Abstract
Increasing evidence suggests that multiple genes in the human leukocyte antigen (HLA) regions play an important role in development of cancers and immunity disorders. However, the biological mechanisms of the HLA associations are not well understood. We recently conducted a survey of all genome-wide association studies (GWAS) with significant findings in the HLA regions and concluded that diseases such as cancer and immune disorders are more likely to be associated with genetic variants located in the HLA regions than other diseases. This finding is suggestive for testing a hypothesis of a common etiology of infectious tumors and other immunity diseases.
The human leukocyte antigen (HLA) complex is the human counterpart to the major histocompatibility complex (MHC). The HLA genes are encoded in a cluster on the short arm of chromosome 6, which encodes the family of HLA antigens. In humans, the MHC is divided into Classes I, II, and III. The A, B, C, E, F, and G genes belong to MHC Class I, whereas the 6 D genes belong to Class II. Most clinically relevant HLA molecules locate in the Class I and II regions. The Class III includes genes coding a few secreted proteins with immune functions, including inflammation-related molecules such as tumor necrosis factor-α (TNF-α).
To date, accumulative evidence suggests that multiple genes in the HLA regions are likely to play an important role in development of cancer and immunity disorders. However, the biological mechanisms of the HLA associations remain to be elucidated. Multiple whole genome association studies either confirmed signals in the HLA regions that were reported previously or established new location for nasopharyngeal carcinoma (NPC) which was not reported earlier [e.g., the HLA-F region reported in a genome-wide association (GWA) study in a Taiwanese population[1]]. Despite the confirmed associations, there is little understanding of the relationship between the genetic variants discovered in the 6p21.3 region and of whether these “significant” variants work independently or additively[1],[2]. Although most previous studies are limited by both the resolution of the genetic map and sample size, there are obvious lessons to be learned from statistical signals detected by multiple GWA studies while limiting disease types by infections tumors or immune disorders.
We recently conducted a survey of all GWA studies reported genome-wide significant findings in the HLA region and concluded that diseases such as cancer and immune disorders are more likely to be associated with genetic variants located in the HLA regions than other diseases. In Figures 1 and 2, we highlighted the peaks of all GWA studies conducted between 2008 and 2010 on immunity disorders and infectious tumors, respectively. The main goal of this mini-review is to discuss the potential relationships of these findings generated from the large-scale GWA studies of infectious tumors and immunity disorders[1]–[44].
Figure 1. Twelve immune diseases and disorders included in all genome-wide association (GWA) studies conducted between 2008 and 2010. The Y-axis indicates the names of the genes, and X-axis indicates the genomic locations. The dark vertical bars indicate the location of the HLA-genes in relations to the association signals. The diseases are expressed using different shapes. Different studies are labeled with colors. For instance, celiac disease is expressed using a red triangle, lupus is expressed using a purple dot, and leprosy is expressed using a blue triangle.
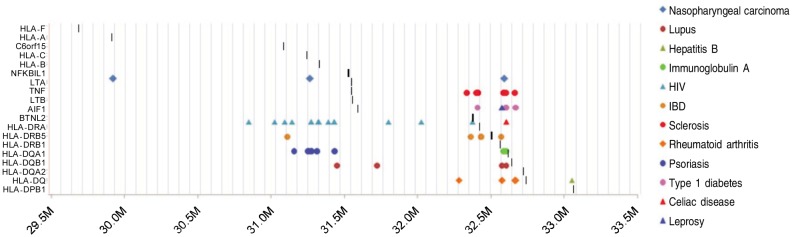
Figure 2. Twelve infectious tumors included in all GWA studies conducted between 2008 and 2010. The Y-axis indicates the names of the genes, and X-axis indicates the genomic locations. The dark vertical bars indicate the location of the HLA-genes in relations to the association signals. Eight diseases are expressed using different shapes and separate studies in the same disease are labeled using colors.
