Abstract
It has long been known that many proteins require folding via molecular chaperones for their function. Although it has become apparent that folding imposes constraints on protein sequence evolution, the effects exerted by different chaperone classes are so far unknown. We have analyzed data of protein interaction with the chaperones in Saccharomyces cerevisiae using network methods. The results reveal a distinct community structure within the network that was hitherto undetectable with standard statistical tools. Sixty-four yeast chaperones comprise ten distinct modules that are defined by interaction specificity for their 2,691 interacting proteins. The classes of interacting proteins that are in turn defined by their dedicated chaperone modules are distinguished by various physiochemical protein properties and are characterized by significantly different protein expression levels, codon usage, and amino acid substitution rates. Correlations between substitution rate, codon bias, and gene expression level that have long been known for yeast are apparent at the level of the chaperone-defined modules. This indicates that correlated expression, conservation, and codon bias levels for yeast genes are attributable to previously unrecognized effects of protein folding. Proteome-wide categories of chaperone–substrate specificity uncover novel hubs of functional constraint in protein evolution that are conserved across 20 fungal genomes.
Keywords: codon usage, community structure, networks, protein folding
Introduction
Chaperones (Ellis 1987), also called heat shock proteins (HSPs), are essential in all living cells as they assist protein folding, prevent protein aggregation, and play a crucial role in survival under stress conditions (Young et al. 2004). Manipulation of chaperone expression has revealed that chaperones have an additional role as capacitors of phenotypic variation (Fares et al. 2002; Queitsch et al. 2002; Rutherford 2003). Inhibition of Hsp90 chaperone function in Arabidopsis thaliana exposes genotype-independent phenotypic variation in a similar manner to growth under heat stress conditions (Queitsch et al. 2002). Increasing the expression level of the GroEL (Hsp60) chaperone confers improved fitness in Escherichia coli under high mutational loads (Fares et al. 2002). Chaperones can thus buffer the effects of slightly deleterious mutations, presumably by compensating for decreased protein structure stability of mutated proteins (Fares et al. 2002; Queitsch et al. 2002; Rutherford 2003).
Protein interaction with the chaperones for folding impacts the evolvability of substrate proteins (Rutherford 2003; Tokuriki and Tawfik 2009). Overexpression of GroEL/GroES can double the number of accumulating mutations in GroEL substrates in vitro (Tokuriki and Tawfik 2009). Furthermore, the amino acid substitution rate of proteins that depend upon the GroEL for folding in E. coli is higher than that of GroEL-independent proteins (Bogumil and Dagan 2010). Here, we study the impact of protein interaction with chaperones on whole-genome evolutionary dynamics. To address this question, we used a network approach to analyze an extensive data set of chaperone–protein interactions assembled by screening for chaperone-associated protein complexes in yeast (Gong et al. 2009). The chaperone repertoire in the Saccharomyces cerevisiae proteome consists of 69 molecular chaperones and their co-chaperones, most of which are known to assist the folding or unfolding of proteins in the cell; other chaperones assume diverse cellular functions including translocation across membranes and stabilizing protein–protein interactions (Voos and Röttgers 2003; Young et al. 2004; Kampinga and Craig 2010). The majority of nascent polypeptides in the yeast protein-folding pathway interact with the ribosome-associated complex (RAC) that includes a member of the Hsp70 family and a co-chaperone from the Hsp40 family (J-proteins) (Young et al. 2004; Kampinga and Craig 2010). Some proteins also interact with one or more of the following chaperone classes: prefoldin (PFD), TriC (CCT), and Hsp90 (Young et al. 2004). Most of the proteins encoded in the yeast genome (3,595 of 5,880) interact with at least one chaperone, many of them (2,952) with two or more chaperones (Gong et al. 2009). The present networks uncover hitherto unrecognized modular interactions between chaperone families and their interacting proteins.
