Abstract
Single-stranded DNA oligonucleotides (ODNs) can be used to direct the exchange of nucleotides in the genome of mammalian cells in a process known as gene editing. Once refined, gene editing should become a viable option for gene therapy and molecular medicine. Gene editing is regulated by a number of DNA recombination and repair pathways whose natural activities often lead to single- and double-stranded DNA breaks. It has been previously shown that introduction of a phosphorotioated ODN, designed to direct a gene-editing event, into cells results in the activation of γH2AX, a well-recognized protein biomarker for double-stranded DNA breakage. Using a single copy, integrated mutant enhanced green fluorescent protein (eGFP) gene as our target, we now demonstrate that several types of ODNs, capable of directing gene editing, also activate the DNA damage response and the post-translational modification of proliferating cell nuclear antigen (PCNA), a signature modification of replication stress. We find that the gene editing reaction itself leads to transient DNA breakage, perhaps through replication fork collapse. Unmodified specific ODNs elicit a lesser degree of replication stress than their chemically modified counterparts, but are also less active in gene editing. Modified phosphothioate oligonucleotides (PTOs) are detrimental irrespective of the DNA sequence. Such collateral damage may prove problematic for proliferation of human cells genetically modified by gene editing.
Keywords: DNA damage, gene editing, replication fork stalling
Introduction
Single-stranded DNA oligonucleotides (ODNs) can direct specific changes in the nucleotide sequence of DNA in a process known as gene editing. This technique has begun to emerge as a useful genetic tool for gene therapy. An increased understanding of the mechanism of action of gene editing has accelerated the progression toward application. Reaction parameters such as strand bias, suppression by certain mismatch repair proteins and the involvement of DNA replication in producing altered bases have been heavily investigated.1,2 While the focus of most workers remains, appropriately, on improving and stabilizing the frequency with which gene editing takes place (see ref. 3 for review), we have been examining DNA damage and its downstream consequences associated with the editing reaction.4,5 The development of double-stranded DNA damage appears tied to the slower progression of genetically altered cells through S phase.4,5,6,7,8,9,10,11,12,13,14,15 The slowing of DNA replication and the creation of strand breaks may be related, perhaps by the collapse of stalled replication forks during the gene editing process, a phenomenon known to occur under normal growth conditions when replication forks are impeded.16,17,18,19
Optimization of the ODN-induced gene editing reaction has led to the identification of factors that significantly enhance or impede the achievement of high levels of correction. Proteins involved in recombinational repair and regulation, which are elevated during S phase, are largely supportive of the gene editing reaction.4,20 Additionally, replication has been shown to positively influence the gene editing reaction. Slowing of replication, and the subsequent increase of the proportion of targeted cells in S phase, results in elevated correction levels; whereas prohibiting S-phase progression abolishes correction.3,10,11,12,13 Furthermore, the specific ODN has been shown to incorporate into the genome of corrected cells.14,15 The summation of current knowledge in the field thus far suggests a possible mechanism of ODN-integration in the context of replication. The associated DNA damage response seen in this context implicates replication stress as an outcome of gene editing and thus the incorporation of phosphorothioate ODNs may inhibit fork progression. In this article, we examine DNA damage and DNA breakage in greater detail with a particular emphasis on the specificity of the ODN and its chemical compositions as it relates to genotoxicity. To our surprise, we find that replication fork collapse21 can occur in the presence of electroporated ODNs even in the absence of gene editing activity. These results suggest that DNA damage induced by all forms of ODNs is a global effect of gene editing. Phosphorothioate linkages on the standard workhorse vector used to attain the highest frequencies of gene editing exacerbate these genotoxic effects.
Results
Gene editing in the HCT116-19 cell system
The model system employed herein is the human cell line HCT116 with a single copy of integrated mutant eGFP gene (HCT116-19). Correction of the single-base nonsense mutation by specific ODNs results in the production of fluorescent eGFP protein, allowing phenotypic correction to be monitored via flow cytometry. As shown in Figure 1, the mutant eGFP gene contains a stop codon (TAG) in place of the wild-type tyrosine codon (TAC). The standard workhorse ODNs, 72NT, 72NT-U, 72NT-PM are 72-mers that are complementary to the nontranscribed (G->C) strand of the eGFP gene. The perfect-match ODN, 72NT-PM complements and binds to the nontranscribed strand, but does not create a mismatched base pair. The asterisks indicate three-phosphorothioate bonds on either end of the ODN. 72NT-U is the same length and sequence as 72NT, but does not have any phosphorothioate bonds (unmodified). The 72NS ODN has no known complementarity to the target region or the human genome as accessed via BLAST (NIH) and BLAT (UCSC) analyses.5
Figure 1.
Model system for gene editing in mammalian cells. The target enhanced green fluorescent protein (eGFP) sequence contains a TAG stop codon in place of the wild-type TAC tyrosine codon (bold and underlined). The mutant eGFP that is produced is truncated and nonfluorescent. Specific DNA oligonucleotides (ODNs) were designed that can hybridize to the nontranscribed (NT) strand (72NT, 72NT-U, 72NT-PM). 72NT-PM is a perfect-match ODN that is designed to bind the target NT strand but not elicit a base-exchange event. 72NS is a random sequence ODN bearing no complementarity to the eGFP gene. *Phosphorothioate linkages and the bold capitalized letter indicates the target base.
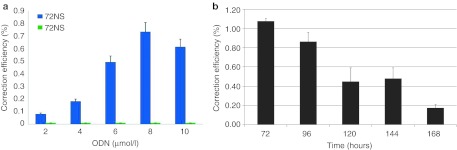
The experimental strategy of ODN-induced gene editing in this system has been previously reported and widely reviewed.22,23,24 Briefly, cells (2.5 × 106) are electroporated with ODN after 24 hours of synchronization with aphidicolin, a drug that reversibly synchronizes cells in early S phase. Synchronization and release has been shown to enhance correction efficiency, most likely because it elevates the number of cells passing through in S phase as a population in the presence of the ODN.4,6,9,10,22,24,25 Extensive washing of the cells removes effective concentrations of aphidicolin, which as a reagent does not influence the gene editing reaction (see ref. 4). Cells are allowed to recover after electroporation in full growth media for the specified time frame before analysis. When 72NT is electroporated into the cells at various levels, a dose response of gene editing is observed (Figure 2a). In contrast, 72NS does not support correction of the eGFP mutation as predicted and previously reported.5 These data validate the assay system used in this paper and display the specificity of the gene editing reaction.
