Abstract
Small noncoding RNAs (ncRNAs) have been shown to guide epigenetic silencing complexes to target loci in human cells. When targeted to gene promoters, these small RNAs can lead to long-term stable epigenetic silencing of gene transcription. To date, small RNAs have been shown to modulate transcriptional gene silencing (TGS) of human immunodeficiency virus type 1 (HIV-1) as well as several other disease-related genes, but it has remained unknown as to what extent particular chemistries can be used to generate single-stranded backbone-modified oligonucleotides that are amenable to this form of gene targeting and regulation. Here, we present data indicating that specific combinations of backbone modifications can be used to generate single-stranded antisense oligonucleotides that can functionally direct TGS of HIV-1 in a manner that is however, independent of epigenetic changes at the target loci. Furthermore, this functionality appears contingent on the absence of a 5′ phosphate in the oligonucleotide. These data suggest that chemically modified oligonucleotide based approaches could be implemented as a means to regulate gene transcription in an epigenetically independent manner.
Keywords: Ago-1, antisense non-coding RNA, DNMT3a, epigenetic, HDAC-1, HIV-1, LNA, oligonucleotide, TGS, transcription
Introduction
Transcriptional gene silencing (TGS) results when small or long noncoding RNAs (ncRNAs) target epigenetic modifications to genetic loci containing gene promoters (reviewed in ref. 1,2). TGS with small ncRNAs was first observed in plants and shown to result in stable long-term gene silencing and was shown to be mechanistically linked to RNA-directed DNA methylation of the gene promoter.3,4 To date, the ability of ncRNAs to direct TGS has been shown in a variety of organisms such as yeast (Schizosaccharomyces pombe), flies (Drosophila), worms (Caenorhabditis elegans), and human cells (reviewed in ref. 5,6). In human cells, the induction of TGS requires a relatively short duration of exposure to a small ncRNA, which can include small interfering RNA (siRNA), short hairpin RNA (shRNA), or single-stranded antisense RNAs (asRNA).7 Initial exposure to the small ncRNA results in histone modifications,8,9 followed later, in some cases, by DNA methylation at the targeted loci.10,11 This form of silencing can be long-lasting and heritable as the epigenetic marks are passed on to daughter cells and thus retained through cell division.10 Notably, TGS has been observed to be functional in vivo suggesting therapeutic relevance.12,13,14
Studies of canonical small antisense ncRNAs targeted to gene promoters have revealed the involvement of proteins: Argonaute 1 (Ago-1), histone deacetylase-1 (HDAC-1), and DNA methyltransferase 3a (DNMT3a)7,11,15,16 all of which are required for the induction of TGS in human cells. The promoter-targeted ncRNAs are able to target the promoter region by recognizing a low-copy promoter-associated RNA that spans the promoter and may act as a scaffold for the recruitment of the transcriptional silencing complex containing HDAC-1, Ago-1, DNMT3a, and possibly several other unknown proteins.9,17,18,19,20 Recent observations have indicated that one class of endogenous RNAs, long antisense ncRNAs, are functionally capable of directing TGS in human cells21,22,23 and discussed in detail in 2,24,25. These long antisense ncRNAs are presumed to function by guiding epigenetic regulatory protein complexes to the particular targeted loci, though studies delineating this mechanism are limited.21,22,23 Interestingly suppression of these endogenous ncRNAs can lead to derepression and transcriptional gene activation.2,26
Although much information has been gleaned regarding the ability of ncRNAs to modulate gene transcription in human cells, it remains unknown as to what extent small synthetic oligonucleotide variants with various backbone chemical modifications can be utilized to exploit the endogenous TGS pathway and epigenetically regulate gene transcription. Of particular interest is that synthetic oligonucleotides may exhibit increased stability when compared to endogenous ncRNA and thus may prove to be an advantageous means of inducing long-term TGS. Data presented here suggests that particular backbone modifications are tolerated and can functionally direct TGS of HIV-1 in human cells but that this method, depending on the modification, does not appear to utilize the same protein components as endogenously expressed asRNAs or siRNAs in directing TGS. This research addresses an understudied area that may one day lend itself to the development of oligonucleotide-mediated therapeutics.
Results
Chemically modified antisense oligonucleotides effectively suppress LTR transcription
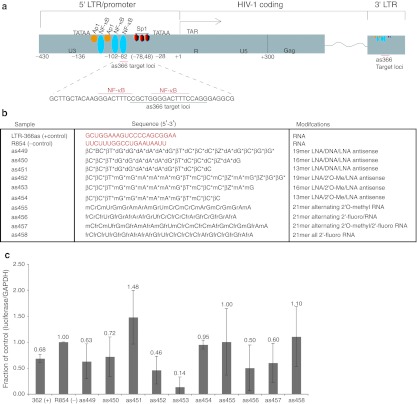
Targeting of an actively transcribed gene promoter by antisense ncRNA is becoming recognized as an endogenous mode of transcriptional gene regulation in human cells.21,22,27,28 Here, we target the LTR-366 site in the human immunodeficiency virus type 1 (HIV-1) long terminal repeat (LTR) (Figure 1a). The small asRNA LTR-366 has previously been found to direct TGS of HIV-1.7,9,11,29 The target site for LTR-366 overlaps a unique nuclear factor-κB (NF-κB) doublet in the LTR, a critical element in HIV-1 transcription (Figure 1a). Although this site has been shown to be susceptible to small asRNA- and siRNA-mediated TGS in vivo,30 the use of modified oligonucelotides has not been explored. In order to determine the ability of chemically modified antisense oligonucleotides to selectively direct TGS of HIV-1, several variants with backbone chemical modifications of the LTR-366 asRNA were synthesized as single-stranded antisense oligonucleotides (Figure 1b). These backbone-modified antisense oligonucleotides were transfected into TZM-bl cells containing an integrated HIV-1 LTR-driven luciferase reporter construct that can be transcriptionally activated by the expression of the viral Tat protein. Using this reporter system, the top suppressive candidates were determined (Figure 1c). From this initial screen of backbone-modified asRNA, we were able to discern several variants of the LTR-366 asRNA that were capable of suppressing Tat-mediated expression of luciferase in TZM-bl cells to an equal or higher efficiency than the unmodified asRNA 366 control. These variants contained 2′OMe and 2′F (as457), intermittently spaced 2′F (as456) and specific LNA–2′ OMe combinations (as452 and as453). The LTR-366 asRNA variant containing continuous 2′F substitutions was ineffective at suppressing luciferase expression (Figure 1a).