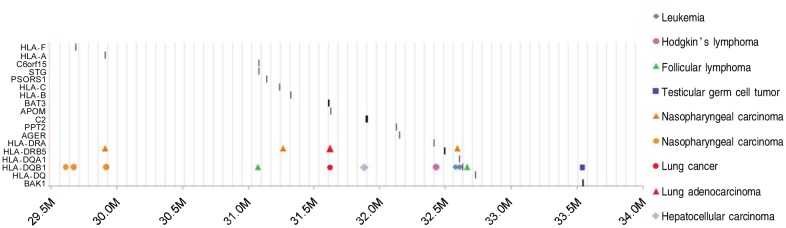
We also reviewed all the GWA peaks occurred in the HLA regions across several types of cancers. As shown in Figure 2, NPC, variants associated with Hodgkin's disease, follicular lymphoma, and chronic lymphocytic leukemia (CLL) revealed in a region of less than 0.25 megabase.
Results indicated that a total of 12 GWA studies of immune disorders revealed positive association signals in 6p21.3 regions which include leprosy, psoriasis, rheumatoid arthritis (RA), type I diabetes, NPC, celiac disease (CD), systemic lupus erythematosus (SLE), inflammatory disease (IBD), multiple sclerosis (MS), immunoglobulin deficiency, and HIV infection. Some of these findings can be implied clinically. For instance, the HLA gene testing is now recommended for CD patients, as most patients with CD carry the HLA-DQ alleles (shown as a red triangle in Figure 1) associated with CD. On the other hand, the presence of CD symptoms alone may not perfectly predict the odds of an individual which might have CD. Therefore, individuals with potential CD in high risk groups, such as first-degree relatives of CD patients, or those with other autoimmune diseases, could consider genetic diagnostic testing. As indicated in Figure 1, the variants associated with SLE, RA, IBD, and type I diabetes locate relatively close to each other, which further justify the rationale of considering other immune disorders in proband's relatives while selecting an individual for genetic testing.
To deepen our understanding of HLA-related associations, a natural further step would be to sequence the genomic region that is in close proximity of the significantly associated single-nucleotide polymorphisms (SNPs) revealed by GWA studies. The cost of whole genome sequencing (WGS) may soon become affordable and the WGS approach will enable the discovery of both common and rare variants in the genome. The WGS approach will provide opportunities to analyze the genetic contribution made by rare variants and less common variants or by joint effect and to analyze the common and rare variants in the near future. Before the WGS data become available, some investigators might be interested in conducting meta-analysis using disease phenotypes that can be defined as a “cluster of phenotypes”. For instance, one may wish to include infectious tumors such as NPC, Hodgkin's disease as well as some types of non-Hodgkin's lymphomas as a “cluster” because they share a known viral etiology related to the infection of Epstein-Barr virus (EBV). Because nearly all patients with NPC are EBV-positive, and some patients with lymphoma tend to be EBV-positive, it can be hypothesized that individuals who carry specific HLA alleles or haplotypes with weakened ability to present EBV antigens to the immune system might have higher likelihood to developing NPC, and Hodgkin's or some types of non-Hodgkin's lymphoma[45]–[55].
Needless to say, the “clustered” meta-analysis should be carried out with caution and need to take population structure into account. Further, researchers can combine data on diseases such as lupus, inflammatory bowl diseases, psoriasis and celiac diseases to conduct a meta-analysis to test whether these immune disorders share a common genetic etiology. Meanwhile, we are holding hope for the gains from upcoming WGS studies. WGS results are likely to allow us to construct more reliable haplotypes in the HLA region and, therefore, increase the power of identifying genetic factors for immune-related diseases or infectious tumors. More importantly, the high resolution of the WGS approach may allow us to test whether the genetic variants in closely related regions carry independent or additive effects. Novel findings are expected to have significant implications for functional studies and for sequencing-based application in personalized medicine.
We would also like to note that 5 disorders listed in Figures 1 and 2 are related to EBV infection. EBV is a ubiquitous virus which infects 90% of the human adult population. It is now believed that at least 90% of EBV infections are transmitted orally through exchange of infected saliva. Although many tissues and organs of the body may become infected with EBV, the primary ones include the epithelial cells of the nasopharynx and the B lymphocytes of the immune system. It is well known that EBV is an important risk factor for NPC. And EBV has long been the suspected cause of Hodgkin's disease. More recently, EBV has been implicated in autoimmune diseases such as RA, SLE, and MS. This survey indicates that findings from GWA studies on the above mentioned diseases revealed multiple potential genetic variants in the 6p21.3 region although among these signals, some are in close proximity and others are farther apart. The current finding is not a proof for a common root for development of immune diseases but is suggestive for testing a hypothesis of a common etiology of different disorders. We will certainly make a great effort in these highlighted regions by linking infections tumors and/or immune disorders into one cluster. This effort may lead to more comprehensive understanding of the biological nature of infectious tumor and immune disorders.