Figure 2.
Gene editing in HCT116-19 cells. (a) HCT116-19 cells were synchronized with aphidicolin for 24 hours and then washed prior to the introduction of the indicated amount of DNA oligonucleotides (ODN); 72NT or 72NS. Gene editing activity was measured by the emergence of enhanced green fluorescent protein (eGFP) by flow cytometry analysis, 24 hours after the addition of the ODN. Correction efficiency (%) was determined as the percent of eGFP positive cells in the overall viable population. (b) HCT116-19 cells were synchronized for 24 hours and electroporated using standard conditions with 8 µmol/l 72NT. Time points were taken at 72, 96, 120, 44, and 168 hours respectively for analysis by FAC of eGFP (corrected cell) expression. Correction percentage was determined as the number of eGFP positive cells divided by the number of live cells in the population.
Figure 2b illustrates one of the barriers to the implementation of gene editing as a therapeutic option; the progressive loss of percentage of corrected cells. This phenomenon has been widely reported (see refs. 5,6,7,25,26 and references within) with reduced correction efficiencies easily observed at 96 hours. Here, we simply extend the time points at which evaluation for correction efficiencies were undertaken. The results of this work herein may offer one solution to this problem.
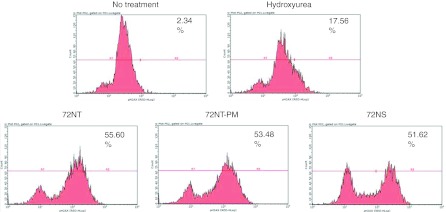
Recently, we observed double-strand break formation in response to the addition of ODNs designed to direct gene editing events.5 All three types of ODNs used in this study, 72NT, 72NS, and 72NT-PM, induced chromosomal damage albeit to various degrees. Double-stranded DNA breakage can result from stress on the replication fork caused by an impediment to S-phase progression. Ferrara and Kmiec.4 showed that the introduction of ODNs into the cell in a gene editing reaction activates Chk1 and Chk2 and in turn, progression through S phase is slowed. A hallmark event that correlates with both DNA damage and the slowing of cells through S phase is the phosphorylation of H2AX.17,25,26,27 One might predict that H2AX activation will also be observed under our standard reaction conditions. Phosphorylation of H2AX at serine 139 by ATM is widely accepted as a cellular response to DNA damage. Thus, we electroporated 72NT, 72NT-PM, and 72NS into synchronized HCT116-19 cells and measured H2AX phosphorylation, 24 hours later. The results are displayed in Figure 3: all three ODNs induce phosphorylation of H2AX as judged by treatment with fluorescently conjugated antibodies directed against H2AX-r and visualized by fluorescence-activated cell sorting (FACS). As a positive control, hydroxyurea is used to induce this H2AX activation because it causes replication fork collapse. The three ODNs, independent of target specificity, induce a very consistent shift in the cellular status of H2AX with activation peaks partitioned to the right of the profile. These data align with our earlier hypothesis that the amount of ODNs required to direct gene editing is at a level that induces a significant DNA damage response in the cell.26
Figure 3.
Fluorescence-activated cell sorting (FACS) analysis of the phosphorylation of H2AX. HCT116-19 cells synchronized with 2 µmol/l aphidicolin 24 hours before the addition of 2 mmol/l hydroxyurea (HU) or 4 µmol/l 72NT-PM, 72NT, or the 72NS DNA oligonucleotides (ODN). The reaction was allowed to proceed for 24 hours after which time the cells were processed for staining with antibodies directed against pH2AX. Analyses took place on a Guava EasyCyte 5HT Flow Cytometer (see Materials and Methods section) and phosphorylation is assessed by the egree of shift of the cell population to the right. The number in the upper right hand corner indicates the percentage of cells scoring positive for H2AX activation.
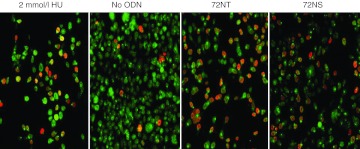
If the targeting reaction is associated with a slowing of S-phase progression,4 and this slowing is attributable to the ODN causing DNA breakage, vis-à-vis replication fork stress, we would predict that a higher degree of DNA breakage would be seen during S phase. It has been previously demonstrated that the proliferating cell nuclear antigen (PCNA) changes cellular location throughout the cell cycle.4,28,29 During S phase, PCNA is distinctly nuclear, while during the rest of the cell cycle, PCNA appears more diffuse throughout the cell; this relocalization can be detected with immunofluorescence. It has been demonstrated that concurrent staining of γH2AX and punctate PCNA is indicative of DNA damage within cells positioned in S phase.30,31 In our experimental approach, HCT116-19 cells were synchronized in early S phase with aphidicolin, treated with ODN, and then allowed to recover for 20 hours before staining for PCNA (green) and γH2AX (red). This delay enables the dispersal of any lingering effects of the synchronization process. hydroxyurea, known to cause replication fork collapse, and thereby double-strand break formation exclusively during S phase, served as a positive control for the punctate localization of nuclear PCNA and γH2AX. As can be seen in Figure 4, treatment of synchronized cells with an ODN (72NT) designed to modify the genomic sequence results in DNA damage during S phase; the appearance of distinct localization of punctate PCNA and γH2AX positive cells. Cells treated with a nonspecific ODN (72NS) however also show γH2AX positive cells at a level comparable to 72NT; the γH2AX positive cells also appear to have punctate, S phase-specific PCNA staining. In the absence of ODN, few cells exhibit H2AX activated staining. Thus, target specificity is not a differential factor in the DNA damage response during gene editing when cells are positioned in S phase.