Figure 1.
Antisense oligonucleotide targeting of HIV-1 5′ LTR-mediated transcription. (a) Schematic representation depicting the HIV-1 5′ LTR and the previously determined as366 small RNA target site. Several transcription factor-binding sites are also shown. (b) Backbone-modified oligonucleotides generated against as366 target site. Note the various backbone modifications are: [2′4′]-locked ribonucleotide (β), 2′-O-methyl-ribonucleotide (m), and 5-methyl-cytosine (Z), phosphorothioate linkage (*). (c) Various backbone-modified single-stranded oligonucleotides, designed to target the LTR-366 site, were cotransfected into TZM-bl indicator cells with a Tat expressing plasmid and screened for effects on luciferase expression 48 hours post-transfection. The average from a duplicate sampling of a single experiment are shown with the respective ranges. GAPDH, glyceraldehyde 3-phosphate dehydrogenase; HIV-1, human immunodeficiency virus type 1; LNA, locked nucleic acid; LTR, long terminal repeat; NF-κB, nuclear factor-κB.
LTR-targeted as452 and as453 direct TGS in variant specific and unique manner
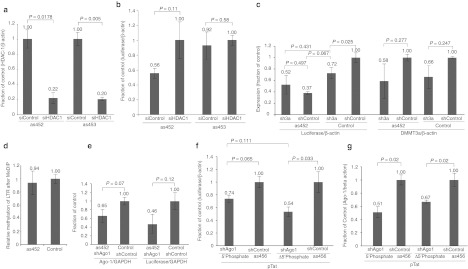
The observed suppression of luciferase expression by the various LTR-366 asRNA variants were indicative of potential TGS, but it is also plausible that the oligonucleotides were functional via interactions with the 3′ LTR-366, which is also expressed in frame with the luciferase transcript and contains the LTR-366 site. In order to determine whether or not the observed antisense oligonucleotide suppression of HIV-1 involves TGS, we suppressed the expression of HDAC-1, Ago-1, and DNMT3a, protein components, which have previously been shown to be required for si/shRNA-directed TGS of HIV-1.9,11,30,31 The ability of the two most promising LTR-366 asRNA variants, as452 and as453, to suppress LTR-driven luciferase expression was examined in conjunction with HDAC-1 knockdown. The as452 and as453 antisense oligonucleotides differ only in that three nucleotides from the 3′ end (βZ*βG*βG) have been removed in as453 (19mer to 16mer, Figure 1b). The removal of these three locked nucleic acid (LNA) nucleotides appeared to affect the pathway used as only as452 seemed to require the action of HDAC-1 (Figure 2a,b). Owing to this observation, that only as452 appeared to require the action of HDAC-1 to function, we surmised that as452 is functional in suppressing HIV-1 LTR transcription in a TGS-based manner. As such as452 was the candidate oligonucleotide explored further.
Figure 2.
Mechanistic assessment of antisense oligonucleotide targeting of the HIV-1 LTR. (a,b) The role of HDAC-1 in oligonucleotide mediated TGS of HIV-1. TZM-bl cells were treated with siRNAs to suppress HDAC-1 or siRNA R854 (control) (100 nmol/l). Twenty-four hours later the cultures were transfected with as452 or as453 (100 nmol/l). The cultures were collected 48 hours later and either (a) HDAC-1 expression or (b) LTR-mediated expression of luciferase determined by qRT-PCR. (c) The role of DNMT3a in oligonucleotide-mediated TGS of HIV-1. TZM-bl cells were transfected with control or DNMT3a suppressing shRNA expressing plasmids. Twenty-four hours later the cultures were transfected with as452 or the R854 (control) (100 nmol/l). The cultures were collected 48 hours later and either luciferase or DNMT3a expression determined by qRT-PCR. (d) Relative level of methylation of the 5′ LTR target region 366 following treatment with pTatDSred and subsequent transfection with as452 or R854 (control) (100 nmol/l). Cells were collected 72 hours later and assayed using MeDIP and qPCR performed specifically for the region of interest. (e) The role of Ago-1 in as452-mediated TGS. TZM-bl cells were transfected with pTatDSred for 24 hours before transfection with either as452 or R854 (control) (100 nmol/l) and a plasmid containing a shRNA targeting Ago-1 (shAgo-1) or the same plasmid without insert (shControl). Cells were collected 72 hours later and either luciferase or Ago-1 expression assayed using qPCR. (f,g) The effects of 5′ phosphate additions on Ago-1 mediated TGS. TZM-bl cells were transfected with pTatDSred with shControl or shAgo-1. Twenty-four hours later the cultures were transfected with either as452 or R854 (control) (100 nmol/l) and cellular RNAs collected 72 hours later and luciferase or Ago-1 expression determined by qRT-PCR (f and g, respectively). For a–g, the averages of triplicate treated cultures are shown with the standard error of the mean, with the exception being e, which shows the standard deviations; P values from a paired t-test are also shown. Ago-1, argonaute 1; DNMT3a, DNA methyltransferase 3a; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; HDAC-1, histone deacetylase-1; HIV-1, human immunodeficiency virus type 1; LTR, long terminal repeat; MeDIP, methylated-DNA immunoprecipitation; qPCR, quantitative PCR; qRT-PCR, quantitative reverse transcription-PCR; shRNA, short hairpin RNA; siRNA, small interfering RNA; TGS, transcriptional gene silencing.
