytes in NHPs. Finally, in order to determine the translational potential of RNAi-mediated silencing in immune cells, we explored leukocyte gene silencing in NHPs. We first performed a nonterminal study in cynomolgus macaques and examined efficacy of a single dose per formulation and one time point of sampling post dose. To avoid confounding effects due to leukocyte redistribution and trafficking in the days following LNP-siRNA administration, we replicated our mouse protocol and collected blood 1 hour post-i.v. injection followed by 3 days of in vitro culture and analysis of CD45 protein cell surface expression. We tested CD45 or luciferase siRNA formulated in KC2 (3 mg/kg) or C12-200 (1 mg/kg) and compared pre- and post injection levels of CD45 expression on blood monocytes. In every animal dosed with CD45 siRNA, but not with control siRNA, we found CD45 protein reductions of ~40–50% (Figure 7a). Encouraged by these results, we next monitored organ resident leukocyte silencing in cynomolgus macaques. The same doses and formulations were used, namely CD45 or luciferase siRNA formulated in KC2 (3 mg/kg) or C12-200 (1 mg/kg). Three days after injection we isolated leukocytes from blood, bone marrow, peritoneal cavity, liver, and spleen to compare surface CD45 expression in animals injected with active versus control siRNA. We observed 30–60% silencing in liver, blood, spleen, and bone marrow-derived cells of monocyte/macrophage lineage (n = 3 per group, Figure 7c). Interestingly, peritoneal cavity myeloid cells did not demonstrate any detectable silencing (Figure 7c). Dot plots from representative animals and overlaid histograms from each animal are shown in Supplementary Figure S6, demonstrating that there is a significant number of cells with diminished CD45 staining. In this experiment, we could not compare pre and post dose levels; therefore we analyzed group averages with expected and significant variability in nongenetically identical animals. Despite this limitation, robust silencing of CD45 was observed in organ resident and blood circulating myeloid cells. It is worth mentioning that in cynomolgus macaques we were able to detect silencing of CD45 in circulating cells up to 3 days after the injection, which indicates potent and durable silencing in the central compartment of myeloid cells ready for recruitment to inflammatory sites.
Figure 7.
Silencing CD45 in vivo in peripheral blood monocytes/macrophages from nonhuman primate (NHP). Blood was drawn from each of the 12 experimental NHPs 1 hours prior and 1 hour after bolus administration of 3 mg/kg of small interfering RNA (siRNA) in KC2 and 1 mg/kg of C12-200. Leukocytes were seeded on plastic tissue culture plates and adherent cells were cultured for 3 days. Then harvested cells were stained for CD31, CD11b, and CD45 and analyzed by flow cytometry. (a) Representative profiles of CD45 staining in CD31/CD11b double-positive population are shown for both lipid nanoparticles (LNPs), blue profiles correspond to after CD45 siRNA dose, whereas black—for before dosing. (b) Average percent silencing in the total CD31/CD11b double-positive population as calculated from the shift in mean fluorescence intensity (MFI) in the surface of the cells before and after in vivo treatment with CD45 siRNA. (c) Summary of % CD45 knockdown in organ resident CD11b+ cells of monocyte/macrophage lineage in NHP dosed with 3 mg/kg of KC2 or 1 mg/kg of C12-200 3 days after dose. Bars represent average MFI value from three animals in the CD45 siRNA-treated group as it related to the average MFI of the Luc siRNA-treated group, plotted with SEM. Significant knockdown is seen in all organs analyzed except for peritoneal cavity. Respective histograms and representative dot plots from where the data is derived are shown in Supplementary Figure S6.