Figure 4.
Colocalization of PCNA and H2AX-r during gene editing. Confocal image of HCT116-19 cells synchronized with 2 µmol/l aphidicolin 24 hours beforeo the addition of 2 mmol/l Hydroxyurea, 72NT or 72NS, all at 8 µmol/l final concentration. After recovery, cells were stained for γ H2AX (red) and PCNA (green diffuse and punctate) to identify activated cells and viewed under confocal microscopy at 20× magnification.
Termini modification of oligonucleotides can affect the degree of DNA breakage during gene editing
Historically, most ODNs used for gene editing have been designed with modifications that prevent nuclease degradation in the cell. These ODNs direct the highest levels of nucleotide exchange. As such, the ODNs used in the previous experiments contain three nonhydrolyzable phosphorothioate linkages on both termini. Recently, a number of independent labs demonstrated that unmodified ODNs support the gene editing reaction with a reduced level of cellular toxicity compared to ODNs containing the phosphorothioate modifications.32,33,34,35,36 Specifically, Aarts and te Riele35 showed that in post-delivery analyses of the unmodified ODN, low levels of DNA breakage are observed as judged by the neutral COMET assay. These observations, coupled with our own new findings surrounding the collateral damage of modified ODNs, prompted us to examine the activity of the unmodified 72-mer (72NT-U) in our gene editing assays.
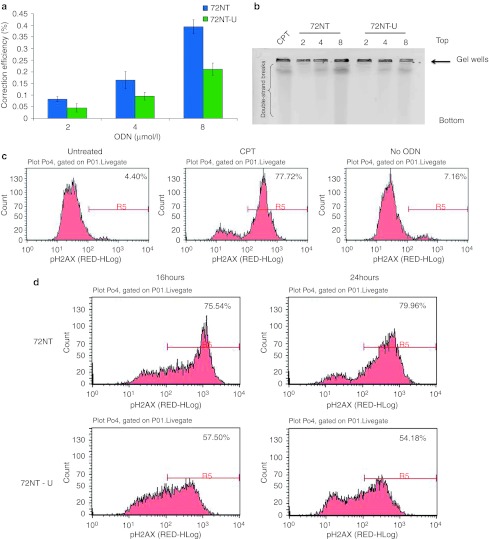
Synchronized HCT116-19 cells were treated with increasing amounts of 72NT or 72NT-U and we assessed the correction efficiencies of the two ODNs after 24 hours of recovery via flow cytometry. As seen in Figure 5a, the cells treated with unmodified ODN exhibit half as many correction events as cells treated with the modified, nuclease-protected ODN; data that are consistent with previous observations.5 Such results have made workers in the field a bit skeptical about using unmodified ODNs for gene editing. It is now clear however, that the collateral damage observed in cells targeted with phosphothioate oligonucleotides (PTOs) may be significant enough to seek alternative forms of ODNs. To this end, we evaluated genotoxic effects of 72NT-U on HCT116 cells as a function of gene editing in a similar fashion to the study conducted on 72NT. We had previously observed double-strand breaks in cells undergoing gene editing with PTO-ODNs.5 Now we asked if double-strand breaks are present when unmodified ODNs are used. We analyzed cells treated with increasing amounts of different ODNs by pulsed-field gel electrophoresis after 24 hours of reaction time. As seen in Figure 5b, both types produce the DNA streaking indicative of double-strand break formation and as the level of ODN increases, a dose response is observed in both cases. However, clearly a lower level of breakage is seen in the samples of cells treated with the unmodified ODN, 72NT-U. This result aligns with the notion that 72NT-U has a lower genotoxic effect on cells targeted for gene editing. To examine the collateral damage in cells treated with 72NT-U more fully, the levels of H2AX activation, via phosphorylation, in response to the introduction of 72NT and 72NT-U, respectively were visualized. A more tepid response to 72NT-U than to 72NT is observed (Figures 5c and d). Data reveal that 20–25% less of the cell population score positive for H2AX-γ as judged by fluorescent antibody binding and FACS-analyses. Thus, one important marker for the DNA damage response, H2AX-r, is present in fewer cells when 72NT-U is used for gene editing. In addition, the degree of damage response is less when 72NT-U is used as opposed to 72NT. These data might suggest that not only do more cells possess activated H2AX, many of them achieve a larger extent of damage when 72NT is used.
Figure 5.
Correction efficiency and DNA damage effects of the integrated enhanced green fluorescent protein (eGFP) gene as analyzed by flow cytometry and pulsed-field gel electrophoresis (PFGE). (a) HCT116-19 cells were synchronized with 2 µmol/l aphidicolin for 24 hours prior to electroporation with 2, 4, or 8 µmol/l of 72NT or 72NT-unm. After a 24-hour recovery period, cells were analyzed via flow cytometry analysis with propidium iodide being used to determine cell viability levels. Correction efficiency (CE) was determined as the percent of GFP positive cells out of the live cell population. *P < 0.05. (b) PFGE analysis of HCT116-19 cells treated with 2, 4, or 8 µmol/l 72NT or 72NT-unm for 24 hours. CPT (300 nmol/l for 24 hours) treatment is used as a positive control. The top and bottom of the gel are indicated as is the position of the loaded well. (c). Fluorescence-activated cell sorting (FACS) analyses of H2AX phosphorylation with samples of HCT116 cells treated with 30 nmol/l (CPT) or not treated (NT) or electroporated without DNA oligonucleotides (ODN). Electroporation only, also serves as a negative control. (D). All cell samples were treated with 8 µmol/l 72NT or 8 µmol/l 72NT-U, respectively. Cells were recovered for 16 hours or 24 hours in a 48-well plate. Cells were harvested by trypsinization, fixed, permeabilized, and stained with pH2AX, antibodies respectively and DNA damage analysis carried out by FACS. Phosphorylation was measured by the shift, to the right, in the histograms. The number in the upper right hand corner of each panel indicates the percentage of cells scoring positive for activation or staining with the H2AX-r antibody.