Previous work with small ncRNA-directed TGS in human cells has demonstrated a requirement for DNMT3a (reviewed in ref. 6). DNMT3a has been previously reported to form a cellular epigenetic remodeling complex with Ezh2 and HDAC-1.32 To determine whether there is a requirement of DNMT3a in as452-mediated TGS of HIV-1 LTR activity, we suppressed DNMT3a expression by shRNA and assayed as452 suppression of luciferase. The suppression of DNMT3a, while also having an off-target effect on endogenous LTR-mediated expression of luciferase, demonstrated a modest reduction in as452-mediated TGS of LTR expressed luciferase (Figure 2c). This reduction was not highly significant, however, previous studies have observed that a longer periods of targeting are required for extensive DNA methylation (~10 days of sustained treatment),10 indicating that DNMT3a involvement may be reduced in the time frame used for these experiments or that as452 does not exert its effects through DNMT3a. Therefore, in order to investigate further whether methylation of cytosine to 5-cytosine by DNMT3a plays a role in as452 suppression of the LTR-366; methylated-DNA immunoprecipitation (MeDIP) was performed with an antibody specific to 5-methylcytosine. Treatment of cells with either as452 or the control R854 did not appear to modulate DNA methylation of the targeted LTR-366 region ~72 hours post-treatment (Figure 2d); indicating that DNMT3a, and de novo DNA methylation, does not play an appreciable role in the mechanism of as452-directed silencing. Taken together these data suggest that as452 appears to modulate TGS via an HDAC-1 dependent but DNMT3a independent manner.
Ago-1 has also been found to be required for siRNA-directed TGS in human cells.16,33 In order to confirm whether or not as452 was operative in a manner that required Ago-1; we knocked down the expression of Ago-1 using shRNA. The suppression of Ago-1 did not appear to affect as452-mediated suppression of LTR expressed luciferase (Figure 2e). Unlike endogenously processed RNA; the chemically modified oligonucleotides contain no 5′ phosphate group. It has recently been proposed that a 5′ phosphate on the guide strand is essential for proper loading of microRNA into Ago in Drosophila melanogaster.34 To determine whether the 5′ phosphate was important, we selected the as456 variant, which is an RNA-based oligonucleotide that contains a alternating 5′ OH, 2′ fluoro and hydroxyl groups (Figure 1b). Two variants of as456 were generated, either a 5′ T4 polynucleotide kinase phosphorylated or 5′ unphosphorylated variant. Both of these variants suppressed LTR expression of luciferase equally well (Figure 2f) but the 5′ phosphorylated as456 appeared more susceptible to the suppression of Ago-1 than did the 5′ unphosphorylated variant (Figure 2f,g). Taken together these data suggest that as452, which lacks a 5′ phosphate, directs TGS in an HDAC-1 dependent, but DNMT3a and Ago1 independent manner.
as452 is tolerant of mistmatches and does not cause an interferon response
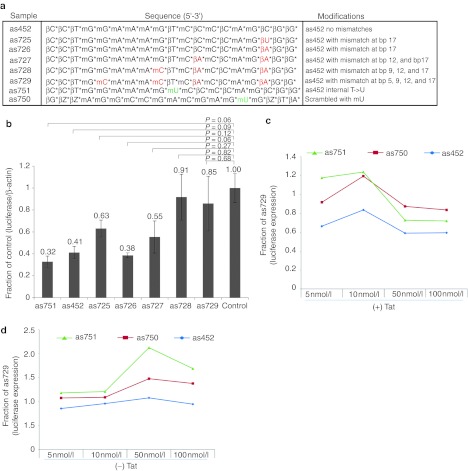
In order to determine the specificity of as452-mediated LTR-366 targeting, we generated various backbone- and nucleotide-modified variants of as452 (Figure 3a). Initial screening of these variants ability to suppress luciferase expression revealed that up to two mismatches (as725–as727, Figure 3a) were tolerated within the original as452 sequence as demonstrated by an appreciable efficacy in suppression. The addition of a third mismatch in as452 ablated its ability to suppress the LTR-366 target (as728 and as729) (Figure 3b). Interestingly, a simple change in the as452 sequence (variant as751) from a 2′-4′ LNA thymine to a 2′-O-methyl uracil demonstrated an increased efficacy of suppression (Figure 3b). Moreover, the suppression instilled in the LTR by the action of as452 appeared Tat dependent at 50–100 nmol/l (Figure 3c), and only as452 remained relatively inert when contrasted with scrambled and as751 (Figure 3d). These data indicate (i) that scrambled controls are not as suitable for determining functionality as mock-treated cells as they appear to instill enhanced biological noise, and (ii) that backbone- and nucleotide-modified variants of as452 targeting the HIV-1 LTR-366 can accommodate two mutations while retaining a relevant level of efficacy compared to as452. This observation may be therapeutically significant as it suggests TGS-active modified oligonuleotides may be able to retain suppression despite up to two mutations which in the case of HIV-1 may arise at the target site.35
Figure 3.
Tolerance of mismatches in antisense oligonucleotide-mediated TGS of HIV-1. (a) Several variations of the TGS inducing as452 antisense oligonucleotide were developed, containing specific variations. (b) The as452 variant antisense oligonucleotides were assessed for their ability to suppress HIV-1 LTR expressed luciferase in TZM-bl cells. (c,d) Dilution curve analysis of as452, as751, and scrambled controls standardized to the as729 control in (c) pTatDSRed or (d) untreated TZM-b1 cells. The cultures were assessed for luciferase protein expression 72 hours post-treatment. For a and b, the averages from triplicate transfected cultures are shown with the standard error of the means and the P values from a paired t-test. HIV-1, human immunodeficiency virus type 1; LTR, long terminal repeat; TGS, transcriptional gene silencing.
