Abstract
Electroporation (EP) of mammalian tissue is a technique that has been used successfully in the clinic for the delivery of genetic-based vaccines in the form of DNA plasmids. There is great interest in platforms which efficiently deliver RNA molecules such as messenger RNA and small interfering RNA (siRNA) to mammalian tissue. However, the in vivo delivery of RNA enhanced by EP has not been extensively characterized. This paper details the optimization of electrical parameters for a novel low-voltage EP method to deliver oligonucleotides (both DNA and RNA) to dermal tissue in vivo. Initially, the electrical parameters were optimized for dermal delivery of plasmid DNA encoding green fluorescent protein (GFP) using this novel surface dermal EP device. While all investigated parameters resulted in visible transfection, voltage parameters in the 10 V range elicited the most robust signal. The parameters optimized for DNA, were then assessed for translation of successful electrotransfer of siRNA into dermal tissue. Robust tagged-siRNA transfection in skin was detected. We then assessed whether these parameters translated to successful transfer of siRNA resulting in gene knockdown in vivo. Using a reporter gene construct encoding GFP and tagged siRNA targeting the GFP message, we show simultaneous transfection of the siRNA to the skin via EP and the concomitant knockdown of the reporter gene signal. The siRNA delivery was accomplished with no evidence of injection site inflammation or local tissue damage. The minimally invasive low-voltage EP method is thus capable of efficiently delivering both DNA and RNA molecules to dermal tissue in a tolerable manner.
Keywords: dermal, DNA vaccine, electroporation, electrotransfer, gene transfer, plasmid DNA, small interfering RNA
Introduction
Small interfering RNAs (siRNAs) have recently demonstrated their potential as novel therapeutics due to their ability to induce robust, sequence-specific gene silencing in cells.1,2 However, several challenges, particularly related to delivery, surrounding the use of siRNA in vivo still need to be optimized before the full potential of therapeutic RNA interference is realized. Due to the polyanionic nature of the molecule, the cellular uptake of naked siRNA on its own is extremely low. These molecules are also highly susceptible to degradation by enzymes both in tissue and intracellularly as well as systemically in the blood. Therefore, to become an effective clinical tool, an efficient, targeted, and tolerable in vivo delivery method must be identified. Using siRNA to induce RNA interference could become a promising therapeutic for treatment of many disorders, such as some cancers and many viral and genetic diseases. Although some trials involving siRNA have entered the clinic, delivery of siRNA, particularly locally for dermal applications, remains a key challenge. Indeed, the potential for clinical success of siRNA therapeutics hinges on an efficient and targetable delivery system.
In addition to improvements in the stability of the molecule which can be achieved through chemical modification, multiple delivery strategies for siRNA have also been attempted. These include both physical and carrier-mediated methods. Examples of drug delivery through carrier-mediated methods include the traditional liposomal methods such as lipid nanoparticles,3 as well as ultrasound and microbubble delivery.4
Electroporation (EP) represents a physical method to temporarily increase the permeability of a target tissue to macromolecules. EP involves the application of brief electrical pulses that result in the creation of aqueous pathways within the lipid bilayer membranes of mammalian cells. This transient perturbation of the lipid bilayer allows the passage of large molecules, including nucleic acids, across the cell membrane which otherwise is less permeable. As such, EP increases the uptake of macromolecules delivered to their target tissue. EP has now entered the clinic as an enabling technology for the delivery of DNA vaccines and immune therapies. The technology has proven to be a safe and effective DNA delivery method in multiple trials delivering an array of genetic vaccines.5,6,7
We have previously reported on a subcutaneous DNA EP delivery device with a penetration depth of 3 mm (CELLECTRA-3P)8 as well as three intramuscular DNA delivery devices, the Medpulser,9 ELGEN,5 and CELLECTRA-5P10,11,12 devices (Inovio Pharmaceuticals, San Diego, CA). These devices cover injection depths between 3 and 25 mm, typically suited for subcutaneous and/or intramuscular vaccine delivery using distinct EP array configurations. Extensive studies have highlighted the critical requirement for optimization of both the EP device and the parameters used for successful delivery which additionally are dependent on both the target tissue and the DNA construct to be delivered.8 Combining optimized EP delivery methods with optimized constructs has led to the successful translation of DNA vaccination into a clinical setting.5,13,14,15
While the body of evidence for both preclinical and clinical success of enhanced DNA delivery by EP is strong, the characterization of EP-enhanced RNA delivery is not as clear. However, electrically assisted delivery of siRNA has been documented in a variety of tissues including the cornea,16 solid tumors,17,18,19 joints,20 muscle,21 brain tissue,22 and skin.23 Certainly DNA and RNA differ both physically and electrochemically. These differences affect the dielectric properties of the molecules and as such could impact their ability to be successfully transfected in vivo using EP. The basis for EP is the application of an external electrical field which results in a significant increase in the permeability of the cell plasma membrane. Therefore the size, structure, and charge of the molecule to be transfected could significantly affect the ability of that molecule to permeate and/or interact with the membrane. Plasmid DNA, generally ranging in size from 3–5 kb, exists in solution primarily in supercoiled form (75–80%). Supercoiled or covalently closed-circular DNA is a relatively large (3–5 kb; 2–4 Mda) contorted and double-stranded molecule adopting a highly compacted structure.24 The structure of siRNA on the other hand is a short (usually 21-nts) double-stranded RNA with 2-nt 3′ overhangs on either end. Clearly, the optimal transfer dynamics of these two nucleotide molecules could be widely disparate.
Here we investigated the delivery of siRNA to skin and optimized the EP conditions for delivery using a surface EP (SEP) device with a novel 4 × 4 minimally invasive needle array.25 This SEP device differs from other EP devices in that the electrodes are minimally invasive (scratch the skin surface but do not penetrate the skin).