Replication fork progression is inhibited significantly by the presence of 72NT in the cell, but less so by 72NT-U
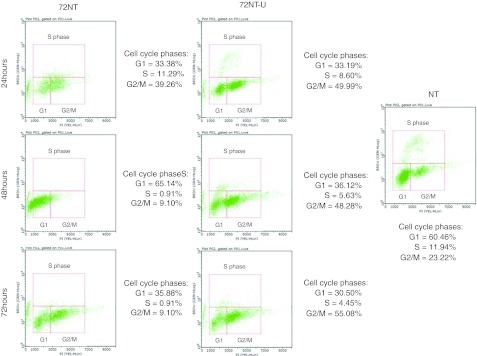
Based on the data presented above, one would predict that an enhanced level of DNA damage response and replication stress induced by 72NT might account, in part, for the observed genotoxicity in targeted cells. This outcome might arise from the cessation of movement or stalling of replication forks as a function of the presence of 72NT. To measure the status of replication fork activity, we analyzed BrdU incorporation in treated cells by flow cytometry. Synchronized cells were released for 4 hours, at which time an ODN, 72NT, or 72NT-U, was delivered into the cells by electroporation; BrdU incorporation was then measured at 24, 48, and 72 hours, respectively. This assay provides a view of the number of actively replicating forks; it is not a measure of the number of cells in S phase per se. Here, we used a FlowCollect Biovariate Cell Cycle kit (Millipore, Temecula, CA) which identifies cells bearing actively replicating forks. The results are presented in Figure 6. In the untreated control panel, we observed a standard cell distribution with each phase of the cycle represented and S phase quantitated at ~12%. The addition of 72NT causes, over the time course of the experiment, a virtual loss of any cells bearing actively replicating DNA forks. At 48 hours, the cells are piling up in G1 and by 72 hours, the total viable cell number has actually diminished dramatically; large percentages are nonviable at this time point. In contrast, cells treated with 72NT-U display a more modest reduction in the number of actively replicating DNA forks. There is clearly a detrimental effect with both ODNs, but cells treated with 72NT-U appear to induce a slower progression into G2/M but a significant number of cells remain active for replication. Thus, it appears that 72NT impacts active replication fork activity more dramatically than 72NT-U, perhaps by inducing replication fork collapse and ultimately double-stranded DNA breakage (see Figure 5b).
Figure 6.
BrdU uptake in cells treated with 72NT or 72NT-U. Synchronized cells were released for 4 hours prior to the electroporation of 72NT or 72NT-U. The cells were allowed to recover for the indicated times prior to being treated with BrdU for 60 minutes. Cells undergoing active replication were identified with the FlowCellect Bivariate Cell Cycle Kit for DNA Replication (Millipore). This kit employs a conjugated Anti-BrdU Alex-Flour 488 antibody mixed with propidium iodide (see Materials and Methods section). The cells were then analyzed on a Guava EasyCyte 5HT Flow Cytometer. The population of cells in each phase of the cell cycle is demarcated and the percentage of each population is presented at the right of each graph. NT; is the control of cells not treated with DNA oligonucleotides (ODN).
Discussion
We and others have demonstrated that single- and double-stranded DNA breaks appear as a function of ODN-directed gene editing, raising concern about secondary effects and collateral damage in the genome.5,6 We demonstrate here that transient double-stranded DNA breaks appear during the gene editing reaction; breakage is seen to arise independent of the homology to the target site. These data suggest that, in general, ODNs activate the DNA damage response pathway and inhibit the progression of cells through S phase. Chromosomal breakage occurs on a global level, but the extent of cellular disruption appears to be related to chemical modifications.
Previous data suggest that certain proteins, important in the gene editing reaction, are activated during S phase.6,8,10 Correction efficiencies are higher in replicating cells than in cells not allowed to progress through S phase.10,11,12,13,25,34 Proteins involved in homologous recombinational repair regulate the overall reaction with regard to the degree of correction.4,32,37 In contrast, proteins involved in nonhomologous end joining, appear to have little direct effect on the correction mechanism.20 These data, along with evidence of ODN integration into the genome,14,15 suggest a mechanism wherein the ODN aligns in homologous register with its target sequence during replication. Once incorporated, the mismatch repair system addresses the mismatched base pair through recognition by MSH2/MSH6/MLH1 etc. This complex signals the nuclease ExoI to remove a short stretch surrounding the mismatched base pair.38 A modified ODN would be resistant to removal by ExoI, thereby causing fork movement to be slowed or stopped. Under these circumstances, fork collapse can occur and a double-stranded DNA break can then ensue. Our previous data,5 suggest that some of these breaks are repaired and the modified ODN incorporates into the double helix resulting in a new nucleotide in the genome. In the specific reaction, the degree of breakage seems to be related to the amount of 72NT that is designed to anneal to the target site. Dilution experiments5 show that ds breaks correlate to the level of specific ODN present in a gene editing reaction. This idea must be understood within the context of the HCT116 cell line used in this study. These cells lack some of the MMR proteins that form the complex initializing ExoI entry. Since PCNA and RFC are present, there may be enough degeneracy in the signals so that once the PTO-ODN aligns in homologous register, ExoI may be present to some degree to initialize nuclease activity. But again, HCT116 cells lack MLH1 and thus the attraction of ExoI to the target site could be problematic. Alternatively, other nuclease activities associated with polymerase editing functions could be involved directly with the PTO-ODN. Considering, however, that gene editing has been shown to be more active on the lagging strand of a replication fork,39 it is not unlikely that nuclease activity associated with the processing of Okazaki fragments cold be involved in the reaction. Taken together, we now suggest that DNA damage and double-strand DNA breakage can develop as a function of the ODN interacting with and inhibiting the progression of the replication machinery.
