Abstract
Ligand-dependent activity has been engineered into enzymes for purposes ranging from controlling cell morphology to reprogramming cellular signaling pathways. Where these successes have typically fused a naturally allosteric domain to the enzyme of interest, here we instead demonstrate an approach for designing a de novo allosteric effector site directly into the catalytic domain of an enzyme. This approach is distinct from traditional chemical rescue of enzymes in that it relies on disruption and restoration of structure, rather than active site chemistry, as a means to achieve modulate function. We present two examples, W33G in a β-glycosidase enzyme (β-gly) and W492G in a β-glucuronidase enzyme (β-gluc), in which we engineer indole-dependent activity into enzymes by removing a buried tryptophan sidechain that serves as a buttress for the active site architecture. In both cases we observe a loss of function, and in both cases we find that the subsequent addition of indole can be used to restore activity. Through detailed analysis of β-gly W33G kinetics we demonstrate that this rescued enzyme is fully functionally equivalent to the corresponding wild-type enzyme. We then present the apo and indole-bound crystal structures of β-gly W33G, which together establish the structural basis for enzyme inactivation and rescue. Finally, we use this designed switch to modulate β-glycosidase activity in living cells using indole. Disruption and recovery of protein structure may represent a general technique for introducing allosteric control into enzymes, and thus may serve as a starting point for building a variety of bioswitches and sensors.
Introduction
The use of small molecules to modulate the activity of re-engineered enzymes is a powerful approach that can be used to control cell shape, signal transduction, growth, and survival1–4. The design strategy for introducing pharmacological control into enzymes underlying each of these successful examples has relied on the modularity of protein domains5. By creating a fusion protein from a naturally allosteric protein domain and a separate catalytic domain, ligand binding can – in select cases –affect enzyme activity1–4,6. The infeasibility of predicting the detailed mechanism by which effector binding produces altered activity, however, has required screening of multiple domain arrangements in order to identify connections that lead to allosteric regulation of enzyme activity1–4,6. In the fortuitous case of a naturally allosteric single-domain enzyme, elucidating the structural basis for regulation can enable introduction of cysteine residues such that activity is dependent on redox conditions that induce disulfide bond formation7. Another approach involves building new catalytic activity into a naturally allosteric (non-catalytic) domain8, but challenges associated with computational enzyme design9 have limited this strategy to a single successful example to date.
Rather than co-opt a natural allosteric transition, we have instead developed a technique for engineering de novo allosteric control into a protein domain that is not naturally allosteric. Our approach is predicated on the observation that cavity-forming mutations can be complemented by binding of small hydrophobic ligands10,11, and we use this starting point as a means to control protein function. Our approach consists of removing a buried structural element in the enzyme, which in turn distorts the active site geometry and results in loss of function; subsequent exogenous replacement of the complementary small molecule can then be used to restore structure, and hence activity (Figure 1a). The ability of an effector ligand to restore protein activity will require not simply binding, but also precise rescue of structure. Previous attempts to rescue cavity-forming mutants with activating ligands have included a zinc finger transcription factor12 and a hormone-receptor pair13. Each of these studies identified a rescuing ligand by screening restricted compound libraries, and demonstrated that the selected ligand restored partial activity. In neither case was activity fully restored, however; we propose that complete rescue requires a ligand with exquisite structural complementarity unlikely to be found in a screening library of limited size. We expect that complete rescue of function will require exact ligand-cavity complementarity, which we attain by rationally matching the effector ligand to the structure of the deleted moiety.
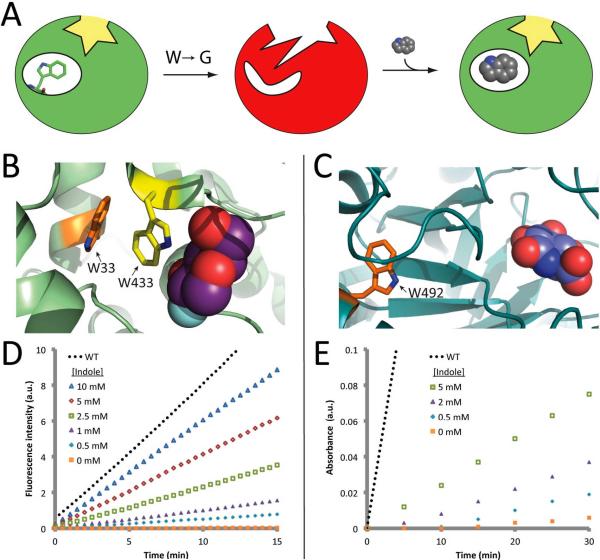
Figure 1. Indole rescue.