Initially, the electrical parameters were optimized using plasmid DNA encoding green fluorescent protein (GFP). These parameters were then assessed for translation to electrotransfer of siRNA using a fluorescently tagged siRNA for detection. We then investigated the delivery of tagged siRNA to skin through histological methods and additionally assessed the ability of EP to enhance targeted gene knockdown in dermal tissue.
In summary, this study sought to answer two fundamental questions: first, can EP be used to efficiently deliver siRNA in vivo in a functionally relevant manner and second, whether parameters optimized for DNA transfection may also be used for RNA delivery, especially at the low-voltage settings?
Results
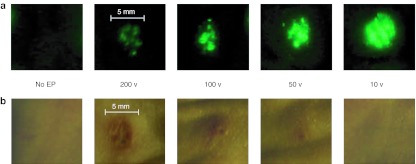
A range of voltage parameters results in successful expression of GFP in dermal tissue. We have previously reported on the development of the 4 × 4 SEP25 where we optimized critical parameters such as electrical pulse pattern, needle array geometry and electrode configuration, depth of penetration, and operating conditions under constant current or constant voltage configurations. With the early development accomplished, we sought to further explore effective voltage parameters for the SEP device, and to this end conducted reporter gene (plasmid-expressing GFP) expression and localization studies in guinea pig skin following EP with the SEP device. Separate skin sites on the flank of a hairless guinea pig were injected with 50 µl of GFP plasmid (@ 1 mg/ml concentration) and immediately pulsed (single 100 ms pulse) using the SEP set at a series of voltage parameters ranging between 10–200 V (Figure 1a). Robust GFP transfection was seen using the 10- and 50-V parameters, with the 10-V treatment appearing stronger and more reproducible than the 50-V treatment. The skin area transfected following the 10-V treatment corresponded in size (~4 mm2) to the surface area of the SEP device and the injection bubble size (4 mm diameter). Although GFP transfection was detected following the 100- and 200-V parameters, unexpectedly, the signal was significantly weaker than the 10- and 50-V treatments and appeared far less reproducible over multiple treatment sites. GFP transfection was visible 8 hours following treatment and peaked at 3 days. In contrast, minimal or no GFP transfection was detectible following GFP plasmid injection alone.
Figure 1.
A range of voltage parameters results in delivery of DNA plasmid expressing GFP to guinea pig skin using the SEP device. Guinea pig skin was injected intradermally with 100 µg of DNA plasmid expressing GFP (pgWIZ-GFP) and either left untreated or immediately pulsed with the SEP device at a voltage setting of 200, 100, 50 or 10 V for one pulse. Skin biopsies were harvested 48 hours later and observed by fluorescent microscopy (a) for determination of GFP positive signal or bright light (b) for determination of local treatment site reaction. Bar is included for approximation of site reaction size. GFP, green fluorescent protein; SEP, surface electroporation.
Direct contact between the skin and electrical devices could result in tissue damage. To assess the effect of voltage parameters on dermal integrity, the guinea pig skin was assessed for signs of tissue damage, including redness, swelling and/or burning following the treatments detailed in Figure 1a across several voltage settings (Figure 1b). At 48 hours post-treatment, skin sites pulsed with 200, 100, and 50 V all showed signs of redness and swelling which reduced in severity with a reduction in voltage. Not surprisingly, the 200 and 100 voltage treatments showed significant surface burning of the skin resulting in scabbing and inflammation. Only the no EP, injection only control group and the 10 V treatments showed no visible signs of tissue damage. Thus the lower voltage parameters resulted in not only a more efficient delivery of DNA, but also led to a more tolerable EP procedure.
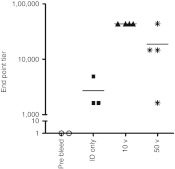
Lower voltage parameters result in higher antibody titers to NP protein. Although establishing the expression patterns for a plasmid reporter gene allowed us to gain insight into parameter optimization, a key determinant for assessing efficient EP is induction of immunogenicity. As such, we then sought to establish the optimal parameters leading to a functional immune response. Since significant tissue damage resulted from pulsing the SEP at 200 and 100 V for intradermal delivery, the immune study was conducted with the 50- and 10-V parameters. Hartley guinea pigs were immunized with plasmid DNA encoding for NP influenza antigens. Matched NP antigen from Puerto Rico/39 strain was optimized, synthesized, and then cloned into the backbone of a mammalian expression vector, pMB76.5, which has been used in previous human clinical trials. Animals were immunized intradermally with 100 µg of DNA at week 0 and were boosted at week 3 with the same amount of DNA at week 3. Groups of guinea pigs (four/group EP or three/group injection only) were electroporated either with the SEP device at the 50 or 10 V setting or left as injection only without EP. As seen in Figure 2, at week 7, robust antibody titers were generated by both EP conditions, although the titers in the 10-V treatment group were double (~60, 000) compared to those in the 50-V group (~30, 000). The titers in the injection only group were approximately tenfold less than the EP groups. Interestingly, the reproducibility of the titers generated was consistently better for the 10-V group over the 50-V group as determined by the scatter of the plotted titers.
Figure 2.
Lower voltage parameters result in higher antibody titers to NP protein in guinea pigs. Guinea pig skin was injected intradermally with 100 µg of DNA plasmid-expressing NP and either left untreated (ID only) or immediately pulsed with the SEP device at a voltage setting of 50 or 10 V. Animals were immunized at day 0 and day 21. Blood was harvested weekly and analyzed for the presence of binding antibodies against NP by ELISA. Data shown here is from day 49. ELISA, enzyme-linked immunosorbent assay; ID, intradermal; SEP, surface electroporation.
A range of voltage parameters results in successful delivery of tagged siRNA to dermal tissue. In a bid to establish effective voltage parameters for siRNA delivery by the SEP device, we carried out a localization study using a fluorescently tagged siRNA in guinea pig skin following SEP at defined parameters. First we investigated whether the parameters that resulted in effective plasmid delivery were similar for optimal siRNA delivery. Separate skin sites on the flank of Hartley guinea pigs were injected with 50 µl of siRNA-tagged with Alexa-488 (@ 2 mg/ml concentration) and immediately pulsed (single 100 ms pulse) using the SEP set at a series of voltage parameters ranging between 10–200 V (Figure 3). Two days post-treatment, the animals were euthanized and 8 mm skin biopsies were removed and visualized under a fluorescent microscope. Strong Alexa signal was detected using all voltage parameters, with the 10-V treatment appearing to elicit the strongest signal. Minimal or no Alexa signal was detectible following injection of the tagged siRNA with no EP.