We have used modified ODNs (phosphorothioate) because they are resistant to nuclease activity and typically provide higher correction levels as compared to unmodified ODNs.15,35,36 Pulsed-field gel electrophoresis analyses herein revealed that double-strand break formation is present in cells treated with either modified or unmodified ODN, but the unmodified ODN induces less breakage. This difference in DNA breakage levels can be attributed to the degradation of the unmodified ODN. If we once again refer to the Aarts and te Riele model38 described above, one can imagine that an unmodified ODN incorporated near or at a replication fork would be, in most cases, efficiently removed by nuclease activity from some molecular process going on in the region. Thus, fewer forks stall and only a small amount of double-stranded DNA breakage would then be generated. This removal would also account for the lower number of successful gene editing events when unmodified ODNs are used. Modified ODNs may resist removal by long enough to direct the nucleotide exchange but can concurrently cause double-strand breakage by inducing fork collapse.
Our observations of gene editing-induced DNA damage lead us to believe that replication fork collapse is the most likely cause of DNA breakage, and perhaps the overall DNA damage response. By several benchmarks used in our system, DNA damage response and localization of PCNA occur at roughly the same level as seen when either 72NT or 72NT-PM is used as the targeting ODN. This is likely due to the overwhelming number of DNA free ends that enter the nucleus when the ODN is delivered by electroporation. Free ends ignite a DNA damage response which may, by itself, cause a stalling of replication forks and a wide-spread slow-down in S-phase progression. Fork collapse and double-strand breakage is likely under these circumstances even without gene editing activity because some forks simply don't recover after the DNA damage response pathway is activated.
Thus, we believe that two distinct pathways account for DNA damage and double-strand breakage observed as a function of gene editing. As reported previously (and extended herein),5 specific or nonspecific ODN can induce a DNA damage response with the activation of H2AX, etc. But, the specific ODN (here 72NT) appears to induce DSBs at a level that is higher than its nonspecific counterpart. In this paper, we suggest two origins for the evolution of DSBs. The first is a relatively succinct and elegant fork collapse induced by the homologous alignment of the ODN to the target site. Inhibition of normal MMR activity by the modified ODN leads ultimately to the stalling of fork movement followed by development of double- strand breaks. This view is supported by a recent RNAi screen by Aarts and te Riele.40 Using an RNAi approach, they conducted a screening of proteins implicated in the ODN-induced gene editing reaction on a mouse embryonic stem cell system. Interestingly, knockdown of TLS proteins, involved in overcoming replication stress, resulted in a considerable depreciation of gene editing. The second pathway is more global in nature, based on a mass-action effect of large levels of ODNs, which overwhelms the cell. The presence of so many DNA free ends induces a protective response in a somewhat artifactual way that can also lead indirectly to DSB. Hence, there may be a specific mechanism of double-strand breakage buried under a global nonspecific reaction, which by itself can lead to collateral damage.
Clearly, replication activity is least affected when unmodified ODNs are used for gene editing, although there is some negative impact. Induction of the DNA damage response pathway and double-strand breaks do occur, but appear to be more moderate. Unmodified ODNs, however, direct <50% of the amount of gene editing seen with modified ODNs, again due in all likelihood to their efficient removal from the target site after alignment and incorporation. Thus, it may come down to a choice between efficiency of correction and the induction of collateral damage, a cost benefit analysis for consideration as gene editing moves closer to therapeutic application.
Materials and Methods
Cell line and culture conditions. HCT116 cells were acquired from American Type Cell Culture (Manassas, VA). The integrated HCT116 clone 19 (HCT116-19) was created by integrating a pEGFP-N3 vector (Clontech, Palo Alto, CA) containing a mutated enhanced green fluorescent protein (eGFP) gene, as described by Hu et al.21 The mutated eGFP gene has a nonsense mutation at position +67 resulting in nonfunctional eGFP protein. For these experiments, HCT116-19 cells were cultured in McCoy's 5A Modified medium (Sigma-Aldrich, St Louis, MO) supplemented with 10% fetal bovine serum, 2 mmol/l L-glutamine, and 1% penicillin/streptomycin. Cells were maintained at 37 °C and 5% CO2.
Oligonucleotide designs and eGFP targeting. ODNs were synthesized by Integrated DNA Technologies (Coralville, IA). The correcting oligonucleotide is a 72-mer that complements the nontranscribed strand of the target mutant eGFP gene. It has a centrally positioned mismatch that directs conversion of the mutant stop codon to the wild-type eGFP tyrosine, thereby allowing expression of functional eGFP. A perfect-match ODN was also designed as a 72-mer to target the nontranscribed strand, but would not induce a correction event. A nonspecific ODN bears a DNA sequence that has no homology to the target gene. Each ODN, unless otherwise noted, has three-phosphorothioate linkages on either end to help prevent nuclease degradation: 72NT-U does not have any phosphorothioate linkages.
Before eGFP targeting, cells were treated with aphidicolin for 24 hours in complete growth medium (unless otherwise noted). Cells were then trypsinized and harvested by centrifugation. Cells were resuspended to a concentration of 2.5 × 107 cells/ml in serum-free medium and 100 µl transferred to a 4-mm gap cuvette (BioExpress, Kaysville, UT). The respective ODN was added to a final concentration of 8 µmol/l (unless otherwise noted) and the cells were electroporated (250 V, 13 ms, 2 pulses, 1-second interval) using a BTX Electro Square Porator ECM 830 (BTX Instrument Division, Holliston, MA). The electroporated cells were then transferred to a 100-mm dish and allowed to recover in complete growth medium for 24 hours (unless otherwise noted) at 37 °C.