Discussion
Bringing RNAi-mediated silencing in leukocytes toward the clinic. Macrophages are an important therapeutic cellular target in many diseases, but so far developing systemic siRNA treatment aimed at these cells has been difficult. In this study, we provide evidence that LNP-mediated siRNA delivery to macrophages and dendritic cells can be used as a robust therapeutic platform. Similar LNP formulations are already in clinical trials or in late stage preclinical development for transthyretin amyloidosis, hypercholesterolemia, and liver cancer.9
We investigate the ability of two types of liposomal formulations KC2 (an ionizable lipid) and C12-200 (a cationic lipid) to mediate silencing in myeloid cells.8 We demonstrate dose-dependent silencing for both liposome types with an in vivo ED50 of 0.2–0.5 mg. The high in vivo potency of these formulations makes them potentially applicable for clinical use and is a major improvement on the dose levels used by Basha et al. where a 3 mg/kg dose of a KC2-based formulation was employed.21 In the mouse system, systemic delivery of LNP-encapsulated siRNA results in silencing in multiple cell types (macrophages, dendritic cells, B cells) and anatomic sites (peripheral blood, peritoneal cavity, spleen, bone marrow, liver). In tissue resident macrophages, such as those found in the peritoneum, CD45 silencing persists for over 2 weeks. We also provide evidence that the observed effects on protein levels are mediated by an RNAi mechanism for multiple gene targets. The formulations used in this study have a relatively simple composition, containing four excipients in addition to the siRNA, and can be manufactured at scale, a crucial attribute for translating LNP-based therapies into the clinic.
Silencing of gene targets in rodent myeloid cells had been reported by multiple groups using a variety of approaches, however translation to higher species has been elusive. Here, we demonstrate target-specific silencing of CD45 in liver, blood, spleen, and bone marrow derived cells of monocyte/macrophage lineage in NHPs for both KC2- and C12-200-based LNPs. Since silencing of CD45 can be detected in circulating myeloid cells 3 days post injection, we have an opportunity for testing multiple formulations in NHPs for further LNP potency improvement. LNPs are highly amenable to attachment of a targeting ligand, as has been recently shown for LNP targeting via a GalNAc ligand for hepatocyte delivery.8 Targeting LNPs to macrophage surface receptors will likely result in an increased cytoplasmic localization of the siRNA by increasing the amount of siRNA taken up by macrophages via nonphagocytic pathways and may allow further lowering of the therapeutic dose.
To further validate the therapeutic potential of this platform we demonstrate that systemic administration of LNP-encapsulated siRNA targeting TNFα results in disease modifying activity in a mouse model of RA. When compared to a previously reported anti-VLA1 antibody treatment, siRNA to TNFα gives a similar level of protection in this immune-inflammatory model. The recent study of Leuschner et al.22 greatly extends the potential therapeutic applications of this platform by demonstrating therapeutic efficacy of anti-CCR2-specific siRNA delivered using a C12-200 LNP. In this study, multiple rodent disease models are interrogated, including models of chronic disease.22 The ability to rapidly select siRNAs to a gene makes the LNP-siRNA platform a powerful tool for rapid validation of therapeutic targets for treating inflammation in vivo.
This process can be further aided by the ability to silence several independent genes by a cocktail of siRNA as is demonstrated here. The cocktail approach allows for studying signaling pathways, unraveling nonredundant signals and mimicking conditions that are believed to affect whole functional groups of genes. In addition, the ability to simultaneously silence several genes paves the way to therapeutic applications that require inhibition of multiple cellular functions.
Delivering siRNA to myeloid cells could provide novel approaches to treating multiple human diseases, such as modulating chronic inflammation in autoimmune disease, protecting against myeloid-tropic viral infections, reprogramming tumor-associated macrophages, restoring functionally insufficient cells, or killing malignantly transformed immune cells.
Materials and Methods
siRNA synthesis and selection. Single-stranded chemically modified RNAs were synthesized at Alnylam Pharmaceuticals (Cambridge, MA) using standard phosphoramidite chemistry. Deprotection and purification of the crude oligoribonucleotides by anion exchange high-performance liquid chromatography were carried out according to established procedures. siRNAs were generated by annealing equimolar amounts of complementary sense and antisense strands. CD45- and luciferase-specific siRNA were described earlier,7 other siRNA used are listed in Supplementary Table S1. siRNA specific to murine CD11b was identified by screening a set of 28 siRNA duplexes designed to target mouse and rat versions of the gene. Best duplexes were selected by transfection of DC2.4 cells and IC50 values determined. siRNA specific to β1 integrin and Rab5c were identified similarly except NIH3T3 fibroblasts were used for screening. A single duplex was selected for each target and synthesized on a larger scale for use in LNP preparation.