Figure 3.
A range of voltage parameters results in successful delivery of tagged siRNA to guinea pig skin using the SEP device. Guinea pig skin was injected intradermally with 100 µg of siRNA-tagged with Alexa 488 and either left untreated or immediately pulsed with the SEP device at a voltage setting of 200, 100, 50 or 10 V for three pulses. Skin biopsies were harvested 48 hours later and observed by fluorescent microscopy for determination of positive signal. Bar is included for approximation of biopsy size. SEP, surface electroporation; siRNA, small interfering RNA.
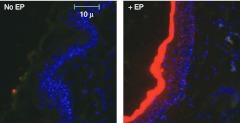
Dermal EP results in robust siRNA-Cy3 signal present 48 hours post-treatment. We next continued the investigation of siRNA delivery enhanced by EP using the 10-V parameters, since this parameter set appeared to have both the optimal delivery efficiency and the least-associated tissue damage. Skin biopsies were taken and histological analysis was carried out to assess delivery efficiency of EP for siRNA at the cellular level. Separate skin sites on the flank of Hartley guinea pigs were injected with 50 µl of siRNA-tagged with Cy3 (@ 2 mg/ml concentration) and either immediately pulsed (three 100 ms pulses) using the SEP set at 10 V or left as injection only controls (Figure 4). Two days post-treatment, the animals were euthanized and 8 mm skin biopsies were removed, fixed, and prepared for paraffin sectioning. Sections were stained with 4′,6-diamidino-2-phenylindole (DAPI) and visualized under a microscope for positive Cy3 signal. At 48 hours, little or no Cy3 signal was detected in the injection only controls. However, robust Cy3 signal was detected in the EP group.
Figure 4.
Dermal electroporation results in robust siRNA-Cy3 signal present 48 hours post-treatment. Guinea pig skin was injected intradermally with 100 µg of siRNA-tagged with Cy3 and either left untreated or immediately pulsed with the SEP device at 10 V for three pulses. Skin biopsies were harvested 48 hours later, fixed in formalin, sectioned, and DAPI stained. Slide sections were observed by fluorescent microscopy for determination of positive signal. DAPI, 4′,6-diamidino-2-phenylindole; SEP, surface electroporation; siRNA, small interfering RNA.
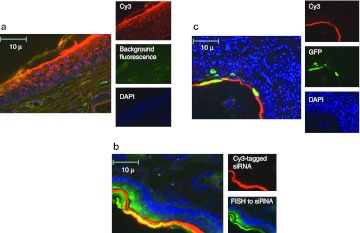
Surface EP results in siRNA-Cy3 signal with confirmation by siRNA-specific FISH, and GFP expression colocalized to the stratum corneum and epithelial cells. We further assessed the localization of the signal to specific layers of the skin using histological techniques (Figure 5). Separate skin sites on the flank of Hartley guinea pigs were injected with 50 µl of siRNA-tagged with Cy3 (@ 2 mg/ml concentration) and immediately pulsed (three 100 ms pulses) using the SEP set at 10 V (Figure 5a). Two days post-treatment, the animals were euthanized and 8 mm skin biopsies were removed and fixed and prepared for paraffin sectioning. Sections were stained with DAPI and visualized under a high power microscope for positive Cy3 signal. The majority of the signal was localized to the upper layers of the epidermis (stratum corneum). However, Cy3 signal was also detected in epidermal cells throughout the stratified layers, from the basement membrane to the surface. No Cy3 signal was detected in the dermis, which is in keeping with the mode of action of this delivery device.
Figure 5.
Dermal electroporation results in siRNA-Cy3 signal with confirmation by siRNA-specific FISH, and GFP expression colocalized to the stratum corneum and epithelial cells. (a) Guinea pig skin was injected intradermally with 100 µg of siRNA-tagged with Cy3 and immediately pulsed with the dermal EP device at 10 V for three pulses. Skin biopsies were harvested 48 hours later, fixed in formalin, DAPI stained, and sectioned. Slide sections were observed by fluorescent microscopy for determination of positive signal. (b) The localization of siRNA was additionally detected by fluroescein-tagged fluorescent in situ hybridization (FISH) to the antisense strand of the Cy3-tagged siRNA in guinea pig skin sections serial to those displayed in part A of this figure. (c) Guinea pig skin was injected intradermally with a mix of 100 µg of siRNA-tagged with Cy3 and 100 µg DNA plasmid-expressing GFP and immediately pulsed with the dermal EP device at 10 V for three pulses. Skin biopsies were harvested 48 hours later, fixed in formalin, sectioned, and DAPI stained. Slide sections were observed by fluorescent microscopy for determination of positive signal. DAPI, 4′,6-diamidino-2-phenylindole; GFP, green fluorescent protein; siRNA, small interfering RNA.
Furthermore, siRNA localization as detected by Cy3 signal was confirmed by fluorescent in situ hybridization (FISH) using probes specific to the antisense strand of the siRNA (Figure 5b). Serial skin sections to those analyzed for Cy3 signal were hybridized to a 5′DIG-labeled, locked nucleic acid (LNA) enhanced anti-antisense siRNA probe and treated with a fluorescein tyramide signal amplifier. Sections were visualized for both positive Cy3 and fluorescein signal. As shown in Figure 5c, there was significant accumulation and clear colocalization of the signals to the stratum corneum. In additon, fluorescein signal was detected to a lesser extent in the epidermal layers, as was seen with Cy3 signal.