Materials and Methods
Data
Data of chaperone interaction repertoire in S. cerevisiae were downloaded from Gong et al. (2009). Amino acid usage data, functional assignment, chromosomal location, frequencies of optimal codons, codon adaptation index (CAI), gravy scores (hydropathy index), and aromaticity scores were obtained from the Saccharomyces Genome Database (Cherry et al. 1997). Protein cellular localization was obtained from Huh et al. (2003) and the Gene Ontology database (Ashburner et al. 2000). Secondary structure of all proteins was inferred using PsiPred (Jones 1999). For the calculation of secondary structure usage, a threshold of probability >0.7 was used. Protein expression data were obtained from Ghaemmaghami et al. (2003). For the statistical analysis, the natural log of protein expression was used. Proteins with no expression level information (107) or with zero expression level (1,665) were omitted from the analysis. All statistical analyses were performed using MatLab Statistics toolbox.
Network Modularity Structure
A division of the nodes in the network into modules was obtained by defining a modularity function of each bipartition of the network, as the number of edges within a module minus the expected number of edges in the module. Maximizing this function over all possible divisions using eigenspectrum analysis yields the optimal division of the network into modules (Newman 2006).
Evolutionary Rate
Positional orthology assignments within 20 fungal proteomes were obtained from Wapinski et al. (2007). Open reading frames lacking orthologs (282 in total) were omitted from the analysis. Multiple alignments of all yeast open reading frames with orthologous sequences were reconstructed with MAFFT (Katoh et al. 2005). Phylogenetic trees were reconstructed with PhyML (Guindon and Gascuel 2003) using the best-fit model as inferred by ProtTest 3 (Darriba et al. 2011) using the Akaike information criterion (Akaike 1974) measure. Distances from the S. cerevisiae proteins to their orthologs were calculated as the sum of branch lengths. To calculate the relative amino acid substitution rates of substrates, we first Z-transformed the distances to the 20 proteomes separately and then averaged the standardized distances over all orthologs.
Results
Modules in the Chaperone–Substrate Interaction Network
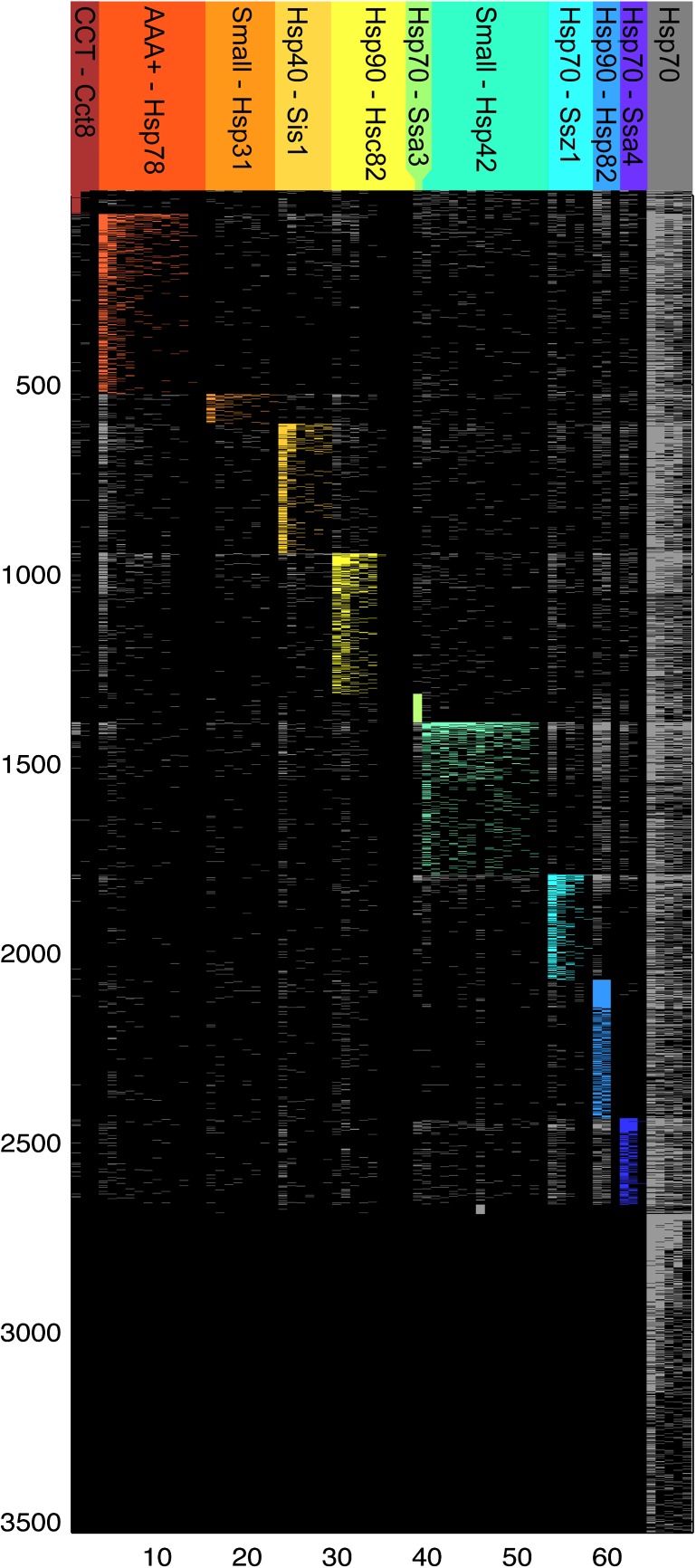