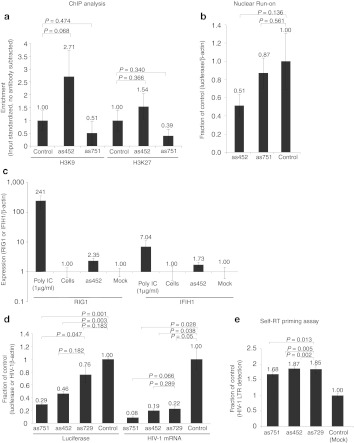
Previous studies have shown that ncRNA-directed TGS causes site-specific epigenetic changes to the ncRNA-targeted loci, such as histone 3 lysine 27 tri-methylation (H3K27me3) and histone 3 lysine 9 di-methylation (H3K9me2) (reviewed in ref. 2,6). These silent state epigenetic changes appear to be required for long-term stable TGS to occur.10 Furthermore, HDAC-1 appeared to be involved in the observed as452-mediated TGS of HIV-1 suggesting the possibility that as452 can target silent state epigenetic changes specifically at the LTR-366 target site. To determine the ability of as452 to target silent state epigenetic changes cultures were treated with either as452 or as751 and assayed for H3K9me2 and H3K27me3 enrichment at the HIV-1 LTR-366 target loci (Figure 1a) by chromatin immunoprecipitation (ChIP). The only notable enrichment observed was for H3K9me2 in the as452-treated cultures (Figure 3c). This observation suggests that backbone-modified as452-directed TGS is due to epigenetic changes at the LTR-366 target site but also that this effect is sequence-specific as as751 did not cause methylation in the same region (Figure 4a). When compared to previous studies, the relative enrichment of H3K9me2 observed at the as452 target site in the LTR-366 was notably lower.9,10,11,23,31 Therefore, in order to confirm that the epigenetic changes caused by as452 were correlative with a reduction in the amount of transcribed gene, nuclear run-on was performed. The as452 exhibited a significant reduction in the amount of transcribed RNA when compared to as751 and mock-treated cells (Figure 4b). Supporting the notion that as452 is suppressing LTR-366 activity in a transcriptional manner and that as751 may be functional in transcriptionally independent manner.
Figure 4.
Effects of antisense oligonucleotides on epigenetic modulation of HIV-1 expression. (a,b) TZM-bl cells were transfected with pTatDSred and 24 hours later transfected with HIV-1 LTR-targeted antisense oligonucleotides, as452, as751 or R854 (control) (100 nmol/l). Cultures were assessed for (a) H3K9me2 and H3K27me3 enrichment at the LTR by ChIP or (b) by nuclear run-on for transcription of luciferase relative to β-actin. (c) Type-I interferon analysis in antisense oligonucleotide-treated TZM-bl cells. Both RIG-1 and IFIH1 expression was assessed by qRT-PCR from as452-treated TZM-bl cells relative to mock controls, Poly I:C treated, or untreated cells. The averages are shown with the respective standard deviations. (d) TZM-bl cells were infected with HIV-1 (HX10, MOI = 0.1) and then 24 hours later transfected with antisense oligonucleotides (100 nmol/l, RNAiMax). The cultures were assessed 72 hours later for either luciferase or HIV-1 expression by qRT-PCR. (e) Culture RNAs from d were assessed by reverse transcription in the absence of a primer to determine possible self-priming capabilities as an indication of 3′ LTR targeting. For a, b, d, and e the averages of triplicate treated cultures are shown with the standard error of the mean (a, d, and e) or standard deviations (b only) and the P value from a paired t-test. ChIP, chromatin immunoprecipitation; HIV-1, human immunodeficiency virus type 1; LTR, long terminal repeat; MOI, multiplicity of infection; mRNA, messenger RNA; qRT-PCR, quantitative reverse transcription-PCR.
The experiments presented here are introducing synthetic chemically modified oligonucleotides to cells, therefore there is a risk of inducing a type-I interferon response. Type-I interferon responses involve increased RIG1 and IFIH1 expression in response to foreign nucleic acids.36 To determine the activation of interferon-related pathways by as452, we compared the TLR3 activation in TZM-b1 by treatment with as452 or the known activator Poly I:C. A modest increase in RIGI and IFIH1 was observed in the as452-treated cells; however, it was not greater than the variation observed between as452 treated, mock treated, and untreated cells (Figure 4c). These data suggest that the previously observed suppression modulated by as452 does not appear to involve activation of the type-I interferon pathway.
as452 and as751 block LTR expression in a TGS and PTGS manner respectively
The above studies were performed in a TZM-bl reporter cell line and therefore, the ability of as452 to functionally suppress infectious HIV-1 remained unclear. To determine the efficacy of as452-mediated TGS in the context of an ongoing HIV-1 infection; TZM-bl cells were infected with HIV-1 and subsequently treated with as452, as751 or the respective controls (as729 and R854, Figures 1a and 3a). Both LTR-expressed luciferase and HIV-1 messenger RNA (mRNA) expression were suppressed in HIV-1–infected TZM-bl cells in a manner concordant with those values observed in TZM-bl cells containing the integrated HIV-1 LTR-driven luciferase reporter activated by viral Tat protein expression (Figure 4d).
There was however a discrepancy observed between the suppression of luciferase, an indication of LTR-366 specific targeting, and HIV-1 in these cells (Figure 4d). Both 5′ and 3′ LTRs contain the same sequence, which is the target for LTR-366 (Figure 1a), and therefore full-length HIV-1 can be effectively targeted by as452 and as751 in the viral 3′LTR/mRNA. One plausible explanation for the previously observed differences between as452- and as751-based suppression might be that the backbone-modified oligonucleotides are more suppressive of viral transcripts than the integrated LTR. In theory, the backbone-modified oligonucleotide molecules might function in a bipartite manner such that in the context of an ongoing HIV-1 infection they might feasibly be inducing both TGS and also some level of either mRNA binding/blocking or post-transcriptional gene silencing (PTGS). In order to determine whether or not this is the case total RNA from HIV-1 infected cells treated with as452, as751, as729 or mock treated were reverse transcribed in the absence of any primers and subsequently analyzed by quantitative PCR (qPCR) with primers specific for the LTR-366 loci. The notion being that if the backbone-modified oligonucleotides are binding the viral mRNA, a signal above the mock-treated cells would be evident. All of the oligonucleotide-treated cultures demonstrated appreciable increases above the mock-treated HIV-1 infected control suggesting that there is some binding of the various backbone-modified oligonucleotides to the HIV-1 mRNA (Figure 4e). Taken together these data suggest that the variant as452 is functional in suppressing HIV-1 in a bipartite manner whereas as751 and as729, which contains four mismatches, suppresses primarily through viral mRNA binding.