(A) Mutation of a remote “buttressing” tryptophan leads to structural disruption of catalytic geometry, resulting in loss of activity. Subsequent addition of exogenous indole restores structure, and thus activity. (B) Crystal structure of β-gly with a substrate analog (2-deoxy-2-fluoro-glucose, purple spheres)16, showing the “buttressing” tryptophan (Trp33, orange) and a nearby tryptophan (Trp433, yellow). (C) Crystal structure of β-gluc17 with a substrate analog (glucaro-δ-lactam, blue spheres) showing the “buttressing” tryptophan (Trp492, orange). (D) Indole-dependent activity of β-gly W33G, measured spectrofluorometrically using 100 μM fluorescein di-β-D-galactopyranoside (FDG) as a substrate. (E) Indole-dependent activity of β-gluc W492G, measured spectrophotometrically using 40 mM 2-nitrophenyl-β-D-glucopyranoside (ONPGlc) as a substrate. No indole was included in data for either wild-type enzyme shown here.
Though this approach may in principle be applied to a deleted structural element corresponding to sidechains from multiple amino acid residues, in this study we focus on mutation of a single buried tryptophan residue to glycine (W→G). If this tryptophan sidechain is serving as a “buttress” to prevent collapse of the enzyme active site, its removal will lead to distortion of the catalytic geometry and thus loss of enzyme activity. Replacement of the buttress via addition of exogenous indole – perfectly complementing the deleted sidechain – should restore the original protein conformation, and thus rescue activity. Critically, the mutation site need not be at the active site: provided the disruption of structure resulting from a remote mutation is relayed to the active site, the recovery of structure upon complementation is expected to similarly be transduced to the active site. This approach therefore represents a rational strategy for designing remote de novo allosteric effector binding sites into proteins.
Methods
All β-glycosidase kinetic assays were carried out using 58 nM β-gly with fluorescein di-β-D-galactopyranoside (FDG) as substrate in a buffer of 50 mM sodium phosphate pH 6.5 and 5% DMSO at 37 °C, with indole concentrations ranging from 0 to 10 mM. β-glycosidase cleaves FDG twice to yield one molecule of D-galactose and two molecules of fluorescein; we detect the latter by fluorescence with excitation at 485 nm and emission at 528 nm.
All β-glucuronidase kinetic assays were carried out using 3.7 μM β-gluc with 2-nitrophenyl-β-D-glucopyranoside (ONPGlc) in a buffer of 75 mM sodium phosphate pH 7.4, 100 mM NaCl and 5% ethanol at 30 °C, with indole concentrations ranging from 0 to 5 mM. β-glucuronidase cleaves ONPGlc to yield D-glucose and 2-nitrophenol; we detect the latter by absorbance at 405 nm.
Coordinates and structure factors for the apo and indole-bound crystal structures of β-gly W33G have been deposited with the Research Collaboratory for Structural Bioinformatics Protein Data Bank (PDB) with accession codes 4EAM and 4EAN, respectively.
A complete description of methods is available as Supporting Information.
Results
To test the hypothesis that protein function can be modulated in this manner, we selected two divergent enzymes that cleave β-glycosidic bonds. While sequence-based evolutionary methods have proven useful in detecting energetic coupling6,14, we instead relied on protein structure to identify buried tryptophan sidechains that may be serving to buttress the active site. Using available crystal structures15–17, we selected Trp33 from the S. solfataricus β-glycosidase (β-gly) and Trp492 from E. coli β-glucuronidase (β-gluc) for testing this hypothesis (Figure 1b,c). While each sidechain occupies a buried environment, neither makes direct interactions with the substrate: Trp33 of β-gly is a “second-shell” residue about 9 Å from the substrate analog, and Trp492 of β-gluc is even more distant (about 13 Å from the substrate analog). Further, we find using modern packing metrics18 that these two sidechains are among the 10% most tightly-packed buried tryptophan sidechains in the Protein Data Bank19 suggesting their surroundings may be poised to serve as indole binding sites. In essence, selective pressure on the amino acids surrounding each of these tryptophan sidechains has inadvertently evolved an optimal indole binding site given the constraints of the protein architecture required for function.