Flow cytometry. EGFP fluorescence was measured by a BD FACSAria II flow cytometer with FACSDiva (BD Biosciences, San Jose, CA) 24 hours after electroporation with ODN. Cells were harvested by trypsinization, washed once with phosphate-buffered saline (PBS), and resuspended in buffer (0.5% bovine serum albumin, 2 mmol/l EDTA, 2 µg/ml propidium iodide in PBS). Correction efficiency was then calculated as the percentage of eGFP positive cells out of the live cell population.
Fluorescence of eGFP was also measured by FACS analysis using a Guava EasyCyte 5HT Flow Cytometer (Millipore). Cells were harvested by trypsinization, washed once with 1× PBS−/− and resuspended in buffer (0.5% bovine serum albumin, 2 mmol/l EDTA, 2 µg/ml propidium iodide in PBS−/−). eGFP fluorescence was calculated two ways: the percentage of the total live eGFP positive population over the total live population and the percentage of the total eGFP positive population (live + dead) over the total cell population. Error bars are produced from three sets of data points generated over three separate experiments.
Analysis of DNA damage activity. HCT116-19 cells were synchronized with 6 µmol/l aphidicolin for 24 hours, released for 4 hours and targeted with ODN (with electroporation) allowed to recover in a 48-well plate in complete growth media for specific amount of time. Cells were then treated with the FlowCellect DNA Damage Kit (Millipore) specific for phosphorylated Histone H2AX. It uses a fluorescently labeled antibody optimized for analysis using flow cytometry. Briefly, 1 × 106 cells were harvested by trypsinization, washed with 1× wash buffer, fixed for 20 minutes on ice, washed again with wash buffer, and permeabilized for 20 minutes on ice. After permeabilization, 200,000 cells were transferred to a V-bottom 96-well plate, washed with assay buffer and then resuspended in 85 µl of assay buffer. The antibody (pH2AX) was added (5 µl) to the cells and allowed to incubate for one hour at room temperature. After incubation, the cells were washed with assay buffer and then resuspended in 200 µl of assay buffer and analyzed on a Guava EasyCyte 5HT Flow Cytometer (Millipore). The position along the x-axis correlates to the amount of phosphorylated H2AX, with a shift further to the right signifying a positive signal—phosphorylated H2AX. The percentage (out of total population) of phosphorylated H2AX is determined by the gating imposed by the positive and negative controls (nontranscribed and CPT, respectively). Anything under the right gate (R5) is considered to be positive and everything to the left is considered to be negative, according to the manufacturer.
Immunofluorescence. Electroporated cells were immediately plated in 8-well chambers (100,000 cells/well) and allowed to recover for 20 hours. Control cells were treated with 2 mmol/l hydroxyurea, which is known to cause replicated fork collapse and double-strand breaks exclusively in S phase for 20 hours. γH2AX and PCNA: Cells were then washed for 5 minutes at 37 °C in PBS. Cells were fixed in 4% paraformaldehyde for 30 minutes at room temperature. After washing with PBS, cells were permeabilized with 0.5% Triton X-100 in PBS for 10 minutes at room temperature. Cells were incubated in 10% normal goat serum for 10 minutes and then for 30 minutes in a 10% milk solution. γH2AX and PCNA antibodies were added at a dilution of 1:100 in 10% milk and kept at 4 °C overnight (γH2AX: Cell Signaling, Boston, MA; PCNA: Santa Cruz Biotechnology, Santa Cruz, CA). The next day, cells were washed four times for 10 minutes each in 0.1% Triton X-100. The secondary antibodies were then added for 1 hour at room temperature (γH2AX: α-rabbit, goat-Cy3; PCNA: α-mouse, goat-Alexa Fluor 488). Cells were again washed four times for 10 minutes each in 0.1% Triton X-100. Cells were visualized with a BioRad/Zeiss MRC 1,024 on an inverted Nikon Diaphot 300 (Nikon, Tokyo, Japan).
Pulsed-field gel electrophoresis. Targeted cells were allowed to recover for specified times in 60 mm dishes. Cells were harvested by trypsinization and 1 × 106 cells were isolated and pelleted by centrifuging at 1,500 r.p.m. for 5 minutes. Cells were washed once in PBS and resuspended in 50 mmol/l EDTA. Cells were combined in a 1:1 ratio with 1% low melt agarose (GIBCO; Invitrogen, Carlsbad, CA) in 50 mmol/l EDTA and transferred to plug molds. Plugs were allowed to cool at 4 °C for ~30 minutes before being transferred to lysis solution (50 mmol/l EDTA, 1% N-laurosylsarcosine, 1 mg/ml proteinase K). Cells were kept in lysis solution at 50 °C for 24 hours while shaking. Plugs were then washed four times in 1× TE buffer before being inserted into a 1% pulsed field certified agarose gel (Bio-Rad, Hercules, CA). The gel was run for 24 hours using a 120° field angle, 60–240 s switch time, 4 V/cm at 14 °C. The next day the gel was stained for 1 hour in ethidium bromide prior to imaging on an Alpha Innotech Fluorchem Q (Cell Biosciences, Santa Clara, CA).
Measurement of BrdU incorporation by flow cytometry. HCT116 cells were synchronized with 6 µmol/l aphidicolin for 24 hours, released for 4 hours and targeted with 6 µmol/l 72NT, 72NT-U by electroporation and allowed to recover in a 6-well plate in complete growth media for specific amount of time. Cells were then treated with the FlowCellect Bivariate Cell Cycle Kit for DNA Replication Analysis Kit (Millipore). This kit identifies cells undergoing replication in S phase of the cell cycle by employing a directly conjugated Anti-BrdU Alexa Fluor 488 antibody plus propidium iodide, a DNA-binding dye. This combination allows for the bivariate detection in two dimensions without the need for software modules and, therefore, can follow labeled cells through the cell cycle. Briefly, after recovery, cells were labeled with 1× BrdU for 60 minutes and then harvested by trypsinization followed by centrifugation, washed with 1× wash buffer, and fixed for 20 minutes on ice. After fixation, 250,000 cells were washed again with wash buffer, transferred to a V-bottom 96-well plate, and permeabilized for 20 minutes on ice. The cells were washed with assay buffer and then DNase I was added at a concentration of 300 µg/ml and incubated for one hour at 37 °C. The DNA denaturation reagent was removed by centrifugation and then washed with assay buffer. The cells are then resuspended in 95 µl assay buffer and 5 µl of the anti-BrdU Alexa Fluor 488 antibody was added and incubated on ice for 1 hour. After the incubation, the cells were washed with assay buffer and the DNA was stained with a freshly prepared solution of propidium iodide/RNase and allowed to incubate for 30 minutes at room temperature. The cells were then analyzed on a Guava EasyCyte 5HT Flow Cytometer (Millipore).