In an extensive screening for proteins that interact with each of the 63 chaperones encoded in yeast, Gong et al. (2009) documented a total of 21,687 interactions. The network reconstructed from Gong et al. (2009) data contains 3,595 entities, 3,526 of which are chaperone-interacting proteins (for simplicity termed “substrates” here, yet making no statement about specificity). The remaining 69 entities are chaperones. We designate this as the chaperone-substrate interaction (CSI) network. The network can be fully defined by a matrix, A = [aij]69 × 3,595, with aij = 1 if chaperone i and protein j interact and aij = 0 otherwise. The chaperones and substrates form two disjoint sets of nodes where interactions between substrate nodes are not allowed because the data reflect the interactions of chaperones with substrate proteins but not other possible interactions among the substrate proteins. The network is thus semi-multipartite, with 9,194 edges of CSIs and 332 edges of chaperone–chaperone interactions (fig. 1). Co-chaperones in our network were found to interact almost exclusively with chaperones.
FIG. 1.—
The network of CSIs. A graphic representation of the network with chaperones on the x axis (i = 1 … 69) and substrates on the y axis (j = 1 … 3,595). Cells in the matrix represent a protein–protein interaction between chaperone i and substrate j. The cells are colored by the module color if both substrate and chaperone are included in the module, and in gray otherwise. Cells of noninteracting proteins are colored in black. Hsp70 group includes the five ungrouped chaperones: Ssb1, Ssa1, Sse1, Ssa2, and Ssb2.
The CSI network includes five highly connected Hsp70 chaperones that are linked to almost all substrates in the network (Gong et al. 2009). The remaining 64 chaperones interact with fewer proteins, ranging between 2 and 732 substrates per chaperone. Some chaperones interact with a similar set of substrates, thereby forming communities within the network. We examined the community structure in the network by partitioning it into modules using the modularity optimization method (Newman 2006). For each possible bipartition of the network, a modularity function is defined as the observed number of edges within a community minus the expected number. Maximizing this modularity function using its leading eigenvector yields the modules within the network (Newman 2006). Each module is a community of nodes (chaperones and substrates), and each node is assigned to only one community allowing no multiple assignment of a protein to multiple modules.