We found that both β-gly W33G and β-gluc W492G exhibit strongly diminished β-glycosidase activity relative to the cognate wild-type enzymes. Remarkably, in both cases addition of indole led to dose-dependent recovery of enzyme activity (Figure 1d,e). In contrast, we found that addition of indole up to 15 mM, the solubility limit of indole in aqueous solution, did not restore activity to four separate W→G point mutants of β-gly (W151G, W361G, W425G, W433G) or to a W→G point mutant of β-gluc (W471G) if the mutation was located at the active site. In the presence of 5 mM indole, β-gly W33G led to product formation at a rate 0.58 times that of the corresponding wild-type enzyme without indole (0.34 μM/min vs. 0.58 μM/min, Figure 1d); in the presence of 5 mM indole β-gluc W492G led to product formation at a rate 0.14 times that of the corresponding wild-type enzyme without indole (7.8 μM/min vs. 56 μM/min, Figure 1e). Because rescue of β-gly W33G was more effective than rescue of β-gluc W492G relative to the corresponding wild-type enzyme (at this indole concentration), we selected the former for further characterization. We further observed that at 10 mM indole, β-gly W33G led to product formation at a rate equivalent to the corresponding wild-type enzyme at this indole concentration (0.561 μM/min for W33G vs. 0.544 μM/min for WT), suggesting complete rescue of activity. While the studies below serve to demonstrate that the mechanism of inactivation and rescue for β-gly W33G are as designed (Figure 1a), we note that in the absence of additional experiments we cannot definitively say the same of β-gluc W492G.
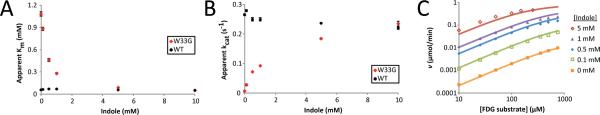
We first explored the mechanism of rescue by examining the rate of product formation as a function of both indole and substrate concentrations. In the simplest allosteric kinetic mechanism (Figure S2), the rate equation predicts that initial velocities as a function of substrate concentration can be fit to a rectangular hyperbola if the concentration of the allosteric modulator is fixed20. Indeed we find this to hold across a wide range of indole concentrations, allowing us to fit “apparent” values of the steady-state Michaelis-Menten kinetic parameters. Individually fitting data collected at each indole concentration, we observe a decrease in apparent Km, from 1080 ± 30 μM without indole to 53 ± 3 μM at 10 mM indole, accompanied by an increase in apparent kcat, from 0.00675 ± 0.00004 s−1 without indole to 0.234 ± 0.009 s−1 at 10 mM indole (Figure 2a,b). By contrast, the kinetic parameters of the wild-type enzyme are nearly indole-independent, with a Km of 58 ± 4 μM and kcat of 0.265 ± 0.002 s−1 without indole, and a Km of 48 ± 1 μM and kcat of 0.221 ± 0.006 s−1 at 10 mM indole, confirming that modulation of β-gly W33G activity indeed relies on the W→G point mutation. Notably, both Km and kcat of β-gly W33G reach the corresponding wild-type values at high indole concentration, suggesting that the rescued holo enzyme is fully functionally equivalent to the wild-type enzyme at high indole concentration. Relative to the apo form of the engineered enzyme, addition of indole leads to a 20-fold decrease in Km and a 39-fold increase in kcat; these two ratios, termed Q and W respectively20, define the allosteric linkage between substrate and allosteric activator. In other words, from the value of Q we infer that the presence of saturating indole enhances substrate binding by −1.8 kcal/mol, or equivalently that the presence of saturating substrate enhances indole binding by −1.8 kcal/mol. Collectively, the strong linkage through both Q and W together produce a 780-fold change in the ratio kcat / Km upon activation by saturating concentrations of indole.
Figure 2. Rescued β-gly W33G is functionally equivalent to the wild-type enzyme.