siRNA formulation in LNPs. LNP siRNA formulations were prepared using either of the two recently described novel lipids DLin-KC2-DMA4 or C12-200.5 The novel lipids, along with the colipids disteroylphosphatidyl choline, cholesterol, and PEG-DMG, were formulated with siRNA using a spontaneous vesicle formation formulation procedure as previously described.4 The LNPs had a component molar ratio of ~50/10/38.5/1.5 (DLin-KC2-DMA or C12-200/disteroylphosphatidyl choline/cholesterol/PEG-DMG). The final lipid:siRNA weight ratio was ~12:1 and 9:1 in the case of DLin-KC2-DMA and C12-200 LNPs, respectively. The LNP-siRNA formulations had mean particle diameters of ~80 nm with >90% entrapment efficiency.
In vitro LNP uptake and microscopy. Primary bone marrow-derived macrophages were isolated using standard methods and cultured in glass bottom 96-well plates (Greiner, Frickenhausen, Germany). Cells were maintained in Dulbecco's modified Eagle's medium (Life Technologies, Grand Island, NY) supplemented with Penicillin, Streptomycin, and fetal bovine serum. Cells were seeded for 5 days in the presence of macrophage colony-stimulating factor (8 ng/ml; Peprotech, Rocky Hill, NJ), then media was completely renewed for the last overnight culture and KC2 or C12-200 liposome formulations of Alexa-647 tagged GFP-targeting siRNA (Alnylam Pharmaceuticals), were added for the indicated time points. Cells were counterstained with Hoechst without fixation and imaged live. All images were acquired using an Opera automated spinning disc confocal system (Perkin Elmer, Waltham, MA) and data analyzed using Acapella Software (Perkin Elmer). Quantification of Alexa-647 labeled siRNA uptake was determined by identifying the number and border of each cell and calculating the number of intracellular siRNA positive spots above a predetermined minimum intensity threshold. The average number of these spots per cell was multiplied by the average intensity value of the indentified spots to determine the average siRNA content per cell. The average content per cell was averaged across 20 fields from three replicate wells. In some experiments, primary macrophages were copulsed with a 1 to 500 dilution of a 2% solution of Alexa-488 latex beads (Invitrogen). Percentage of colocalization of Alexa-647 siRNA and Alexa-488 beads was determined by identifying regions of intracellular siRNA signal similar to above using Acapella software and determining the percent of the total area overlapping with AF488 latex bead signal. Similarly, the colocalization percentage values were an average of % colocalization of siRNA and beads across 20 fields from three replicate wells.
Isolation of lymphocytes from spleen, blood, bone marrow, lymph nodes, peritoneal cavity and liver. Spleens or lymph nodes were minced through a nylon mesh (Cell Strainer; BD Falcon; BD Biosciences, Franklin Lakes, NJ) to obtain single cell suspensions in DMEM, 5% fetal calf serum, and 2 mmol/l L-glutamine. Single-cell suspensions were prepared from bone marrow by flushing femurs with DMEM (containing 5% fetal calf serum). Erythrocytes were lysed by incubating in lysis buffer (140 mmol/l NH4Cl, 17 mmol/l Tris–HCl, pH 7.65) for 3 minutes on ice. Blood was collected in EDTA-containing tubes (BD Biosciences – Pharmingen, San Diego, CA). To isolate blood lymphocytes, 200 µl of blood was underlaid with Ficoll-Paque (GE Healthcare, Piscataway, NJ) and centrifuged at 1,000g at room temperature for 20 minutes. Lymphocytes were collected from the interface. The peritoneal cavity was washed with 3 ml of DMEM, 5% fetal calf serum, and 2 mmol/l L-glutamine to collect peritoneal leukocytes. Following these procedures, lymphocytes were washed twice in DMEM, 5% fetal calf serum by 300g centrifugation at 4 °C and resuspended in PBS/BSA/azide for flow cytometric analysis. Liver lymphocyte isolation was done as described earlier.37