To address whether plasmid DNA and siRNA can colocalize when codelivered, plasmid DNA expressing GFP (100 µg) was mixed with siRNA-tagged with Cy3 (100 µg) and simultaneously injected into the skin of guinea pigs followed by EP using the 10-V settings. The biopsies were prepared as described above but this time visualized for both positive Cy3 signal and positive GFP signal. There was clear colocalization of both signals to the stratum corneum. Both signals were also detected in cells in the epidermis. Although highly likely, we were nevertheless unable to definitively determine visually whether any single cell contained both signals.
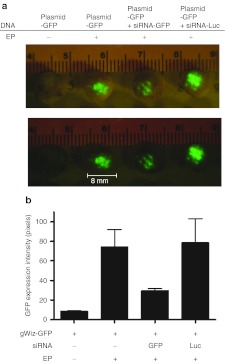
Dermal EP results in targeted reporter gene knockdown in dermal tissue. Having demonstrated successful delivery of tagged siRNA to skin through histological methods, we wanted to assess whether EP-enhanced siRNA delivery would result in targeted gene knockdown in dermal tissue. Separate skin sites on the flank of Hartley guinea pigs were injected with 20 µg siRNA-GFP mixed with 25 µg plasmid-expressing GFP immediately pulsed (three 100 ms pulses) using the SEP device set at 10 V (Figure 6a). The expression of GFP in this group was compared to the plasmid only group. Negative controls of plasmid injection only and plasmid injection plus non-matched (luciferase) siRNA were also included. Representative examples of biopsies are shown here. Twenty-four hours post-treatment, the animal was euthanized and 8 mm skin biopsies were removed. The biopsies (four for each siRNA group and eight from the plasmid only groups) were visualized under fluorescent microscopy and pixel counting software used to quantify the signal (Figure 6b). Visually, there was a clear reduction in the GFP signal in the group where siRNA-GFP was codelivered with the plasmid. The plasmid only and plasmid plus siRNA-Luc (non-matched control) appeared similar. The pixel density quantification (determined from the average of multiple samples) demonstrated an approximate 50% reduction in GFP signal. As is consistent with previous experiments, little or no GFP signal was detected in the plasmid injection alone group.
Figure 6.
Delivery of siRNA by the SEP device results in targeted gene knockdown of reporter gene in dermal tissue. Guinea pig skin was injected intradermally with 25 µg of DNA plasmid-expressing GFP (pgWIZ-GFP) left untreated, 25 µg of DNA plasmid-expressing GFP and immediately pulsed with the SEP device or plasmid mixed with either siRNA-GFP or siRNA-Luc (20 µg) and immediately pulsed with the SEP device. (a) Skin biopsies were harvested 24 hours later and observed under bright light for determination of treatment site location or fluorescent microscopy for determination of GFP positive signal. (b) GFP pixel density was determined using pixel counting software as described (see Materials and Methods). GFP, green fluorescent protein; luc, luciferase; SEP, surface electroporation; siRNA, small interfering RNA.
Discussion
Due to their ability to induce robust, sequence-specific gene silencing in cells, siRNAs1,2 are an exciting and novel therapeutic option for disease targets such as macular degeneration,26 solid tumors,27 and melanoma's. Indeed, clinical programs assessing the safety, tolerability, and pharmacokinetics of targeted siRNA have now been assessed in patients.26,27 However, RNA-based drugs in the absence of an appropriate delivery vehicle or method are hampered in their applicability due to their low cellular uptake and susceptibility to degradation. As such, to become an effective clinical tool, it is important to identify a targeted delivery system that can achieve effective transfection of siRNAs.
Our group at Inovio has focused on the design and development of novel, innovative solutions to key oligonucleotide delivery problems.23,28,29 Since a need exists for more effective delivery platforms for localized in vivo siRNA delivery, we assessed whether our EP platform was a viable solution for local siRNA delivery.
The primary interest for this study was to establish whether EP parameters optimized to deliver plasmid DNA would translate to effective delivery of RNA in vivo. If successful, the ability to deliver distinct nucleic acids would give the EP platform far-reaching therapeutic potential. New EP devices and parameters optimized for efficacy, tolerability, and safety could expand treatment and drug delivery options, especially in the field of RNA therapeutics. Both preclinical and clinical studies have demonstrated that EP, as an effective physical delivery method, can improve both the expression and immunogenicity of DNA vaccines by 100–1,000 fold.
Having previously developed a novel EP device that effectively targets dermal tissue, we were also keen to establish optimal EP parameters for oligonucleotide transfer which could offer both efficient delivery and reduced tissue damage.
Initially, using GFP expression as a readout for intracellular delivery, we determined parameter ranges for plasmid DNA transfection using this EP device, and were able to observe transfection of epidermal tissue over a spectrum of voltage parameters, ranging from 200 V down to 10 V (Figure 1a). Interestingly, GFP transfection at the lowest voltage parameter (10 V) appeared more reproducible and robust in comparison to the 50–200 V parameters. Although 10 V is considered a low-voltage parameter, the actual field strength (a function of the electrode distance) is still 67 V/cm, well within the acceptable range for in vivo EP.30,31 At the higher voltage settings, this field strength increases substantially (200 applied volts = 1,334 V/cm), resulting in significant cell death as observed through histological analysis. This may explain the reduced levels of GFP transfection seen when the applied voltage is increased for the SEP device. We note that other earlier EP devices operate in the 100–200 voltage range for effective gene delivery. However, for those devices, there is typically greater separation between electrodes (0.5 to >1.0 cm) thereby maintaining reasonable field strengths to achieve efficient gene transfer without significant tissue damage. Importantly, efficient transfection was achieved at voltage settings as low as 10 V without any observable tissue damage with the SEP device.