Discussion
Previous studies with duplexed RNAs containing backbone modifications demonstrated that various modifications in the passenger strand of the duplex RNAs such as 2′ fluoro-RNA, 2′ OMe-RNA, and LNA substitutions or combinations of multiple modifications were permissive for siRNA-directed PTGS and recently, TGS and transcriptional gene activation of progesterone.37 Although this study proved highly informative with regards to double-stranded RNA-based approaches to modulating TGS and/or transcriptional gene activation, it has remained unknown as to the efficacy of single-stranded backbone-modified antisense oligonucleotides at directing TGS. The emerging notion that natural antisense transcripts are endogenous effectors driving TGS and epigenetic regulation of genes in human cells suggest small asRNA mimics might prove useful with regards directing TGS in a therapeutic relevant manner (reviewed in ref. 24,25,38). Data provided here indicates that the ability of single-stranded moieties to cause TGS is highly dependent upon sequence length, backbone chemistry, and that different protein pathways may be involved in the process depending on the particular molecule.
Several different molecules were found to be capable of directing suppression of the HIV-1 LTR include intermittent 2′ fluoro, 2′ OMe, and LNA substitutions with the location of the particular modification proving important with regards to TGS function. While the observed suppression was as potent as previous observations of small asRNA or siRNA-directed TGS,9,10,11,29,31 there appeared to be a far less robust recruitment of epigenetic silencing associated changes involved in the process observed here7,11 and these oligonucleotides did not fair as well as LNAs containing 2′-methoxyethyl bases.39 Though these previous studies did not appreciate or assess the LNAs for transcriptional suppression as determined by nuclear run-on or epigenetic changes at the target loci39 as was done here. The lack of robust epigenetic changes could be due to an inability of as452 to efficiently utilize DNMT3a in the targeted silencing. Such an eventuality suggests that a different mechanism of TGS is being used to instill changes at the target loci, similar to observations by others who have targeted siRNAs directly at RNA polymerase II binding sites and TATA which function in an epigenetically independent manner.18,40 Furthermore, observations in as452-treated HIV-1 infected TZM-bl cells indicated that TGS was operative at the LTR, as was evident by the suppression of luciferase expression in these cells, but that as452 is also suppressive by targeting the 3′ LTR in the context of the viral transcript in a manner similar to as751. While TZM-bl cells also contain the 3′ LTR when activated by Tat the level of expression is far lower than when an ongoing viral infection is present, suggesting that the stoichiometry of target to oligonucleotide has an effect on how well TGS is established. These observations also suggest that combinations of as452 and as751 might provide increased efficacy in silencing as they appear to utilize different modes of silencing.
Previous work has shown that synthetic siRNA duplexes with 5′-hydroxyl ends are rapidly phosphorylated inside cells by the cellular kinase Clp141 but it is unknown whether this occurs for single-stranded oligonucleotides. As shown here, the addition of a 5′ phosphate group to a chemically modified as456 altered the TGS pathway from Ago-1 independence to increased Ago-1 dependence. This is in keeping with recent results that show an essential requirement for a 5′ phosphate group in order for proper loading and processing of microRNA and acts as a checkpoint for the microRNA in procession through the RNA-induced silencing complex and subsequent silencing of its target mRNA.34
Although the observations presented here indicate that single-stranded antisense oligonucleotides with particular backbone modifications can be utilized in targeted TGS in human cells there appear to be distinct shortcomings with these moieties. Most notably these molecules do not appear to epigenetically target the LTR-366 loci as profoundly as has been observed previously with other small RNAs targeted to the same LTR-366 loci but expressed either via vector transfection (asRNAs, shRNAs) or synthetically derived transiently transfected siRNAs.7,9,11,29 The lack of robust epigenetic silencing suggests that this form of suppression, although transcriptional in nature, will not be long-lasting and heritable.10 Although the reason for the difference in epigenetic modifications is not entirely clear it is highly plausible that the inherent chemical charges on the backbone of the chemically modified antisense oligonucleotides affect binding interactions with the target loci, which in this case is a promoter-associated transcript,11,17,19 or with those proteins, such as DNMT3a, which is required for ncRNA-directed epigenetic silencing.7,15 Taken together the data provided here suggests that an alternative non-epigenetic–driven mechanism can be utilized by particular chemically modified antisense oligonucleotides to transcriptionally modulate gene expression in human cells. Such a mechanism may prove useful as a means to control gene transcription while minimizing impacts on the endogenous ncRNA pathway in human cells.24,42
Materials and Methods
Cell cultures. TZM-bl cultures were grown at 37 °C in 5% CO2 in Dulbecco's modified Eagle's medium. Transfection of TZM-bl cells with the Tat expressing plasmid pTatDSred43 (1.6 µg/106 cells) was carried out using Lipofectamine 2000 according to the manufacturer's specifications (Life Technologies, Carlsbad, CA). The respective antisense oligonucleotides were provided (Coley/Pfizer, Dusseldorf, Germany) and transfected into TZM-bl cells 24 hours after Tat transfection in a dropwise manner to a final concentration of 100 nmol/l using Lipofectamine RNAiMAX (Life Technologies). The sequences of these various oligonucleotides used in this study are provided in Figures 1 and 2. The cultures were assessed for luciferase expression (primers in Supplementary Table S1), as an indication of LTR transcriptional fidelity at 48 or 72 hours after antisense oligonucleotide treatment and data standardized to the control, RNA 854, giving a value of 1.