Flow cytometry. Fluorescence staining was performed as previously described.38 Antibodies specific for mouse CD45, CD19, CD11b, CD86, B220, MHCII, TCRb (eBioscience, San Diego, CA), or CD11c (BD Biosciences – Pharmingen) were used. Antibodies specific for NHP or human (h) hCD31, hCD11b, CD45, hCD20, hCD3 (BD Biosciences – Pharmingen) or for CD11c (Abcam, Cambridge, MA) were used. Antibodies were conjugated to fluorescein isothiocyanate, phycoerythrin, allophycocyanin (APC), phycoerythrin-Cy7, or biotin. Biotinylated antibodies were detected with streptavidin conjugated to fluorescein isothiocyanate, phycoerythrin, or APC. For intracellular staining cells were resuspended in fixation/permeabilization solution (BD Biosciences – Pharmingen), incubated for 20 minute at 4 °C, washed twice with permeabilization/wash buffer (BD Biosciences – Pharmingen) and stained with TNFα or isotype control Ab (BD Biosciences – Pharmingen) diluted in permeabilization/wash buffer for 30 minutes at 4 °C. Stained cells were analyzed using BD LSRII (BD Biosciences – Pharmingen). Analysis was done using FlowJo software (Tree Star, Ashland, OR).
Animals. All experiments followed institutional, federal, state and local guidelines and were approved by Institutional Animal Care and Use Committee. All the animals were kept in a conventional barrier animal facility with a climate-controlled environment having 12-hour light/dark cycles in polystyrene cages containing wood shavings, fed standard rodent chow and water. Nude mice were purchased from COX-7 (Massachusetts General Hospital, Boston, MA), C57/BL6 mice from Jackson Labs (Bar Harbor, ME). N5RAGE-GFP transgenic mice39 were bred in house as homozygous X C57BL/6, heterozygous animals were used for experiments. For silencing and distribution experiments mice were i.v. bolus injected and sacrificed by CO2 overdose before tissue harvest.
NHP experiments were performed with male cynomolgus monkeys originating from China, 3–6 years of age, weighing 4–8 kg. Treatment of the NHPs was conducted by a Charles River Laboratories (Wilmington, MA). in accordance with the testing facility's standard operating procedure, which adheres to the regulations outlined in the United States Department of Agriculture Animal Welfare Act (9 CFR, Parts 1–3) and the conditions specified in the Guide for the Care and Use of Laboratory Animals (ILAR publication, 1996, National Academy Press). On the day of dosing, all monkeys were given a single 15-minute i.v. infusion of LNP-formulated siRNAs. Blood samples were collected 1 hour predose and 1 hour post the end of the infusion and reached Alnylam Pharmaceuticals by courier within <2 hours post-collection. In the nonterminal study animals were returned to the colony after sample collection. In the terminal study, animals were bled pre- and post-15 minute i.v. infusion, bone marrow and peritoneal lavage samples were collected while the animal is anesthetized with Nembutal; splenic and liver samples were collected post humane sacrifice.
FMT-CT imaging. FMT-CT imaging is a fully quantitative hybrid technique that combines whole body noninvasive fluorescence tomography to determine tissue concentration of AF647-tethered siRNA with high-resolution CT imaging for exact anatomic location of the siRNA.These imaging studies provide the biodistribution of LNP siRNA. Please see Supplementary Materials and Methods for a detailed description of the FMT-CT methods.