Finally, we would like to highlight an impressive study of multinational consortium in which researchers identified more than one million genetic variants in DNA samples from people with HIV, some are considered “super controls” and some are not. The super controls are HIV-positive, but have not taken medicine and have not developed AIDs. The researchers reported more than 300 variations reside in the HLA regions (due to the limited space in the graph, only a portion of 300 signals were included in Figure 1, and more details are referred to the original manuscript) that differed substantially between the super controls and the normal controls. This observation suggests that these variations can change the means that the human immune system recognizes HIV-infected cells via several relevant HLA proteins. This finding speaks for the need of a careful examination of the role of HLA for immune diseases, and also suggests identifying relevant HLA genes which can function either jointly or independently and demonstrating the functional role of HLA genes in recognizing virus-infected cells.
References
- 1.Tse KP, Su WH, Chang KP, et al. Genome-wide association study reveals multiple nasopharyngeal carcinoma-associated loci within the HLA region at chromosome 6p21.3 [J] Am J Hum Genet. 2009;85(2):194–203. doi: 10.1016/j.ajhg.2009.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bei JX, Li Y, Jia WH, et al. A genome-wide association study of nasopharyngeal carcinoma identifies three new susceptibility loci [J] Nat Genet. 2010;42(7):599–603. doi: 10.1038/ng.601. [DOI] [PubMed] [Google Scholar]
- 3.Clancy RM, Marion MC, Kaufman KM, et al. Identification of candidate loci at 6p21 and 21q22 in a genome-wide association study of cardiac manifestations of neonatal lupus [J] Arthritis Rheum. 2010;62(11):3415–3424. doi: 10.1002/art.27658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harley JB, Alarcon-Riquelme ME, Criswell LA, et al. Genome-wide association scan in women with systemic lupus erythematosus identifies susceptibility variants in ITGAM, PXK, KIAA1542 and other loci [J] Nat Genet. 2008;40(2):204–210. doi: 10.1038/ng.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Horn G, Graham RR, Modrek B, et al. Association of systemic lupus erythematosus with c8orf13-blk and itgam-itgax [J] N Engl J Med. 2008;358(9):900–909. doi: 10.1056/NEJMoa0707865. [DOI] [PubMed] [Google Scholar]
- 6.Han JW, Zheng HF, Cui Y, et al. Genome-wide association study in a Chinese Han population identifies nine new susceptibility loci for systemic lupus erythematosus [J] Nat Genet. 2009;41(11):1234–1237. doi: 10.1038/ng.472. [DOI] [PubMed] [Google Scholar]
- 7.Kamatani Y, Wattanapokayakit S, Ochi H, et al. A genome-wide association study identifies variants in the HLA-DP locus associated with chronic hepatitis B in Asians [J] Nat Genet. 2009;41(5):591–595. doi: 10.1038/ng.348. [DOI] [PubMed] [Google Scholar]
- 8.Hirschfield GM, Liu X, Xu C, et al. Primary biliary cirrhosis associated with HLA, IL12A, and IL12RB2 variants [J] N Engl J Med. 2009;360(24):2544–2555. doi: 10.1056/NEJMoa0810440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ferreira RC, Pan-Hammarstrom Q, Graham RR, et al. Association of IFIH1 and other autoimmunity risk alleles with selective iga deficiency [J] Nat Genet. 2010;42(9):777–780. doi: 10.1038/ng.644. [DOI] [PubMed] [Google Scholar]