The apparent Michaelis-Menten parameters (A) Km and (B) kcat are presented as a function of indole for the wild type enzyme and W33G β-gly, measured spectrofluorometrically using FDG as a substrate. Parameters for wild type β-gly are essentially indole independent, and the β-gly W33G parameters approach the equivalent wild type values with increasing indole. Wild type and W33G data points overlap at 10 mM indole. Measurements were carried out in duplicate; error bars represent the standard error of the mean. (C) The measured initial velocities are consistent with a simple allosteric model with one free parameter corresponding to the enzyme-indole dissociation constant.
To further test the appropriateness of this simple allosteric kinetic mechanism (Figure S2) for describing activation of β-gly W33G by indole, we fit the rate equation for this model20 to the complete set of experimentally-determined initial velocities. The limiting values of the Michaelis-Menten kinetic parameters for the apo and holo enzyme were set to those of the W33G and wild-type enzyme in the absence of indole, leaving a single free parameter in the fitting corresponding to the enzyme-indole dissociation constant in the absence of substrate. This simple kinetic model proved sufficient to describe the initial velocity across a broad range of indole and substrate concentrations (Figure 2c). The value of the free parameter in this optimal fit was 15 mM, representing the enzyme-indole dissociation constant in the absence of substrate. From the linkage relationship, this dissociation constant in the presence of saturating substrate drops to 0.75 mM.
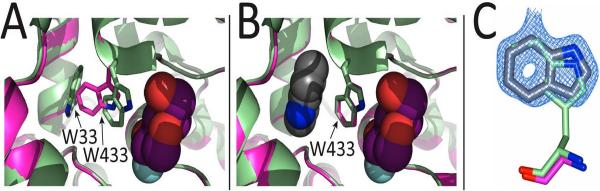
To better understand the detailed basis for β-gly W33G rescue, we next determined the crystal structure of β-gly W33G to 1.7 Å resolution (Table S1). The structural basis for inactivation of β-gly W33G is clearly demonstrated by comparing this crystal structure to that of the wild-type enzyme with a substrate analog bound in the active site (Figure 3a). In the β-gly W33G structure, the cavity resulting from mutation of W33 to glycine is occupied by W433, a nearby tryptophan, which moves away from its original position near the active site. Though no substrate analog is present in our structure, previous studies of wild-type β-gly have highlighted the importance of W433 in forming a key hydrogen bond to substrate hydroxyl group16. Consistent with our analysis of enzyme kinetics, studies of a closely related enzyme found that a synthetic substrate lacking this hydroxyl group exhibits a markedly higher Km than the corresponding natural substrate21.
Figure 3. Structural basis for indole rescue of β-gly W33G.
Crystal structures of apo β-gly W33G and indole-bound β-gly W33G were refined to 1.7 Å resolution and 1.75 Å resolution, respectively. Structures are superposed to the wild type enzyme (pale green)16. A substrate analog (2-deoxy-2-fluoro-glucose, purple spheres) present in the structure of the wild type enzyme is shown to indicate the location of the active site. (A) Crystal structure of apo β-gly W33G (pink). Trp433 occupies the cavity produced by mutation of Trp33 to Gly, causing it to lose contact with the substrate. (B) Crystal structure of holo β-gly W33G (pink). Trp433 reverts to the conformation observed in the wild type enzyme. No other sidechains differ between the holo and apo structures of β-gly W33G. (C) A difference electron density (Fo-Fc) map contoured at 3σ from holo β-gly W33G chain A before inserting the indole. The electron density is consistent with indole (gray) at a location overlapping Trp33 in the wild type enzyme (pale green). The orientation of indole cannot be unambiguously determined at this resolution.
To explore the basis for re-activation of β-gly W33G, we then soaked these crystals with indole and determined the crystal structure to 1.75 Å resolution (Table S1). Dramatically, in this structure we find that W433 has reverted to its original location near the active site (Figure 3b). We further observe clear electron density in the void left by the W33G mutation, which is presumably occupied by indole (Figures 3c and S3). An indole molecule modeled into this void fits the electron density map, which closely aligns with the W33 side chain in the wild-type structure. Aside from very small changes near the W33G mutation, the bound and unbound structures are otherwise identical (0.22 Å Cα RMSD excluding residue 33). The reversion of the indole-bound β-gly W33G structure to match that of the wild-type enzyme is fully consistent with our kinetic characterization showing complete rescue of β-gly W33G at high indole concentration.