Anti-collagen mAb-induced arthritis. Arthrogen-collagen–induced arthritis antibody kits were purchased from Chondrex LLC (Redmond, WA), and arthritis was induced using an established protocol.40,41 Groups of 10 BALB/c mice (CRL) were injected intraperitoneally with a cocktail of five anticollagen type II mAb's (0.75 mg/animal total, Chondrex, catalog #53100) on day 0, followed by intraperitoneal injection of 25 µg lipopolysaccharide on day 3. After 3–4 days, the mice developed swollen wrists, ankles, and digits. 0.5 mg/kg of siRNA in C12-200 formulation was administered i.v. to appropriate groups of mice on day −3, day 0, day +4, and day+7. Anti-VLA1 mAb (100 µg; BD Biosciences Pharmingen; catalog #555000) was administered intraperitoneally on day 0, 4 hours before arthritogenic Ab cocktail. Severity of arthritis in each limb was scored as follows: 0 = normal; 1 = mild redness, slight swelling of ankle or wrist; 2 = moderate swelling of ankle or wrist; 3 = severe swelling including some digits, ankle, and foot; 4 = maximally inflamed. For FMT-CT imaging of arthritis, animals were injected with 2 nmol of the pan-cathepsin protease sensor Prosense-750 (Perkin Elmer) 24 hours prior FMT imaging.36 FMT imaging was done at excitation/emission 750/780 nm for quantitation of protease activity. Please see Supplementary Materials and Methods for a detailed description of the FMT-CT imaging method.
Acknowledgments
We acknowledge Yoshiko Iwamoto, BS (CSB-MGH) for immunohistochemical staining of spleens, Peter Waterman, BS; Brett Marinelli, BS, and Florian Leuschner, MD (CSB-MGH) for help with imaging. We thank Drs Brian Bettencourt and Greg Hinkle for designing initial sets of siRNA and helping with statistical analysis. We greatly acknowledge the small, medium, and large-scale RMA synthesis groups at Alnylam Pharmaceuticals as well as analytical, duplex annealing, and QC groups for synthesizing and characterizing RNAs. We thank Julia Hettinger, Patrick Karper, and Yongfeng Jiang for help with toxicology studies. We are also thankful to Dr John Maraganore for critical reading of the manuscript. This work was in part supported by R01HL096576 (to M.N.) as well as a NIAID contract HHSN266200600012C to Alnylam Pharmaceuticals. T.I.N., A.B., J.W., B.K., M.Z., K.Y., P.G., W.Q., S.M., L.S, R.D., A.A., K.N.J., M.J., M.M., A.d.F., and V.K. are current or former employees of Alnylam Pharmaceuticals. The work of V.M.R., G.B., P.C., R.L.B., K.L., K.W., C.L., R.L., and D.G.A. is supported in part by Alnylam Pharmaceuticals.
Supplementary Material
Distribution of CD45 silencing in multiple cell types and tissues.
Minor changes in activation status of cells engulfing LNP.
Silencing of multiple genes in myeloid cells with LNP siRNA.
Uptake of KC2 and C12-200 formulations in primary macrophages.
5′-RACE amplification of siRNA-specific RNAi-induced break points.
Silencing in organ resident cells of monocyte/macrophage lineage in NHP.
Sequences of siRNAs.
References
- Bumcrot D, Manoharan M, Koteliansky V., and, Sah DW. RNAi therapeutics: a potential new class of pharmaceutical drugs. Nat Chem Biol. 2006;2:711–719. doi: 10.1038/nchembio839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judge AD, Robbins M, Tavakoli I, Levi J, Hu L, Fronda A.et al. (2009Confirming the RNAi-mediated mechanism of action of siRNA-based cancer therapeutics in mice J Clin Invest 119661–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehead KA, Langer R., and, Anderson DG. Knocking down barriers: advances in siRNA delivery. Nat Rev Drug Discov. 2009;8:129–138. doi: 10.1038/nrd2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semple SC, Akinc A, Chen J, Sandhu AP, Mui BL, Cho CK.et al. (2010Rational design of cationic lipids for siRNA delivery Nat Biotechnol 28172–176. [DOI] [PubMed] [Google Scholar]
- Love KT, Mahon KP, Levins CG, Whitehead KA, Querbes W, Dorkin JR.et al. (2010Lipid-like materials for low-dose, in vivo gene silencing Proc Natl Acad Sci USA 1071864–1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann TS, Lee AC, Akinc A, Bramlage B, Bumcrot D, Fedoruk MN.et al. (2006RNAi-mediated gene silencing in non-human primates Nature 441111–114. [DOI] [PubMed] [Google Scholar]