References
- Andersen MS, Sørensen CB, Bolund L., and, Jensen TG. Mechanisms underlying targeted gene correction using chimeric RNA/DNA and single-stranded DNA oligonucleotides. J Mol Med. 2002;80:770–781. doi: 10.1007/s00109-002-0393-8. [DOI] [PubMed] [Google Scholar]
- Igoucheva O, Alexeev V, Anni H., and, Rubin E. Oligonucleotide-mediated gene targeting in human hepatocytes: implications of mismatch repair. Oligonucleotides. 2008;18:111–122. doi: 10.1089/oli.2008.0120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engstrom JU, Suzuki T., and, Kmiec EB. Regulation of targeted gene repair by intrinsic cellular processes. Bioessays. 2009;31:159–168. doi: 10.1002/bies.200800119. [DOI] [PubMed] [Google Scholar]
- Ferrara L., and, Kmiec EB. Targeted gene repair activates Chk1 and Chk2 and stalls replication in corrected cells. DNA Repair (Amst) 2006;5:422–431. doi: 10.1016/j.dnarep.2005.11.009. [DOI] [PubMed] [Google Scholar]
- Bonner M., and, Kmiec EB. DNA breakage associated with targeted gene alteration directed by DNA oligonucleotides. Mutat Res. 2009;669:85–94. doi: 10.1016/j.mrfmmm.2009.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen PA, Solhaug A, Booth JA, Gelazauskaite M., and, Krauss S. Cellular responses to targeted genomic sequence modification using single-stranded oligonucleotides and zinc-finger nucleases. DNA Repair (Amst) 2009;8:298–308. doi: 10.1016/j.dnarep.2008.11.011. [DOI] [PubMed] [Google Scholar]
- Ferrara L, Engstrom JU, Schwartz T, Parekh-Olmedo H., and, Kmiec EB. Recovery of cell cycle delay following targeted gene repair by oligonucleotides. DNA Repair (Amst) 2007;6:1529–1535. doi: 10.1016/j.dnarep.2007.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorpe P, Stevenson BJ., and, Porteous DJ. Optimising gene repair strategies in cell culture. Gene Ther. 2002;9:700–702. doi: 10.1038/sj.gt.3301750. [DOI] [PubMed] [Google Scholar]
- Engstrom JU., and, Kmiec EB. Manipulation of cell cycle progression can counteract the apparent loss of correction frequency following oligonucleotide-directed gene repair. BMC Mol Biol. 2007;8:9. doi: 10.1186/1471-2199-8-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- .Wu XS, Xin L, Yin WX, Shang XY, Lu L, Watt RM.et al. (2005Increased efficiency of oligonucleotide-mediated gene repair through slowing replication fork progression Proc Natl Acad Sci USA 1022508–2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachman EE., and, Kmiec EB. Gene repair in mammalian cells is stimulated by the elongation of S phase and transient stalling of replication forks. DNA Repair (Amst) 2005;4:445–457. doi: 10.1016/j.dnarep.2004.11.007. [DOI] [PubMed] [Google Scholar]
- Olsen PA, Randol M., and, Krauss S. Implications of cell cycle progression on functional sequence correction by short single-stranded DNA oligonucleotides. Gene Ther. 2005;12:546–551. doi: 10.1038/sj.gt.3302454. [DOI] [PubMed] [Google Scholar]
- Olsen PA, Randøl M, Luna L, Brown T., and, Krauss S. Genomic sequence correction by single-stranded DNA oligonucleotides: role of DNA synthesis and chemical modifications of the oligonucleotide ends. J Gene Med. 2005;7:1534–1544. doi: 10.1002/jgm.804. [DOI] [PubMed] [Google Scholar]
- Huen MS, Li XT, Lu LY, Watt RM, Liu DP., and, Huang JD. The involvement of replication in single stranded oligonucleotide-mediated gene repair. Nucleic Acids Res. 2006;34:6183–6194. doi: 10.1093/nar/gkl852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radecke S, Radecke F, Peter I., and, Schwarz K. Physical incorporation of a single-stranded oligodeoxynucleotide during targeted repair of a human chromosomal locus. J Gene Med. 2006;8:217–228. doi: 10.1002/jgm.828. [DOI] [PubMed] [Google Scholar]
- Dubrana K, van Attikum H, Hediger F., and, Gasser SM. The processing of double-strand breaks and binding of single-strand-binding proteins RPA and Rad51 modulate the formation of ATR-kinase foci in yeast. J Cell Sci. 2007;120 Pt 23:4209–4220. doi: 10.1242/jcs.018366. [DOI] [PubMed] [Google Scholar]
- Zou L., and, Elledge SJ. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003;300:1542–1548. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
- Branzei D., and, Foiani M. Maintaining genome stability at the replication fork. Nat Rev Mol Cell Biol. 2010;11:208–219. doi: 10.1038/nrm2852. [DOI] [PubMed] [Google Scholar]
- Branzei D., and, Foiani M. The checkpoint response to replication stress. DNA Repair (Amst) 2009;8:1038–1046. doi: 10.1016/j.dnarep.2009.04.014. [DOI] [PubMed] [Google Scholar]
- Morozov V., and, Wawrousek EF. Single-strand DNA-mediated targeted mutagenesis of genomic DNA in early mouse embryos is stimulated by Rad51/54 and by Ku70/86 inhibition. Gene Ther. 2008;15:468–472. doi: 10.1038/sj.gt.3303088. [DOI] [PubMed] [Google Scholar]