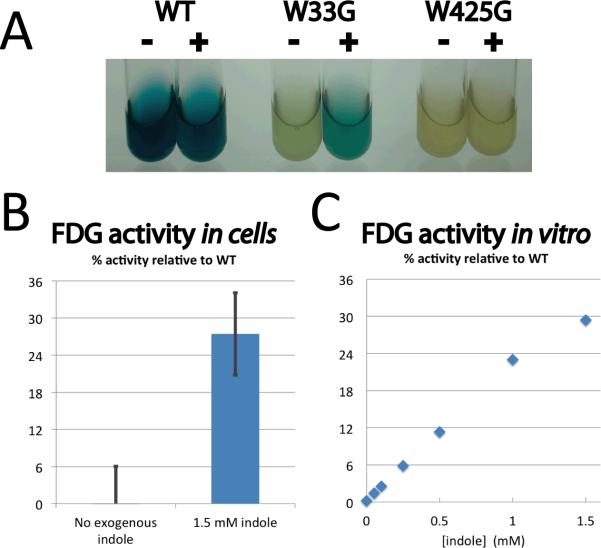
To determine whether our engineered allosteric enzyme is active in living cells, we next grew E. coli cells containing expression plasmids in media supplemented with one of two substrates: either X-gal for qualitative (visual) detection (Figure 4a), or FDG for quantitative (fluorometric) detection (Figure 4b). We found that cell cultures expressing the wild-type enzyme retained β-glycosidase activity in the presence or absence of indole. In contrast β-gly W425G, an active-site mutation that had no detectable activity in vitro (Figure S1), served as a negative control with no detectable activity in cell culture with or without indole. Cell cultures expressing β-gly W33G without the addition of indole exhibited a significant reduction in activity relative to the wild-type enzyme. Notably, activity in cell cultures expressing β-gly W33G could be rescued by addition of exogenous indole to the media (Figure 4a,b). To exclude the possibility that indole was disrupting cells and thus releasing β-gly W33G into the media, we tested for β-glycosidase activity in the media both quantitatively using FDG and qualitatively using X-gal. By partitioning components of these cultures we tracked β-glycosidase activity and found that intracellular activation of β-gly W33G was responsible for product formation (Figure S4). By interpolation from indole dependence of β-gly W33G determined in the analogous in vitro experiment (Figure 4c), we find that addition of 1.5 mM indole to the media leads to intracellular β-gly W33G activation commensurate with approximately 1.35 mM indole in vitro (Figure 4b). The extent of β-gly W33G activation, corresponding to 27% relative to the wild-type enzyme, nearly reaches the maximal level expected at this concentration of exogenous indole.
Figure 4. Indole rescue in living cells.
(A) E. coli cells expressing β-gly supplemented with X-gal substrate for qualitative detection of activity. Addition of indole (indicated with +/−) affects neither wild type β-gly activity (left pair, positive control) nor β-gly W425G activity (right pair, negative control). Only β-gly W33G (center) shows indole-dependent activity. All tubes contain 196 μM X-gal and 1 mM IPTG; those marked with a “+” additionally contain 2 mM indole. (B) Quantification of β-gly activity in E. coli cells supplemented with FDG substrate. Fraction of product formation by β-gly W33G after 30 min is reported relative to wild type β-gly. Four experimental replicates were averaged; error bars represent the standard error. (C) The analogous experiment was carried out in vitro to determine the maximal activity relative to the wild type enzyme that could be expected from addition of 1.5 mM indole to the cell cultures.
Discussion
In both β-gly W33G and β-gluc W492G, the structural disruption arising from the cavity-forming mutations was transduced to the active site leading to diminished function of the engineered apo enzyme. Identifying analogous sites in other enzymes represents a key challenge in extending this approach to introduce allosteric control into other enzymes. Coevolving networks of amino acids have been used to trace energetic “wires” linking remote sites in proteins, which may allow rapid detection of mutations at distant regions that could be used to modulate protein function6,22. Alternatively at the level of structure, complementary predictions may be facilitated by the use of computational tools to predict whether the structural disruption arising from a particular cavity-forming mutation will be transduced to the active site23–27. From a number of studies, the identification of allosteric sites with no known natural effector in a variety of protein families has fueled speculation that energetic coupling of functional sites to remote regions of the protein may be a common phenomenon28. The suggestion that proteins are “primed” to show allosteric behavior implies that the current challenge in engineering switchable enzymes lies not in designing mechanisms for allosteric signal transduction, but rather in incorporating de novo binding sites29. This view is consistent with the results presented here: the effector binding site was rationally designed by chemical rescue of structure, allowing allosteric modulation of enzyme activity to naturally emerge.