- 10.Pereyra F, Jia X, McLaren PJ, et al. The major genetic determinants of HIV-1 control affect HLA class I peptide presentation [J] Science. 2010;330(6010):1551–1557. doi: 10.1126/science.1195271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pelak K, Goldstein DB, Walley NM, et al. Host determinants of HIV-1 control in African Americans [J] J Infect Dis. 2010;201(8):1141–1149. doi: 10.1086/651382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fellay J, Ge D, Shianna KV, et al. Common genetic variation and the control of HIV-1 in humans [J] PLoS Genet. 2009;5(12):e1000791. doi: 10.1371/journal.pgen.1000791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Franke A, Balschun T, Sina C, et al. Genome-wide association study for ulcerative colitis identifies risk loci at 7q22 and 22q13 (IL17REL) [J] Nat Genet. 2010;42(4):292–294. doi: 10.1038/ng.553. [DOI] [PubMed] [Google Scholar]
- 14.Asano K, Matsushita T, Umeno J, et al. A genome-wide association study identifies three new susceptibility loci for ulcerative colitis in the Japanese population [J] Nat Genet. 2009;41(12):1325–1329. doi: 10.1038/ng.482. [DOI] [PubMed] [Google Scholar]
- 15.Barrett JC, Lee JC, Lees CW, et al. Genome-wide association study of ulcerative colitis identifies three new susceptibility loci, including the HNF4A region [J] Nat Genet. 2009;41(12):1330–1334. doi: 10.1038/ng.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Silverberg MS, Cho JH, Rioux JD, et al. Ulcerative colitis-risk loci on chromosomes 1p36 and 12q15 found by genome-wide association study [J] Nat Genet. 2009;41(2):216–220. doi: 10.1038/ng.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Franke A, Balschun T, Karlsen TH, et al. Sequence variants in IL10, arpc2 and multiple other loci contribute to ulcerative colitis susceptibility [J] Nat Genet. 2008;40(11):1319–1323. doi: 10.1038/ng.221. [DOI] [PubMed] [Google Scholar]
- 18.Kugathasan S, Baldassano RN, Bradfield JP, et al. Loci on 20q13 and 21q22 are associated with pediatric-onset inflammatory bowel disease [J] Nat Genet. 2008;40(10):1211–1215. doi: 10.1038/ng.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Radstake TR, Gorlova O, Rueda B, et al. Genome-wide association study of systemic sclerosis identifies CD247 as a new susceptibility locus [J] Nat Genet. 2010;42(5):426–429. doi: 10.1038/ng.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nischwitz S, Cepok S, Kroner A, et al. Evidence for VAV2 and ZNF433 as susceptibility genes for multiple sclerosis [J] J Neuroimmunol. 2010;227(1–2):162–166. doi: 10.1016/j.jneuroim.2010.06.003. [DOI] [PubMed] [Google Scholar]
- 21.Sanna S, Pitzalis M, Zoledziewska M, et al. Variants within the immunoregulatory cblb gene are associated with multiple sclerosis [J] Nat Genet. 2010;42(6):495–497. doi: 10.1038/ng.584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jakkula E, Leppa V, Sulonen AM, et al. Genome-wide association study in a high-risk isolate for multiple sclerosis reveals associated variants in STAT3 gene [J] Am J Hum Genet. 2010;86(2):285–291. doi: 10.1016/j.ajhg.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Genome-wide association study identifies new multiple sclerosis susceptibility loci on chromosomes 12 and 20 [J] Nat Genet. 2009;41(7):824–828. doi: 10.1038/ng.396. [DOI] [PubMed] [Google Scholar]
- 24.De Jager PL, Jia X, Wang J, et al. Meta-analysis of genome scans and replication identify CD6, IRF8 and TNFRSF1A as new multiple sclerosis susceptibility loci [J] Nat Genet. 2009;41(7):776–782. doi: 10.1038/ng.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Comabella M, Craig DW, Camina-Tato M, et al. Identification of a novel risk locus for multiple sclerosis at 13q31.3 by a pooled genome-wide scan of 500,000 single nucleotide polymorphisms [J] PLoS One. 2008;3(10):e3490. doi: 10.1371/journal.pone.0003490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hafler DA, Compston A, Sawcer S, et al. Risk alleles for multiple sclerosis identified by a genomewide study [J] N Engl J Med. 2007;357(9):851–862. doi: 10.1056/NEJMoa073493. [DOI] [PubMed] [Google Scholar]