As seen in Figure 1, transfection of the reporter gene plasmid may be occurring; however, the cells may be unable to recover from the EP treatment and thus are unable to express the plasmid. In support of this theory is the steady increase in tissue damage occurring following EP with the higher voltage parameters. Visible redness, inflammation and in some cases, scabbing was identified following treatments between 50–200 V with the SEP device (Figure 1b). We also note that the presence of hemoglobin quenches the GFP signal.32
Although exploring plasmid reporter gene expression across a wide range of voltage parameters was informative, of greater interest was assessing the voltage parameters which resulted in an immune response. It was clear that the 100 and 200 voltage parameters caused significant tissue damage, and as a result, reduced plasmid expression upon treatment with the SEP device. As such, we limited our assessment to the 50- and 10-V parameters to induce humoral immune responses as compared to intradermal injection alone (Figure 2). Both the 50- and 10-V parameters illicited responses of significant magnitude against the administered antigen. However, the spread of the titers was wider in the 50-V group. Indeed, one animal's response was similar in magnitude to the injection-only control. The reduction in expression due to increased cell damage/death may also explain the attenuated responses and titers. While only one time point is shown here (week 7), the trend towards achieving higher titers with lower voltage parameters was demonstrated at all time points, through study completion (week 12).
Ultimately, our primary question for this study was to establish whether EP would be able to enhance the delivery of siRNA (Figure 3). Again, initial range-finding experiments were conducted using the original DNA-EP parameter set (200–10 V) used in Figure 1. Since siRNAs are considerably smaller than plasmids, we were unsure whether EP parameters optimized for a large circular DNA molecule (~2 kb) would effectively deliver a smaller double-stranded RNA (20–25 nts in length). Using Alexa-488 tagged siRNA, we were able to demonstrate that a large range of voltage parameters (200–10 V) successfully delivered the siRNA to dermal tissue. There appeared to be a trend towards higher delivery efficiency at the lower voltage settings, thus supporting the pattern observed with plasmid delivery. Since the siRNA is tagged and therefore does not require expression for visualization, (unlike the plasmid DNA), the effect of the tissue damage at higher voltages seemed less pronounced in these samples. Since the 10-V parameter appeared to induce the most effective delivery of siRNA, we wanted to further investigate the localization of the delivered siRNA in the dermal tissue by histological techniques. Forty-eight hours following an injection-only delivery displayed no detectable Cy3 siRNA in the skin biopsy. However, robust Cy3 signal was present in the epidermis of the skin following EP-enhanced delivery (Figure 4). At an increased magnification, it was clear that the majority of the Cy3 signal was localized to the stratum corneum (Figure 5a) which is the upper-most stratified layer in the epidermis. However, clear localization of Cy3 could also be seen in epithelial cells in the epidermis, from the basement membrane up. As cells in the epidermis differentiate they move from the lowest layer (basement membrane) to the surface (stratum corneum). The concentrated signal in the stratum corneum could potentially be attributed to cells transfected with Cy3 siRNA that are moving through the natural regenerative cycle. The Cy3 signal remained strong at 48 hours although it was unclear whether the tag was still attached to the siRNA.
In order to confirm the localization of siRNA as detected by Cy3 signal, we employed FISH, visualized by a fluorescein-tagged tyramide signal amplifier, to detect the antisense strand of the siRNA. Analyzing serial sections from the same dermal tissue that was used for Cy3 detection allowed us to determine the extent of signal colocalization. As seen in Figure 5b, there was significant accumulation and clear colocalization of the Cy3 and fluorescein signals to the stratum corneum. Fluorescein signal was detected in the epidermal layers; however, to a lesser extent than the stratum corneum similar to the observations with direct detection of Cy3 signal. These data confirm that the distribution pattern of Cy3 in the skin sections is reflects that of siRNA localization and is not simply detection of free fluorophore.
We were keen to understand whether simultaneous codelivery of fluorescent DNA and RNA molecules would result in colocalization of signals. To investigate this, plasmid-expressing GFP was mixed with Cy3-labeled siRNA and delivered simultaneously to skin using the SEP (Figure 5c). There was clear colocalization of both signals to the stratum corneum and there was also signal in epithelial cells in the epidermis; however, it was difficult to determine whether any single cell contained both GFP and Cy3-siRNA signals. It also appeared that at the 48-hour time point, more cells were Cy3-positive than GFP-positive. This may reflect the kinetics of expression or stability of GFP in the cell.
While the tagged siRNA allowed us to establish the feasibility of siRNA delivery and optimize delivery parameters, the true application would be to demonstrate targeted gene knockdown. Using the plasmid expression of GFP, we demonstrated that matched siRNA simultaneously delivered was able to significantly knockdown reporter gene expression in the skin (Figure 6). As hypothesized, unmatched (siRNA-Luc) had no effect on GFP expression. We believe that this successful demonstration of siRNA delivery to dermal tissue demonstrates that EP-mediated delivery of siRNA is a viable option to enhance gene knockdown in the skin and could offer a clinical platform for therapeutic options in the future. We considered the use of sequential administration of GFP plasmid followed by the administration of siRNA at a subsequent time point (a day or two later to better model a therapeutic effect). However, skin delivery of DNA and siRNA to the animal at the same site presents a significant technical challenge and cannot be adequately controlled. This is because over time the injected liquid (and the plasmid or siRNA) dissipates away from the injection site, the surface of the skin and the underlying dermal layer slide relative to each other, and it is difficult to adequately ensure that the same cells that receive the reporter plasmid are also getting the siRNA molecules. Towards this end, subsequent studies will address the effects of improved EP-mediated siRNA delivery in surface tumor models and investigate the impact on tumor growth. Using a visible tumor model allows us to accurately target siRNA delivery to the tumor target directly.