- Akinc A, Zumbuehl A, Goldberg M, Leshchiner ES, Busini V, Hossain N.et al. (2008A combinatorial library of lipid-like materials for delivery of RNAi therapeutics Nat Biotechnol 26561–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akinc A, Querbes W, De S, Qin J, Frank-Kamenetsky M, Jayaprakash KN.et al. (2010Targeted delivery of RNAi therapeutics with endogenous and exogenous ligand-based mechanisms Mol Ther 181357–1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaishnaw AK, Gollob J, Gamba-Vitalo C, Hutabarat R, Sah D, Meyers R.et al. (2010A status report on RNAi therapeutics Silence 114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P.et al. (2007Transvascular delivery of small interfering RNA to the central nervous system Nature 44839–43. [DOI] [PubMed] [Google Scholar]
- Kim SS, et al. RNAi-mediated CCR5 silencing by LFA-1-targeted nanoparticles prevents HIV infection in BLT mice. Mol Ther. 2010;18:370–376. doi: 10.1038/mt.2009.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SS.et al. (2010Targeted delivery of siRNA to macrophages for anti-inflammatory treatment Mol Ther 18993–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanya S.et al. (2010Targeted delivery of small interfering RNA to human dendritic cells to suppress dengue virus infection and associated proinflammatory cytokine production J Virol 842490–2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peer D.et al. (2008Systemic leukocyte-directed siRNA delivery revealing cyclin D1 as an anti-inflammatory target Science 310627–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aouadi M.et al. (2009Orally delivered siRNA targeting macrophage Map4k4 suppresses systemic inflammation Nature 4581180–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard KA, Paludan SR, Behlke MA, Besenbacher F, Deleuran B., and, Kjems J. Chitosan/siRNA nanoparticle-mediated TNF-alpha knockdown in peritoneal macrophages for anti-inflammatory treatment in a murine arthritis model. Mol Ther. 2009;17:162–168. doi: 10.1038/mt.2008.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawroth I.et al. (2010Intraperitoneal administration of chitosan/DsiRNA nanoparticles targeting TNFa prevents radiation-induced fibrosis Radiother Oncol 97143–148. [DOI] [PubMed] [Google Scholar]
- Robbins M.et al. (2008Misinterpreting the therapeutic effects of small interfering RNA caused by immune stimulation Hum Gene Ther 19991–999. [DOI] [PubMed] [Google Scholar]
- Nguyen DN.et al. (2009Drug delivery-mediated control of RNA immunostimulation Mol Ther 171555–1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geissmann F.et al. (2010Development of monocytes, mAacrophages, and dendritic cells Science 327656–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basha G.et al. (2011Influence of cationic lipid composition of gene silencing properties of lipid nanoparticle formulations of siRNA in antigen-presenting cells Mol Ther 192186–2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leuschner F.et al. (2011Therapeutic siRNA silencing in inflammatory monocytes in mice Nat Biotechnol 291005–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nahrendorf M.et al. (2010Hybrid PET-optical imaging using targeted probes Proc Natl Acad Sci USA 1077910–7915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landesman Y.et al. (2010In vivo quantification of formulated and chemically modified small interfering RNA by heating-in-Triton quantitative reverse transcription polymerase chain reaction (HIT qRT-PCR) Silence 116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas ML. The leukocyte common antigen family. Annu Rev Immunol. 1989;8:339–369. doi: 10.1146/annurev.iy.07.040189.002011. [DOI] [PubMed] [Google Scholar]
- Desjardins M., and, Griffiths J. Phagocytosis: latex leads the way. Curr Opin Cell Bio. 2003;15:498–503. doi: 10.1016/s0955-0674(03)00083-8. [DOI] [PubMed] [Google Scholar]