- 27.Kochi Y, Okada Y, Suzuki A, et al. A regulatory variant in CCR6 is associated with rheumatoid arthritis susceptibility [J] Nat Genet. 2010;42(6):515–519. doi: 10.1038/ng.583. [DOI] [PubMed] [Google Scholar]
- 28.Genome-wide association study of 14,000 cases of seven common diseases and 3000 shared controls [J] Nature. 2007;447(7145):661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Julia A, Ballina J, Canete JD, et al. Genome-wide association study of rheumatoid arthritis in the Spanish population: KIF12 as a risk locus for rheumatoid arthritis susceptibility [J] Arthritis Rheum. 2008;58(8):2275–2286. doi: 10.1002/art.23623. [DOI] [PubMed] [Google Scholar]
- 30.Plenge RM, Seielstad M, Padyukov L, et al. TRAF1-C5 as a risk locus for rheumatoid arthritis—a genome-wide study [J] N Engl J Med. 2007;357(12):1199–1209. doi: 10.1056/NEJMoa073491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Raychaudhuri S, Remmers EF, Lee AT, et al. Common variants at CD40 and other loci confer risk of rheumatoid arthritis [J] Nat Genet. 2008;40(10):1216–1223. doi: 10.1038/ng.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stahl EA, Raychaudhuri S, Remmers EF, et al. Genome-wide association study meta-analysis identifies seven new rheumatoid arthritis risk loci [J] Nat Genet. 2010;42(6):508–514. doi: 10.1038/ng.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ellinghaus E, Ellinghaus D, Stuart PE, et al. Genome-wide association study identifies a psoriasis susceptibility locus at TRAF3IP2 [J] Nat Genet. 2010;42(11):991–995. doi: 10.1038/ng.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Strange A, Capon F, Spencer CC, et al. A genome-wide association study identifies new psoriasis susceptibility loci and an interaction between HLA-C and ERAP1 [J] Nat Genet. 2010;42(11):985–990. doi: 10.1038/ng.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barrett JC, Clayton DG, Concannon P, et al. Genome-wide association study and meta-analysis find that over 40 loci affect risk of type 1 diabetes [J] Nat Genet. 2009;41(6):703–707. doi: 10.1038/ng.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Capon F, Bijlmakers MJ, Wolf N, et al. Identification of ZNF313/RNF114 as a novel psoriasis susceptibility gene [J] Hum Mol Genet. 2008;17(13):1938–1945. doi: 10.1093/hmg/ddn091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cooper JD, Smyth DJ, Smiles AM, et al. Meta-analysis of genome-wide association study data identifies additional type 1 diabetes risk loci [J] Nat Genet. 2008;40(12):1399–1401. doi: 10.1038/ng.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hakonarson H, Grant SF, Bradfield JP, et al. A genome-wide association study identifies kiaa0350 as a type 1 diabetes gene [J] Nature. 2007;448(7153):591–594. doi: 10.1038/nature06010. [DOI] [PubMed] [Google Scholar]
- 39.Liu Y, Helms C, Liao W, et al. A genome-wide association study of psoriasis and psoriatic arthritis identifies new disease loci [J] PLoS Genet. 2008;4(3):e1000041. doi: 10.1371/journal.pgen.1000041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nair RP, Duffin KC, Helms C, et al. Genome-wide scan reveals association of psoriasis with IL-23 and NF-kappaB pathways [J] Nat Genet. 2009;41(2):199–204. doi: 10.1038/ng.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang XJ, Huang W, Yang S, et al. Psoriasis genome-wide association study identifies susceptibility variants within Ice gene cluster at 1q21 [J] Nat Genet. 2009;41(2):205–210. doi: 10.1038/ng.310. [DOI] [PubMed] [Google Scholar]