EP has been previously used in several studies to deliver siRNA to target tissues. Like the Inoue et al. study,23 we also demonstrate successful delivery of siRNA to skin. A distinct difference between the two studies is the animal model used. Mouse skin was used in the Inoue study which also presents a distinct challenge in that because it is very thin, it is difficult to control transfection and gene silencing to the skin; instead it is likely that both skin and the underlying muscle are impacted. In this study, we investigated siRNA transfer in guinea pig skin which represents a more relevant dermatological model due to its similarities in thickness and physiology to human skin. A major advancement in the siRNA delivery protocol that we describe here is related to the device and the optimal EP parameters. Here we determined that low-voltage parameters (10 applied volts or 67 V/cm), which generate minimal current, using our SEP device was optimal at siRNA transfer. Indeed, such parameters result in no discernable tissue damage and as previously reported for delivery of DNA molecules, are highly tolerable.25 The specific design of the device also lends itself to optimized delivery. Due to the grind of the electrodes, no additional conductive gel need to be applied and the device makes only contact with the surface of the skin. Therefore from a patient's perspective, the benefit of combining the SEP device with low-voltage parameters for RNA delivery is increased tolerability. Achieving efficacy and target specificity are crucial elements of this delivery platform.
We not only established that we could enhance RNA delivery to skin by EP and demonstrate reporter gene knockdown, we also established that our low voltage, tolerable SEP parameters appeared to induce the most effective delivery. The EP platform detailed in this study at present is particularly suited to topical RNA delivery applications.
In summary, these data support the idea that RNA delivery can be facilitated by EP, much in the same way that DNA delivery is, and as such, this platform could pave the way for development of targeted RNA-based therapies for local applications.
Materials and Methods
SEP minimally invasive device design. Electrode arrays consisting of a 4 × 4 gold-plated trocar needle of 0.0175 inch diameter at a 1.5 mm spacing were constructed to be used in conjunction either with the ELGEN1000 (Inovio Pharmaceuticals, Blue Bell, PA) pulse generator or a battery powered low voltage circuit.
Plasmid preparation. The gWiz GFP plasmid was purchased from Aldevron (Fargo, ND). The NP plasmid encodes the full-length NP derived from Puerto Rico 8 (H1N1) strain of influenza (accession number: ADY00024.1). The construct had the nuclear targeting signals mutated and was optimized and synthesized by GeneArt (Grand Island, NY) then cloned into the backbone of a mammalian expression vector, pMB76.5. All plasmids were diluted in 1× phosphate-buffered saline before injection.
siRNA preparation. The Cy3-tagged (Quasar570) and Alexa-488-tagged siRNAs were synthesized by Alynlam, Cambridge, MA. RNA oligonucleotides were synthesized using commercially available 5′-O-(4,4′-dimethoxytrityl)-2′-O-t-butyldimethylsilyl-3′-O-(2-cyanoethyl-N,N-diisopropyl) phosphoramidite monomers of uridine (U), 4-N-acetylcytidine(CAc), 6-N-benzoyladenosine (ABz), and 2-N-isobutyrylguanosine (GiBu), according to standard solid phase oligonucleotide synthesis protocols. For sequences containing Quasar570, Quasar 570 amidite (Biosearch Technologies, Novato, CA) was directly coupled during RNA synthesis. After cleavage and deprotection, RNA oligonucleotides were purified by anion-exchange high-performance liquid chromatography and characterized by electrospray mass spectrometry and capillary gel electrophoresis. RNA with phosphorothioate backbone at a given position was achieved by oxidation of phosphite during oligonucleotide synthesis using 0.1 mol/lDDTT (3-(dimethylaminomethylene) amino-3H-1, 2, 4-dithiazole-5-thione). For sequences containing Alexa 488, the conjugation of the dye was done using the Alexa488 succinimidyl ester (Invitrogen, Carlsbad, CA) post-synthetically to the corresponding purified amino-containing RNA precursor.
The GFP-siRNA and Luc-siRNA was purchased from Bioneer, Daejeon, Korea. The sequence of the GFP sense strand was CGAAGGUUAUGUACAGGAA(dTdT) and the sequence of the luciferase sense strand was UUGUUUUGGAGCACGG AAA(dTdT).
The sense strand of the tagged siRNA is 5′-UCGAAGUACUCAGCGUAAG-3′ and the antisense strand is 5′-CUUACGCUGAGUACUUCGA-3′.
Animals. Female Hartley guinea pigs (strain code 051) and female IAF hairless guinea pigs (strain code 161) were purchased from Charles River Laboratories, Wilmington, MA. Guinea pigs (four animals per group for immune study) were housed at BioQuant (San Diego, CA).
All animals were housed and handled according to the standards of the Institutional Animal Care and Use Committee (IACUC).
Preparation of animals. The GFP reporter results observed on the Hartley guinea pigs after hair removal were the same as the results observed on the IAF hairless guinea pigs in the initial plasmid localization experiments. Since hair removal appeared to have no effect on the resulting transfection and due to cost considerations, we chose to carry out the immune study in Hartley guinea pigs. Hartley guinea pigs were shaved and stubble removed by dilapatory cream (Veet) 24 hours before treatment.
DNA/siRNA injections. Guinea pigs were injected intradermally (Mantoux method (needle parallel to skin)–29 gauge insulin needle) with 50 µl of 1× phosphate-buffered saline containing the desired dose of plasmid or siRNA. For the immune study, animals were vaccinated twice 3 weeks apart with 100 µg plasmid at each immunization.
Dermal device EP. Immediately following injection of DNA or siRNA, the dermal device was applied to the site of dermal injection. The array was “wiggled” at the injection site to ensure good contact and electrotransfer achieved through pulse generation either from the Elgen 1000 or a low-voltage battery circuit.
Visualization of GFP reporter gene signal. Skin samples or biopsies were removed postmortem from animals after termination and stored on ice until imaged under an OV 100 imaging microscope (AntiCancer, San Diego, CA) at 480 nm.
Pixel count method. The images were processed using Adobe Photoshop CS5. A “gated region” of electrode contact for pixel analysis was established on the presumption that transfection occurs only where the electric field is applied and that the electric field is formed only where the electrodes are in direct contact with the skin. The distance between the first and fourth electrode in the SEP device is 4.5 mm. The “ruler tool” in Photoshop was used to isolate a 4.5 mm2 region which was defined as ~95 pixels in length. The images obtained from the microscopy were 8 bit RGBs files. Photoshop is able to recognize pixel intensities ranging from 0–255 (darkest–brightest) in three different channels (red, green, blue). Intuitively, positive GFP signal would predominate in the green channel; therefore, pixel analysis was restricted to this channel. The CS5 version of Photoshop is able to automatically calculate mean and median pixel intensity of a selected region. Since the distribution of pixel intensity was not symmetrical in most cases, the median gave a better representation of central tendency for the histogram.