- Fratti RA, Backer JM, Gruenberg J, Corvera S., and, Deretic V. Role of phosphatidylinositol 3-kinase and Rab5 effectors in phagosomal biogenesis and mycobacterial phagosome maturation arrest. J Cell Biol. 2001;154:631–644. doi: 10.1083/jcb.200106049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swirski FK, Weissleder R., and, Pittet MJ. Heterogeneous in vivo behavior of monocyte subsets in atherosclerosis. Arterioscler Thromb Vasc Biol. 2009;29:1424–1432. doi: 10.1161/ATVBAHA.108.180521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swirski FK.et al. (2009Identification of splenic reservoir monocytes and their deployment to inflammatory sites Science 325612–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nahrendorf M, Pittet MJ., and, Swirski FK. Monocytes: protagonists of infarct inflammation and repair after myocardial infarction. Circulation. 2010;121:2437–2445. doi: 10.1161/CIRCULATIONAHA.109.916346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansel KM, Harris, RB, Cyster JG. CXCL13 is required for B1 cell homing, natural antibody production, and body cavity immunity. Immunity. 2002;16:67–76. doi: 10.1016/s1074-7613(01)00257-6. [DOI] [PubMed] [Google Scholar]
- Klein I, Cornejo JC, Polakos NK, John B, Wuensch SA, Topham DJ.et al. (2007Kupffer cell heterogeneity: functional properties of bone marrow derived and sessile hepatic macrophages Blood 1104077–4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soutschek J, Akinc A, Bramlage B, Charisse K, Constien R, Donoghue M.et al. (2004Therapeutic silencing of an endogenous gene by systemic administration of modified siRNAs Nature 432173–178. [DOI] [PubMed] [Google Scholar]
- Mori L, Iselin S, De Libero G., and, Lesslauer W. Attenuation of collagen-induced arthritis in 55-kDa TNF receptor type 1 (TNFR1)-IgG1-treated and TNFR1-deficient mice. J Immunol. 1996;157:3178–3182. [PubMed] [Google Scholar]
- de Fougerolles AR, Sprague AG, Nickerson-Nutter CL, Chi-Rosso G, Rennert PD, Gardner H.et al. (2000Regulation of inflammation by collagen-binding integrins alpha1beta1 and alpha2beta1 in models of hypersensitivity and arthritis J Clin Invest 105721–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nahrendorf M.et al. (2009Hybrid in vivo FMT-CT imaging of protease activity in atherosclerosis with customized nanosensors Arterioscler Thromb Vasc Biol 291444–1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novobrantseva TI, Majeau GR, Amatucci A, Kogan S, Brenner I, Casola S.et al. (2005Attenuated liver fibrosis in the absence of B cells J Clin Invest 1153072–3082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Förster I., and, Rajewsky K. Expansion and functional activity of Ly-1+ B cells upon transfer of peritoneal cells into allotype-congenic, newborn mice. Eur J Immunol. 1987;17:521–528. doi: 10.1002/eji.1830170414. [DOI] [PubMed] [Google Scholar]
- Constien R, Forde A, Liliensiek B, Gröne HJ, Nawroth P, Hämmerling G.et al. (2001Characterization of a novel EGFP reporter mouse to monitor Cre recombination as demonstrated by a Tie2 Cre mouse line Genesis 3036–44. [DOI] [PubMed] [Google Scholar]
- Terato K, Hasty KA, Reife RA, Cremer MA, Kang AH., and, Stuart JM. Induction of arthritis with monoclonal antibodies to collagen. J Immunol. 1992;148:2103–2108. [PubMed] [Google Scholar]
- Terato K, Harper DS, Griffiths MM, Hasty DL, Ye XJ, Cremer MA.et al. (1995Collagen-induced arthritis in mice: synergistic effect of E. coli lipopolysaccharide bypasses epitope specificity in the induction of arthritis with monoclonal antibodies to type II collagen Autoimmunity 22137–147. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Distribution of CD45 silencing in multiple cell types and tissues.
Minor changes in activation status of cells engulfing LNP.
Silencing of multiple genes in myeloid cells with LNP siRNA.
Uptake of KC2 and C12-200 formulations in primary macrophages.
5′-RACE amplification of siRNA-specific RNAi-induced break points.
Silencing in organ resident cells of monocyte/macrophage lineage in NHP.
Sequences of siRNAs.