Structural disruption by mutation of a single buried tryptophan residue to glycine naturally lends itself to the use of indole as its cognate activating effector ligand. However, indole is a bioactive molecule present in many cell types and accordingly this lack of bioorthogonality may prove limiting for certain future applications. Further, the size and chemical characteristics of indole may place intrinsic limits on binding affinity, manifest through the high concentrations of indole required for rescue in the two examples presented here. While 10 mM indole is required to recover β-gly W33G activity equivalent to that of the wild-type enzyme (“complete rescue”), in the case of β-gluc W492G the solubility limit of indole in aqueous solution prevents us from ascertaining whether complete rescue though addition of indole is possible. Even given higher binding affinity for indole, however, the possibility remains that indole rescue may leave behind some unanticipated change to either the structure or the dynamics of the rescued enzyme that renders it not fully equivalent to the corresponding wild-type enzyme.
Chemical rescue of structure need not be limited to this particular mutation, however, or even to cavity-forming mutations at a single site. By identifying constellations of atoms that match a particular compound, precise cavities may be carved out in proteins that will be complemented by larger and more diverse compounds, which in turn may afford enhanced sensitivity and selectivity. As such, we expect that chemical rescue of structure will represent a technique for building switches and sensors that respond to a wide variety of activating effector ligands.
Supplementary Material
Acknowledgements
We thank Ning Zheng, Chet Egan, Eric Deeds, Jacob Corn, Audrey Lamb, Scott Hefty, Aron Fenton, Tanja Kortemme, and Brian Kuhlman for valuable discussions. We thank Marco Moracci for providing the β-gly gene, Bret Wallace and Matt Redinbo for providing the β-gluc gene, Susan Egan for providing the pHG165 vector, the COBRE PPG for assistance in subcloning, and Kevin Battaile for synchrotron data collection. Use of the IMCA-CAT beamline 17-ID at the Advanced Photon Source was supported by the companies of the Industrial Macromolecular Crystallography Association through a contract with Hauptman-Woodward Medical Research Institute. Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. DE-AC02-06CH11357. This work was supported by grants from the National Center for Research Resources (5P20RR017708-07S1 and 5P20RR017708-09) (J.K.), the National Institute of General Medical Sciences (8 P20 GM103420-10) (J.K.), the NIH Dynamic Aspects of Chemical Biology Predoctoral Training Grant 2T32GM008545-17 (K.D.), and the Alfred P. Sloan Fellowship (J.K.).
Footnotes
Supporting Information
A complete description of experimental methods and procedures. Table S1 containing crystallographic data for apo β-gly W33G and indole-bound β-gly W33G. Figure S1 showing indole dependence of activity in vitro for β-gly wild type, W33G, and W425G. Figure S2 showing the simple allosteric scheme used to interpret kinetic data. Figure S3 showing a difference electron density map from holo β-gly W33G chain B. Figure S4 showing fractionation scheme of cell-based X-gal experiment. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- (1).Karginov AV, Ding F, Kota P, Dokholyan NV, Hahn KM. Nat Biotechnol. 2010;28:743. doi: 10.1038/nbt.1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Yeh BJ, Rutigliano RJ, Deb A, Bar-Sagi D, Lim WA. Nature. 2007;447:596. doi: 10.1038/nature05851. [DOI] [PubMed] [Google Scholar]
- (3).Tucker CL, Fields S. Nat Biotechnol. 2001;19:1042. doi: 10.1038/nbt1101-1042. [DOI] [PubMed] [Google Scholar]