Histological analysis. Skin samples or biopsies were removed postmortem from animals after termination and immediately preserved in 10% neutral buffered formalin and processed for histopathological analysis. Appropriate tissues were trimmed, processed, embedded in paraffin, sectioned at ~5 µm, and stained with DAPI. All images were captured on the Zeiss Axiovision Microscope using (Carl Zeiss, Goettingen, Germany) a ×20 objective.
FISH. Skin sections that were serial to those used for histological analysis were processed by FISH using a 5′ DIG-labeled, LNA enhanced probe (Exiqon, Woburn, MA) designed to detect the antisense strand of the Cy3-siRNA. The sequence of the probe is 5′-AACTTACGCTGAGTACTTC-3′.
Briefly, deparaffinized sections underwent a series of blocking steps and were hybridized to the LNA probe overnight at Tm-20 °C. The hybridization temperature was 50 °C. The hybridization buffer is composed of: 5 ml 50% Formamide, 2.5 ml of 20× saline-sodium citrate (SSC), 2 ml of 50% Dextran sulfate, 200 µl of 50× Denhardt's, and 500 µl of 10 mg/ml yeast RNA.
Sections were washed, blocked, and incubated with an anti-DIG-HRP antibody (Perkin Elmer, Waltham, MA). A fluorescein-fluorophore Tyramide Signal Amplification Kit (Perkin Elmer) was used to enhance HRP-generated signal. Skin sections were then stained with DAPI and visualized for both Cy3 signal (directly tagged to siRNA), and fluorescein signal (detection of the AS strand of the Cy3-siRNA). Images were overlaid to determine the extent of colocalization.
Acknowledgments
We thank Janess Mendoza (Inovio) and Maria Yang (Inovio) for technical help with plasmid preparation, assays, and animal care. In addition, we acknowledge Jay McCoy (Inovio) and Steve Kemmerrer (Inovio) for their engineering assistance. N.Y.S., X.S., A.S.K., F.L., K.E.B, and G.K. are current employees of Inovio and as such have financial interest (in the form of salary compensation, stock options and/or stock ownership) in the work described in this manuscript. T.S.Z., A.C., and J.A. are employees of Alnylam Pharmaceuticals and as such have financial interest (in the form of salary compensation, stock options and/or stock ownership) in the work described in this manuscript.
References
- Bumcrot D, Manoharan M, Koteliansky V., and, Sah DW. RNAi therapeutics: a potential new class of pharmaceutical drugs. Nat Chem Biol. 2006;2:711–719. doi: 10.1038/nchembio839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Fougerolles A, Vornlocher HP, Maraganore J., and, Lieberman J. Interfering with disease: a progress report on siRNA-based therapeutics. Nat Rev Drug Discov. 2007;6:443–453. doi: 10.1038/nrd2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akinc A, Querbes W, De S, Qin J, Frank-Kamenetsky M, Jayaprakash KN.et al. (2010Targeted delivery of RNAi therapeutics with endogenous and exogenous ligand-based mechanisms Mol Ther 181357–1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi Y, Kawakami S., and, Hashida M. Strategies for in vivo delivery of siRNAs: recent progress. BioDrugs. 2010;24:195–205. doi: 10.2165/11534450-000000000-00000. [DOI] [PubMed] [Google Scholar]
- Low L, Mander A, McCann K, Dearnaley D, Tjelle T, Mathiesen I.et al. (2009DNA vaccination with electroporation induces increased antibody responses in patients with prostate cancer Hum Gene Ther 201269–1278. [DOI] [PubMed] [Google Scholar]
- Sallberg M, Frelin L, Diepolder HM, Jung MC, Mathiesen I, Fons M.et al. (2010Therapeutic vaccination followed by standard-of-care therapy in patients with chronic hepatitis C: a rapid clearance of viremia Mol Ther 18S110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J, Pankhong P, Shen X, Giffer M, Lee J, Harris D.et al. (2010Phase I safety and immunogenicity of HPV 16 and 18 DNA vaccines delivered via electroporation Mol Ther 18S184 [Google Scholar]
- Hirao LA, Wu L, Khan AS, Satishchandran A, Draghia-Akli R., and, Weiner DB. Intradermal/subcutaneous immunization by electroporation improves plasmid vaccine delivery and potency in pigs and rhesus macaques. Vaccine. 2008;26:440–448. doi: 10.1016/j.vaccine.2007.10.041. [DOI] [PubMed] [Google Scholar]
- Wallace M, Evans B, Woods S, Mogg R, Zhang L, Finnefrock AC.et al. (2009Tolerability of two sequential electroporation treatments using MedPulser DNA delivery system (DDS) in healthy adults Mol Ther 17922–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draghia-Akli R, Khan AS, Brown PA, Pope MA, Wu L, Hirao L.et al. (2008Parameters for DNA vaccination using adaptive constant-current electroporation in mouse and pig models Vaccine 265230–5237. [DOI] [PubMed] [Google Scholar]
- Laddy DJ, Yan J, Khan AS, Andersen H, Cohn A, Greenhouse J.et al. (2009Electroporation of synthetic DNA antigens offers protection in nonhuman primates challenged with highly pathogenic avian influenza virus J Virol 834624–4630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel V, Valentin A, Kulkarni V, Rosati M, Bergamaschi C, Jalah R.et al. (2010Long-lasting humoral and cellular immune responses and mucosal dissemination after intramuscular DNA immunization Vaccine 284827–4836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frelin L, Brass A, Ahlén G, Brenndörfer ED, Chen M., and, Sällberg M. Electroporation: a promising method for the nonviral delivery of DNA vaccines in humans. Drug News Perspect. 2010;23:647–653. doi: 10.1358/dnp.2010.23.10.1513492. [DOI] [PubMed] [Google Scholar]
- Bagarazzi M, Yan J, Shen X, Giffear M, Lee J, Khans A, et al. 2010Immunotherapy of post-LEEP CIN2/3 with HPV 16 and 18 E6/E7 DNA vaccines/electroporation 50th ICAAC Boston, poster G1-202, 12–15 September 2010.