- Kannouche PL, Wing J., and, Lehmann AR. Interaction of human DNA polymerase eta with monoubiquitinated PCNA: a possible mechanism for the polymerase switch in response to DNA damage. Mol Cell. 2004;14:491–500. doi: 10.1016/s1097-2765(04)00259-x. [DOI] [PubMed] [Google Scholar]
- Hu Y, Parekh-Olmedo H, Drury M, Skogen M., and, Kmiec EB. Reaction parameters of targeted gene repair in mammalian cells. Mol Biotechnol. 2005;29:197–210. doi: 10.1385/MB:29:3:197. [DOI] [PubMed] [Google Scholar]
- Parekh-Olmedo H, Ferrara L, Brachman E., and, Kmiec EB. Gene therapy progress and prospects: targeted gene repair. Gene Ther. 2005;12:639–646. doi: 10.1038/sj.gt.3302511. [DOI] [PubMed] [Google Scholar]
- Parekh-Olmedo H., and, Kmiec EB. Progress and prospects: targeted gene alteration (TGA) Gene Ther. 2007;14:1675–1680. doi: 10.1038/sj.gt.3303053. [DOI] [PubMed] [Google Scholar]
- Ferrara L., and, Kmiec EB. Camptothecin enhances the frequency of oligonucleotide-directed gene repair in mammalian cells by inducing DNA damage and activating homologous recombination. Nucleic Acids Res. 2004;32:5239–5248. doi: 10.1093/nar/gkh822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrara, L., and, Kmiec, EB. Targeted gene repair activates Chk1 and Chk2 and stalls replication in corrected cells. DNA Repair (Amst) 2006;5:422–431. doi: 10.1016/j.dnarep.2005.11.009. [DOI] [PubMed] [Google Scholar]
- Engstrom JU., and, Kmiec EB. Manipulation of cell cycle progression can counteract the apparent loss of correction frequency following oligonucleotide-directed gene repair. BMC Mol Biol. 2007;8:9. doi: 10.1186/1471-2199-8-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bravo R., and, Macdonald-Bravo H. Existence of two populations of cyclin/proliferating cell nuclear antigen during the cell cycle: association with DNA replication sites. J Cell Biol. 1987;105:1549–1554. doi: 10.1083/jcb.105.4.1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madsen P., and, Celis JE. S-phase patterns of cyclin (PCNA) antigen staining resemble topographical patterns of DNA synthesis. A role for cyclin in DNA replication. FEBS Lett. 1985;193:5–11. doi: 10.1016/0014-5793(85)80068-5. [DOI] [PubMed] [Google Scholar]
- Shimura T, Torres MJ, Martin MM, Rao VA, Pommier Y, Katsura M.et al. (2008Bloom's syndrome helicase and Mus81 are required to induce transient double-strand DNA breaks in response to DNA replication stress J Mol Biol 3751152–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleker T, Nagai S., and, Gasser SM. Posttranslational modifications of repair factors and histones in the cellular response to stalled replication forks. DNA Repair (Amst) 2009;8:1089–1100. doi: 10.1016/j.dnarep.2009.04.010. [DOI] [PubMed] [Google Scholar]
- Olsen, PA, Randol M, Luna, L, , T., and, Krauss, S. Genomic sequence correction by single-stranded DNA oligonucleotides: role of DNA synthesis and chemical modifications of the oligonucleotide ends. J Gene Med. 2005;7:1534–1544. doi: 10.1002/jgm.804. [DOI] [PubMed] [Google Scholar]
- Andrieu-Soler C, Casas M, Faussat AM, Gandolphe C, Doat M, Tempé D.et al. (2005Stable transmission of targeted gene modification using single-stranded oligonucleotides with flanking LNAs Nucleic Acids Res 333733–3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papaioannou I, Disterer P., and, Owen JS. Use of internally nuclease-protected single-strand DNA oligonucleotides and silencing of the mismatch repair protein, MSH2, enhances the replication of corrected cells following gene editing. J Gene Med. 2009;11:267–274. doi: 10.1002/jgm.1296. [DOI] [PubMed] [Google Scholar]
- Aarts M., and, te Riele H. Subtle gene modification in mouse ES cells: evidence for incorporation of unmodified oligonucleotides without induction of DNA damage. Nucleic Acids Res. 2010;38:6956–6967. doi: 10.1093/nar/gkq589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertoni C, Rustagi A., and, Rando TA. Enhanced gene repair mediated by methyl-CpG-modified single-stranded oligonucleotides. Nucleic Acids Res. 2009;37:7468–7482. doi: 10.1093/nar/gkp757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engstrom JU., and, Kmiec EB. DNA replication, cell cycle progression and the targeted gene repair reaction. Cell Cycle. 2008;7:1402–1414. doi: 10.4161/cc.7.10.5826. [DOI] [PubMed] [Google Scholar]
- Aarts M., and, te Riele H. Progress and prospects: oligonucleotide-directed gene modification in mouse embryonic stem cells: a route to therapeutic application. Gene Ther. 2011;18:213–219. doi: 10.1038/gt.2010.161. [DOI] [PubMed] [Google Scholar]
- Brachman EE., and, Kmiec EB. DNA replication and transcription direct a DNA strand bias in the process of targeted gene repair in mammalian cells. J Cell Sci. 2004;117 Pt 17:3867–3874. doi: 10.1242/jcs.01250. [DOI] [PubMed] [Google Scholar]
- Aarts M., and, te Riele H. Parameters of oligonucleotide-mediated gene modification in mouse ES cells. J Cell Mol Med. 2010;14 6B:1657–1667. doi: 10.1111/j.1582-4934.2009.00847.x. [DOI] [PMC free article] [PubMed] [Google Scholar]