- (4).Guntas G, Mansell TJ, Kim JR, Ostermeier M. Proc Natl Acad Sci U S A. 2005;102:11224. doi: 10.1073/pnas.0502673102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Ostermeier M. Curr Opin Struct Biol. 2009;19:442. doi: 10.1016/j.sbi.2009.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Lee J, Natarajan M, Nashine VC, Socolich M, Vo T, Russ WP, Benkovic SJ, Ranganathan R. Science. 2008;322:438. doi: 10.1126/science.1159052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Nomura AM, Marnett AB, Shimba N, Dotsch V, Craik CS. Nat Struct Mol Biol. 2005;12:1019. doi: 10.1038/nsmb1006. [DOI] [PubMed] [Google Scholar]
- (8).Korendovych IV, Kulp DW, Wu Y, Cheng H, Roder H, Degrado WF. Proc Natl Acad Sci U S A. 2011;108:6823. doi: 10.1073/pnas.1018191108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Baker D. Protein Sci. 2010;19:1817. doi: 10.1002/pro.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Eriksson AE, Baase WA, Wozniak JA, Matthews BW. Nature. 1992;355:371. doi: 10.1038/355371a0. [DOI] [PubMed] [Google Scholar]
- (11).Das A, Wei Y, Pelczer I, Hecht MH. Protein Sci. 2011;20:702. doi: 10.1002/pro.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Lin Q, Barbas CF, 3rd, Schultz PG. J Amer Chem Soc. 2003;125:612. doi: 10.1021/ja028408e. [DOI] [PubMed] [Google Scholar]
- (13).Guo Z, Zhou D, Schultz PG. Science. 2000;288:2042. doi: 10.1126/science.288.5473.2042. [DOI] [PubMed] [Google Scholar]
- (14).Lockless SW, Ranganathan R. Science. 1999;286:295. doi: 10.1126/science.286.5438.295. [DOI] [PubMed] [Google Scholar]
- (15).Gloster TM, Roberts S, Perugino G, Rossi M, Moracci M, Panday N, Terinek M, Vasella A, Davies GJ. Biochemistry. 2006;45:11879. doi: 10.1021/bi060973x. [DOI] [PubMed] [Google Scholar]
- (16).Gloster TM, Roberts S, Ducros VM, Perugino G, Rossi M, Hoos R, Moracci M, Vasella A, Davies GJ. Biochemistry. 2004;43:6101. doi: 10.1021/bi049666m. [DOI] [PubMed] [Google Scholar]
- (17).Wallace BD, Wang H, Lane KT, Scott JE, Orans J, Koo JS, Venkatesh M, Jobin C, Yeh LA, Mani S, Redinbo MR. Science. 2010;330:831. doi: 10.1126/science.1191175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Sheffler W, Baker D. Protein Sci. 2009;18:229. doi: 10.1002/pro.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Bernstein FC, Koetzle TF, Williams GJ, Meyer EF, Jr., Brice MD, Rodgers JR, Kennard O, Shimanouchi T, Tasumi M. J Mol Biol. 1977;112:535. doi: 10.1016/s0022-2836(77)80200-3. [DOI] [PubMed] [Google Scholar]
- (20).Reinhart GD. Arch Biochem Biophys. 1983;224:389. doi: 10.1016/0003-9861(83)90225-4. [DOI] [PubMed] [Google Scholar]
- (21).Namchuk MN, Withers SG. Biochemistry. 1995;34:16194. doi: 10.1021/bi00049a035. [DOI] [PubMed] [Google Scholar]
- (22).Reynolds KA, McLaughlin RN, Ranganathan R. Cell. 2011;147:1564. doi: 10.1016/j.cell.2011.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Machicado C, Bueno M, Sancho J. Protein Eng. 2002;15:669. doi: 10.1093/protein/15.8.669. [DOI] [PubMed] [Google Scholar]
- (24).Demerdash ON, Daily MD, Mitchell JC. PLoS Comput Biol. 2009;5:e1000531. doi: 10.1371/journal.pcbi.1000531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Dixit A, Verkhivker GM. PLoS Comput Biol. 2011;7:e1002179. doi: 10.1371/journal.pcbi.1002179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Kidd BA, Baker D, Thomas WE. PLoS Comput Biol. 2009;5:e1000484. doi: 10.1371/journal.pcbi.1000484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Laine E, Goncalves C, Karst JC, Lesnard A, Rault S, Tang WJ, Malliavin TE, Ladant D, Blondel A. Proc Natl Acad Sci U S A. 2010;107:11277. doi: 10.1073/pnas.0914611107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Hardy JA, Wells JA. Curr Opin Struct Biol. 2004;14:706. doi: 10.1016/j.sbi.2004.10.009. [DOI] [PubMed] [Google Scholar]
- (29).Wright CM, Heins RA, Ostermeier M. Curr Opin Chem Biol. 2007;11:342. doi: 10.1016/j.cbpa.2007.04.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.