ChIP analysis. ChIPs were performed as follows; TZM-bl cells (106/10 cm plate) were transfected with 1.6 µg pTatDSred. Twenty-four hours later TZM-bl cells were transfected with either as452 or as751 (final concentration 100 nmol/l, Lipofectamine 2000 (Life Technologies)). Seventy-two hours after oligonucleotide transfection the cells were cross-linked and ChIP performed as described in 11,17 with the following modification: chromatin was sonicated with a Misonix S-4000 using 2 × 90 seconds pulses at amplitude 4 with 120 seconds rest in between. The immuno-precipitated DNA was recovered by phenol/chloroform extraction and ethanol precipitation. DNA was then analyzed by qPCR using indicated primers (Supplementary Table S1) (Kapa Sybr Fast; Kapa Biosystems).
Nuclear run-on. Nuclear run-on was performed as described previously (Hawkins and Morris 2010). The protocol was modified to include the use of bioitin-16-UTP (Trilink, San Diego, CA). Biotinylated RNA was pulled down using Dynal C1 beads (Dynal; Life Technologies). cDNA copies of the RNA were generated using M-MLV reverse transcriptase (Life Technologies) and then analyzed by qPCR using the indicated primers (Kapa Sybr Fast; Kapa Biosystems). Data was standardized to β-actin expression and then normalized to mock control values.
qRT-PCR analysis of gene expression. RNA was extracted (RNeasy; Qiagen, Valencia, CA) using the QiaCube (Qiagen) and DNase treated (TURBO DNase; Life Technologies), reverse transcribed using nonspecific primers (Reverse Transcription Core Kit; Eurogentec, San Diego, CA, and analyzed by qPCR using indicated primers (Kapa Sybr Fast; Kapa Biosystems) (Supplementary Table S1). Results from qPCR were normalized first to internal glyceraldehyde 3-phosphate dehydrogenase (GAPDH) (primers reported in ref. 7,10,11,17) or β-actin levels (Supplementary Table S1) and then expressed as fractions of control values.
Suppression of HDAC-1 and DNMT3a. TZM-bl cells were plated in a 12-well plate (5 × 104/well) and transfected with pTatDSred (80 ng). For DNMT3a suppression, Mission shRNA plasmids targeted to DNMT3a or an irrelevant control (300 ng) (Mission shRNAs; Sigma-Aldrich, St Louis, MO, a gift from Carol Kreider) were cotransfected with pTatDSred. HDAC-1 expression was suppressed by cotransfection of an HDAC-targeted siRNA (Hawkins and others 2009) or the appropriate irrelevant control siRNA (R854)(final concentration 100 nmol/l) 24 hours later the cultures were transfected with the HIV-1 targeted as452 or control (final concentration of 100 nmol/l). Forty-eight hours post-transfection the cells were collected and luciferase, DNMT3a, or HDAC-1 expression determined by quantitative reverse transcription-PCR (qRT-PCR) using DNMT3a, HDAC-1 forward/reverse primers (Supplementary Table S1).
MeDIP. TZM-bl cells were TZM-bl cells were cotransfected with pTatDSred and 24 hours later the cultures were transfected with either as452 or the RNA control R854 (final concentration 100 nmol/l). The MeDIP assay was performed as described in 44 was performed with the following changes. AluI was replaced was MseI, 1 µg of digested DNA was used and samples were incubated overnight with a monoclonal antibody against 5-methylcytidine (Active Motif, Carlsbad, CA). Data was normalized against input DNA with primers targeting the 366 region (362 primers, see Supplementary Table S1) and GAPDH promoter and then normalized to R854 to compare relative methylation levels of the target region.
Ago-1 knockdown. TZM-bl cells were plated (5 × 104 cells/well) and transfected (Lipofectamine 2000; Life Technologies) with pTatDSred (80 ng) and a Mission shAgo-1 or Control plasmid (100 ng), respectively (Mission shRNAs; Sigma-Aldrich, a gift from Carol Kreider). Twenty-four hours later the cultures were transfected with either as452 or the RNA control R854 (final concentration 100 nmol/l). The cultures were assessed for Ago-1 and luciferase expression at 48 hours after antisense oligonucleotide treatment.
5′ phosphate labelling of as456. An RNA-based oligonucleotide targeted against the 366 region (as366) containing alternating 2′ fluoro and 2′ hydroxyl groups in addition to a 5′ OH was purchased from Integrated DNA Technologies (Coralville, IA). The oligonucleotide was 5′ end-labeled using T4 PNK (New England Biolabs, Ipswich, MA) for 30 minutes at 37 °C, purified via phenol/chloroform extraction, ethanol precipitated and resuspended in 10 mmol/l Tris-Cl, pH 8.5. The 5′ phosphate labeled as366 and non-labeled as366 concentration was determined using spectrofluorimetry. These oligonucelotides were then used as described in the method for Ago-1 knockdown.
as452 mutation analysis. TZM-bl cells (5 × 104/well in 12-well plate) were transfected with 80 ng pTatDSred. The following day the cultures were transfected using RNAiMAX with the following antisense oligonucleotides, as452, as725, as726, as727, as728, as729, and as751 (100 nmol/l), targeted to the HIV-1 366 site.9,29 Forty-eight hours post-transfection, luciferase expression was measured by qRT-PCR and normalized to β-actin expression. The averages of triplicate treated cultures are shown with the standard error of the means and P values from a paired t-test.
Detection of antisense oligonucleotide binding HIV-1 transcripts. TZM-bl cells were infected with HIV-1 (HX10, multiplicity of infection = 0.1) and 24 hours later transfected with various antisense oligonucleotides (100 nmol/l, RNAiMax; Life Technologies). The cultures were collected and assessed 72 hours later for luciferase or HIV-1 expression by qRT-PCR. To determine whether the antisense oligonucleotides were binding the 3′ LTR of HIV-1, a total of 400 ng of culture RNA was isolated, DNase treated and reverse transcribed in the absence of any primer. The resultant cDNAs were used in qPCR for HIV-1 with primers p128 and p129 (Supplementary Table S1).