- 42.Dubois PC, Trynka G, Franke L, et al. Multiple common variants for celiac disease influencing immune gene expression [J] Nat Genet. 2010;42(4):295–302. doi: 10.1038/ng.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van Heel DA, Franke L, Hunt KA, et al. A genome-wide association study for celiac disease identifies risk variants in the region harboring IL2 and IL21 [J] Nat Genet. 2007;39(7):827–829. doi: 10.1038/ng2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang FR, Huang W, Chen SM, et al. Genomewide association study of leprosy [J] N Engl J Med. 2009;361(27):2609–2618. doi: 10.1056/NEJMoa0903753. [DOI] [PubMed] [Google Scholar]
- 45.Hildesheim A, Apple RJ, Chen CJ, et al. Association of hla class I and II alleles and extended haplotypes with nasopharyngeal carcinoma in Taiwan [J] J Natl Cancer Inst. 2002;94(23):1780–1789. doi: 10.1093/jnci/94.23.1780. [DOI] [PubMed] [Google Scholar]
- 46.Lin HJ, Cherng JM, Hung MS, et al. Functional assays of hla a2-restricted epitope variant of latent membrane protein 1 (lmp-1) of Epstein-Barr virus in nasopharyngeal carcinoma of southern china and taiwan [J] J Biomed Sci. 2005;12(6):925–936. doi: 10.1007/s11373-005-9017-y. [DOI] [PubMed] [Google Scholar]
- 47.Sengupta S, den Boon JA, Chen IH, et al. Genome-wide expression profiling reveals EBV-associated inhibition of MHC class I expression in nasopharyngeal carcinoma [J] Cancer Res. 2006;66(16):7999–8006. doi: 10.1158/0008-5472.CAN-05-4399. [DOI] [PubMed] [Google Scholar]
- 48.Butsch Kovacic M, Martin M, Gao X, et al. Variation of the killer cell immunoglobulin-like receptors and HLA-C genes in nasopharyngeal carcinoma [J] Cancer Epidemiol Biomarkers Prev. 2005;14(11 Pt 1):2673–2677. doi: 10.1158/1055-9965.EPI-05-0229. [DOI] [PubMed] [Google Scholar]
- 49.Diepstra A, Niens M, Vellenga E, et al. Association with hla class I in Epstein-Barr-virus-positive and with hla class III in Epstein-Barr-virus-negative Hodgkin's lymphoma [J] Lancet. 2005;365(9478):2216–2224. doi: 10.1016/S0140-6736(05)66780-3. [DOI] [PubMed] [Google Scholar]
- 50.Niens M, Jarrett RF, Hepkema B, et al. HLA-A*02 is associated with a reduced risk and HLA-A*01 with an increased risk of developing EBV* Hodgkin lymphoma [J] Blood. 2007;110(9):3310–3315. doi: 10.1182/blood-2007-05-086934. [DOI] [PubMed] [Google Scholar]
- 51.Rickinson AB, Moss DJ. Human cytotoxic t lymphocyte responses to Epstein-Barr virus infection [J] Annu Rev Immunol. 1997;15:405–431. doi: 10.1146/annurev.immunol.15.1.405. [DOI] [PubMed] [Google Scholar]
- 52.Skibola CF, Bracci PM, Halperin E, et al. Genetic variants at 6p21.33 are associated with susceptibility to follicular lymphoma [J] Nat Genet. 2009;41(8):873–875. doi: 10.1038/ng.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gavioli R, Kurilla MG, de Campos-Lima PO, et al. Multiple HLA A11-restricted cytotoxic T-lymphocyte epitopes of different immunogenicities in the Epstein-Barr virus–encoded nuclear antigen 4 [J] J Virol. 1993;67(3):1572–1578. doi: 10.1128/jvi.67.3.1572-1578.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Burrows JM, Burrows SR, Poulsen LM, et al. Unusually high frequency of Epstein-Barr virus genetic variants in papua new guinea that can escape cytotoxic T-cell recognition: Implications for virus evolution [J] J Virol. 1996;70(4):2490–2496. doi: 10.1128/jvi.70.4.2490-2496.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.de Campos-Lima PO, Gavioli R, Zhang QJ, et al. HLA-A11 epitope loss isolates of Epstein-Barr virus from a highly A11+ population [J] Science. 1993;260(5104):98–100. doi: 10.1126/science.7682013. [DOI] [PubMed] [Google Scholar]