- Ottensmeier CH, Low L, Mander A, Tjelle T, Campos-perez J, Williams T.et al. (2008DNA fusion gene vaccination, delivered with or without in vivo electroporation: a potent and safe strategy for inducing antitumor immune response in prostate cancer AACR Meeting Abstract 2843
- Hao J, Li SK, Liu CY., and, Kao WW. Electrically assisted delivery of macromolecules into the corneal epithelium. Exp Eye Res. 2009;89:934–941. doi: 10.1016/j.exer.2009.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai N, Kishida T, Hartmann G, Katoh N, Imanishi J, Kishimoto S.et al. (2010Mitf silencing cooperates with IL-12 gene transfer to inhibit melanoma in mice Int Immunopharmacol 10540–545. [DOI] [PubMed] [Google Scholar]
- Golzio M, Mazzolini L, Paganin-Gioanni A., and, Teissié J. Targeted gene silencing into solid tumors with electrically mediated siRNA delivery. Methods Mol Biol. 2009;555:15–27. doi: 10.1007/978-1-60327-295-7_2. [DOI] [PubMed] [Google Scholar]
- Dharmapuri S, Aurisicchio L, Biondo A, Welsh N, Ciliberto G., and, La Monica N. Antiapoptotic small interfering RNA as potent adjuvant of DNA vaccination in a mouse mammary tumor model. Hum Gene Ther. 2009;20:589–597. doi: 10.1089/hum.2008.210. [DOI] [PubMed] [Google Scholar]
- Nakagawa S, Arai Y, Mori H, Matsushita Y, Kubo T., and, Nakanishi T. Small interfering RNA targeting CD81 ameliorated arthritis in rats. Biochem Biophys Res Commun. 2009;388:467–472. doi: 10.1016/j.bbrc.2009.06.150. [DOI] [PubMed] [Google Scholar]
- Takayama K, Suzuki A, Manaka T, Taguchi S, Hashimoto Y, Imai Y.et al. (2009RNA interference for noggin enhances the biological activity of bone morphogenetic proteins in vivo and in vitro J Bone Miner Metab 27402–411. [DOI] [PubMed] [Google Scholar]
- Molotkov DA, Yukin AY, Afzalov RA., and, Khiroug LS.2010Gene delivery to postnatal rat brain by non-ventricular plasmid injection and electroporation J Vis Exp43). [DOI] [PMC free article] [PubMed]
- Inoue T, Sugimoto M, Sakurai T, Saito R, Futaki N, Hashimoto Y.et al. (2007Modulation of scratching behavior by silencing an endogenous cyclooxygenase-1 gene in the skin through the administration of siRNA J Gene Med 9994–1001. [DOI] [PubMed] [Google Scholar]
- Vologodskii AV, Levene SD, Klenin KV, Frank-Kamenetskii M., and, Cozzarelli NR. Conformational and thermodynamic properties of supercoiled DNA. J Mol Biol. 1992;227:1224–1243. doi: 10.1016/0022-2836(92)90533-p. [DOI] [PubMed] [Google Scholar]
- Broderick KE, Shen X, Soderholm J, Lin F, McCoy J, Khan AS.et al. (2011Prototype development and preclinical immunogenicity analysis of a novel minimally invasive electroporation device Gene Ther 18258–265. [DOI] [PubMed] [Google Scholar]
- Kaiser PK, Symons RC, Shah SM, Quinlan EJ, Tabandeh H, Do DV.et al. (2010RNAi-based treatment for neovascular age-related macular degeneration by Sirna-027 Am J Ophthalmol 15033–39.e2. [DOI] [PubMed] [Google Scholar]
- Davis ME, Zuckerman JE, Choi CH, Seligson D, Tolcher A, Alabi CA.et al. (2010Evidence of RNAi in humans from systemically administered siRNA via targeted nanoparticles Nature 4641067–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broderick KE, Kardos T, McCoy JR, Fons MP, Kemmerrer S., and, Sardesai NY. Piezoelectric permeabilization of mammalian dermal tissue for in vivo DNA delivery leads to enhanced protein expression and increased immunogenicity. Hum Vaccin. 2011;7 suppl.:22–28. doi: 10.4161/hv.7.0.14559. [DOI] [PubMed] [Google Scholar]
- Lin F, Shen X, McCoy JR, Mendoza JM, Yan J, Kemmerrer SV.et al. (2011A novel prototype device for electroporation-enhanced DNA vaccine delivery simultaneously to both skin and muscle Vaccine 296771–6780. [DOI] [PubMed] [Google Scholar]
- Rabussay D. Applicator and electrode design for in vivo DNA delivery by electroporation. Methods Mol Biol. 2008;423:35–59. doi: 10.1007/978-1-59745-194-9_3. [DOI] [PubMed] [Google Scholar]
- Tjelle TE, Rabussay D, Ottensmeier C, Mathiesen I., and, Kjeken R. Taking electroporation-based delivery of DNA vaccination into humans: a generic clinical protocol. Met hods Mol Biol. 2008;423:497–507. doi: 10.1007/978-1-59745-194-9_39. [DOI] [PubMed] [Google Scholar]
- Contag CH., and, Bachmann MH. Advances in in vivo bioluminescence imaging of gene expression. Annu Rev Biomed Eng. 2002;4:235–260. doi: 10.1146/annurev.bioeng.4.111901.093336. [DOI] [PubMed] [Google Scholar]