Dilution analysis of oligonucleotides. TZM-bl cells (1.2 × 104/well in 96-well plate) were transfected with 20 ng pTatDSred. Six hours later the media was replaced and cells were transfected with oligonucleotides as452, as729, as750, as751, each at concentrations of 5 nmol/l, 10 nmol/l, 50 nmol/l, and 100 nmol/l (RNAiMAX; Life Technologies). Seventy-two hours later, 100 µl of ONE-Glo Luciferase Assay reagent was added to each well (Promega, Madison, WI). The plate was incubated at room temperature for 10 minutes, and luminescence intensity was measured using a Clarity Microplate Luminometer (BioTek, Winooski, VT). Luminosity measurements were taken at 1 second/well.
Acknowledgments
Funding for this project was the result of NIH R01 AI084406 and a specific funded project from Coley/Pfizer to K.V.M. We thank Carol Kreider, of Sigma-Aldrich, for providing the shRNA construct used to suppress DNMT3a, Ago-1 and Barbora Malecova for performing several experiments required for this body of work. This is TSRI manuscript 21490. The authors declared no conflict of interest.
Supplementary Material
Oligonucleotide primers used in ChIP and qPCR analysis.
References
- Matzke MA., and, Birchler JA. RNAi-mediated pathways in the nucleus. Nat Rev Genet. 2005;6:24–35. doi: 10.1038/nrg1500. [DOI] [PubMed] [Google Scholar]
- Morris KV. RNA-directed transcriptional gene silencing and activation in human cells. Oligonucleotides. 2009;19:299–306. doi: 10.1089/oli.2009.0212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzke MA, Primig M, Trnovsky J., and, Matzke AJ. Reversible methylation and inactivation of marker genes in sequentially transformed tobacco plants. EMBO J. 1989;8:643–649. doi: 10.1002/j.1460-2075.1989.tb03421.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassenegger M, Heimes S, Riedel L., and, Sänger HL. RNA-directed de novo methylation of genomic sequences in plants. Cell. 1994;76:567–576. doi: 10.1016/0092-8674(94)90119-8. [DOI] [PubMed] [Google Scholar]
- Hawkins PG., and, Morris KV. RNA and transcriptional modulation of gene expression. Cell Cycle. 2008;7:602–607. doi: 10.4161/cc.7.5.5522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malecová B., and, Morris KV. Transcriptional gene silencing through epigenetic changes mediated by non-coding RNAs. Curr Opin Mol Ther. 2010;12:214–222. [PMC free article] [PubMed] [Google Scholar]
- Weinberg MS, Villeneuve LM, Ehsani A, Amarzguioui M, Aagaard L, Chen ZX.et al. (2006The antisense strand of small interfering RNAs directs histone methylation and transcriptional gene silencing in human cells RNA 12256–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DH, Villeneuve LM, Morris KV., and, Rossi JJ. Argonaute-1 directs siRNA-mediated transcriptional gene silencing in human cells. Nat Struct Mol Biol. 2006;13:793–797. doi: 10.1038/nsmb1142. [DOI] [PubMed] [Google Scholar]
- Suzuki K, Juelich T, Lim H, Ishida T, Watanebe T, Cooper DA.et al. (2008Closed chromatin architecture is induced by an RNA duplex targeting the HIV-1 promoter region J Biol Chem 28323353–23363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins PG, Santoso S, Adams C, Anest V., and, Morris KV. Promoter targeted small RNAs induce long-term transcriptional gene silencing in human cells. Nucleic Acids Res. 2009;37:2984–2995. doi: 10.1093/nar/gkp127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner AM, De La Cruz J., and, Morris KV. Mobilization-competent Lentiviral Vector-mediated Sustained Transcriptional Modulation of HIV-1 Expression. Mol Ther. 2009;17:360–368. doi: 10.1038/mt.2008.268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Peng C, Li B, Wang F, Zhou C, Hong D.et al. (2012Transcriptional gene silencing of HPV16 E6/E7 induces growth inhibition via apoptosis in vitro and in vivo Gynecol Oncol 124296–302. [DOI] [PubMed] [Google Scholar]
- Perrone L, Devi TS, Hosoya KI, Terasaki T., and, Singh LP. Inhibition of TXNIP expression in vivo blocks early pathologies of diabetic retinopathy. Cell Death Dis. 2010;1:e65. doi: 10.1038/cddis.2010.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turunen MP, Lehtola T, Heinonen SE, Assefa GS, Korpisalo P, Girnary R.et al. (2009Efficient regulation of VEGF expression by promoter-targeted lentiviral shRNAs based on epigenetic mechanism: a novel example of epigenetherapy Circ Res 105604–609. [DOI] [PubMed] [Google Scholar]
- Jeffery L., and, Nakielny S. Components of the DNA methylation system of chromatin control are RNA-binding proteins. J Biol Chem. 2004;279:49479–49487. doi: 10.1074/jbc.M409070200. [DOI] [PubMed] [Google Scholar]
- Janowski BA, Huffman KE, Schwartz JC, Ram R, Nordsell R, Shames DS.et al. (2006Involvement of AGO1 and AGO2 in mammalian transcriptional silencing Nat Struct Mol Biol 13787–792. [DOI] [PubMed] [Google Scholar]
- Han J, Kim D., and, Morris KV. Promoter-associated RNA is required for RNA-directed transcriptional gene silencing in human cells. Proc Natl Acad Sci USA. 2007;104:12422–12427. doi: 10.1073/pnas.0701635104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napoli S, Pastori C, Magistri M, Carbone GM., and, Catapano CV. Promoter-specific transcriptional interference and c-myc gene silencing by siRNAs in human cells. EMBO J. 2009;28:1708–1719. doi: 10.1038/emboj.2009.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross JP, Suetake I, Tajima S., and, Molloy PL. Recombinant mammalian DNA methyltransferase activity on model transcriptional gene silencing short RNA-DNA heteroduplex substrates. Biochem J. 2010;432:323–332. doi: 10.1042/BJ20100579. [DOI] [PubMed] [Google Scholar]
- Chu Y, Yue X, Younger ST, Janowski BA., and, Corey DR. Involvement of argonaute proteins in gene silencing and activation by RNAs complementary to a non-coding transcript at the progesterone receptor promoter. Nucleic Acids Res. 2010;38:7736–7748. doi: 10.1093/nar/gkq648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris KV, Santoso S, Turner AM, Pastori C., and, Hawkins PG. Bidirectional transcription directs both transcriptional gene activation and suppression in human cells. PLoS Genet. 2008;4:e1000258. doi: 10.1371/journal.pgen.1000258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W, Gius D, Onyango P, Muldoon-Jacobs K, Karp J, Feinberg AP.et al. (2008Epigenetic silencing of tumour suppressor gene p15 by its antisense RNA Nature 451202–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins PG., and, Morris KV. Transcriptional regulation of Oct4 by a long non-coding RNA antisense to Oct4-pseudogene 5. Transcription. 2010;1:165–175. doi: 10.4161/trns.1.3.13332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris KV. Non-coding RNAs, epigenetic memory and the passage of information to progeny. RNA Biol. 2009;6:242–247. doi: 10.4161/rna.6.3.8353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris KV. Long antisense non-coding RNAs function to direct epigenetic complexes that regulate transcription in human cells. Epigenetics. 2009;4:296–301. doi: 10.4161/epi.4.5.9282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahlestedt C. Natural antisense and noncoding RNA transcripts as potential drug targets. Drug Discov Today. 2006;11:503–508. doi: 10.1016/j.drudis.2006.04.013. [DOI] [PubMed] [Google Scholar]
- Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D.et al. (2009Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals Nature 458223–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F.et al. (2010Long noncoding RNA as modular scaffold of histone modification complexes Science 329689–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K, Shijuuku T, Fukamachi T, Zaunders J, Guillemin G, Cooper D.et al. (2005Prolonged transcriptional silencing and CpG methylation induced by siRNAs targeted to the HIV-1 promoter region J RNAi Gene Silencing 166–78. [PMC free article] [PubMed] [Google Scholar]
- Turner AM, Ackley AM, Matrone MA, Morris KV. Characterization of an HIV targeted transcriptional gene silencing RNA in primary cells. Hum Gene Ther. 2012;22:1–11. doi: 10.1089/hum.2011.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagishi M, Ishida T, Miyake A, Cooper DA, Kelleher AD, Suzuki K.et al. (2009Retroviral delivery of promoter-targeted shRNA induces long-term silencing of HIV-1 transcription Microbes Infect 11500–508. [DOI] [PubMed] [Google Scholar]
- Viré E, Brenner C, Deplus R, Blanchon L, Fraga M, Didelot C.et al. (2006The Polycomb group protein EZH2 directly controls DNA methylation Nature 439871–874. [DOI] [PubMed] [Google Scholar]
- Kim DH, Villeneuve LM, Morris KV, Rossi JJ. Argonaute-1 directs siRNA-mediated transcriptional gene silencing in human cells. Nat Struct Mol Biol. 2006;13:793–797. doi: 10.1038/nsmb1142. [DOI] [PubMed] [Google Scholar]
- Kawamata T, Yoda M., and, Tomari Y. Multilayer checkpoints for microRNA authenticity during RISC assembly. EMBO Rep. 2011;12:944–949. doi: 10.1038/embor.2011.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerhout EM, Ooms M, Vink M, Das AT., and, Berkhout B. HIV-1 can escape from RNA interference by evolving an alternative structure in its RNA genome. Nucleic Acids Res. 2005;33:796–804. doi: 10.1093/nar/gki220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlee M, Barchet W, Hornung V., and, Hartmann G. Beyond double-stranded RNA-type I IFN induction by 3pRNA and other viral nucleic acids. Curr Top Microbiol Immunol. 2007;316:207–230. doi: 10.1007/978-3-540-71329-6_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts JK, Yu D, Charisse K, Montaillier C, Potier P, Manoharan M.et al. (2010Effect of chemical modifications on modulation of gene expression by duplex antigene RNAs that are complementary to non-coding transcripts at gene promoters Nucleic Acids Res 385242–5259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faghihi MA., and, Wahlestedt C. Regulatory roles of natural antisense transcripts. Nat Rev Mol Cell Biol. 2009;10:637–643. doi: 10.1038/nrm2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beane RL, Ram R, Gabillet S, Arar K, Monia BP., and, Corey DR. Inhibiting gene expression with locked nucleic acids (LNAs) that target chromosomal DNA. Biochemistry. 2007;46:7572–7580. doi: 10.1021/bi700227g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janowski BA, Kaihatsu K, Huffman KE, Schwartz JC, Ram R, Hardy D.et al. (2005Inhibiting transcription of chromosomal DNA with antigene peptide nucleic acids Nat Chem Biol 1210–215. [DOI] [PubMed] [Google Scholar]
- Weitzer S., and, Martinez J. The human RNA kinase hClp1 is active on 3' transfer RNA exons and short interfering RNAs. Nature. 2007;447:222–226. doi: 10.1038/nature05777. [DOI] [PubMed] [Google Scholar]
- Morris KV. The emerging role of RNA in the regulation of gene transcription in human cells. Semin Cell Dev Biol. 2011;22:351–358. doi: 10.1016/j.semcdb.2011.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unwalla HJ, Li MJ, Kim JD, Li HT, Ehsani A, Alluin J.et al. (2004Negative feedback inhibition of HIV-1 by TAT-inducible expression of siRNA Nat Biotechnol 221573–1578. [DOI] [PubMed] [Google Scholar]
- Weber M, Davies JJ, Wittig D, Oakeley EJ, Haase M, Lam WL.et al. (2005Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells Nat Genet 37853–862. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Oligonucleotide primers used in ChIP and qPCR analysis.