The result uncovered ten modules that include a total of 64 chaperones and 2,691 substrates, along with 843 lesser (residual) modules that contain a single protein each. The network groups co-chaperones into modules based on their experimental interaction data with the chaperones (Gong et al. 2009). The modules furthermore group together chaperones that interact frequently with common substrates as well as those substrates. Five Hsp70 chaperones were not grouped into the ten main modules, forming five single-chaperone modules (Ssa1, Ssa2, Ssb1, Ssb2, and Sse1) (fig. 1). These chaperones are characterized by a promiscuous substrate binding and have many substrates in common (Gong et al. 2009). The remaining 838 singleton modules include proteins that interact solely with the five promiscuous chaperones. We designate the ten main modules by their most connected chaperone. The modules contain between 1 (Hsp70-Ssa3) and 14 (Small-Hsp42) chaperones. The number of substrates folded by each module ranges from 65 (CCT-Cct8) to 485 (AAA+-Hsp78) (supplementary table 1, Supplementary Material online).
The RAC-induced association of Hsp70 family chaperones and J-proteins (Hsp40 family) is clearly evident in the CSI network. For example, the Hsp70-Ssb1 chaperone interacts with 1,044 substrates in total. Of those, 585 (56%) are shared with Hsp40-Ydj1, 483 (46%) with Hsp70-Ssz1, 281 (27%) substrates are shared with Hsp40-Sis1, and 92 (9%) are shared with Hsp40-Zuo1 (Gong et al. 2009). Chaperones Ssb1, Zuo1, and Ssz1 are members of the yeast ribosomal chaperones triad that is anchored to the ribosome and interacts with nascent polypeptides (Gautschi et al. 2001; Conz et al. 2007). No in vivo interactions between Ssb1 and the Hsp40 chaperones Ydj1 or Sis1 have been verified experimentally. Nevertheless, in vitro studies showed that both Ydj1 or Sis1 interact with Ssb1 to determine its specificity for substrate polypeptides (Shorter and Lindquist 2008). The high frequency of common substrates among these chaperones in the Gong et al. (2009) data might indicate that they are associated also in vivo. Three modules (Small-Hsp42, Hsp90-Hsp82, and CCT-Cct8) contain only an Hsp40 chaperone lacking the obligatory partner from Hsp70 family. However, all substrates in these modules also interact with one or more of the five ungrouped promiscuous Hsp70 chaperones. Two modules, Hsp70-Ssa3 and Hsp70-Ssa4, include only an Hsp70 chaperone lacking an Hsp40 partner. Substrates in those two modules interact with various Hsp40 chaperones and with the Ydj1, which has no substrate specificity (Kampinga and Craig 2010), as the most common interactor. Two modules include members of both TriC and PFD chaperone families, whereas three modules include only a TriC chaperone and one module only a PFD chaperone (supplementary table 1, Supplementary Material online).
Members within the modules are not restricted to a certain cellular localization (supplementary fig. 1, Supplementary Material online). This result conforms with the high abundance of interactions between chaperones and substrates that are localized in different cell compartments as reported in various protein–protein interaction databases (70% in Gong et al. (2009) data used here, 66% in BioGrid [ver. 3.1.77], Stark et al. 2006, and 67% in Strings [ver. 8.3], Szklarczyk et al. 2011). This indicates that protein folding and function do not always occur in the same compartment. Module Hsp90-Hsc82 is, however, enriched with chaperones localized in the mitochondrion (5 of 9; supplementary table 1, Supplementary Material online). The module includes Hsp60 and Hsp10 that interact to fold proteins in the mitochondrion (Rospert et al. 1993). These two chaperones are homologous to the eubacterial GroEL/GroES chaperonin system (Gupta 1995). Furthermore, the Hsp70 (Ssc1) and Hsp40 (Mdj2) chaperones in this module are known to be localized in the mitochondrion (supplementary table 1, Supplementary Material online) (Huh et al. 2003). Notably, the Hsp90-Hsc82 module is lacking both PFD and TriC chaperones, which are homologous to archaeal chaperones (Hartl and Hayer-Hartl 2009). The chaperone repertoire of this module suggests that it is of mitochondrial origin, reflecting a functional eubacterial unit within the yeast proteome (Esser et al. 2004).
Module Expression and Biochemical Properties
Substrate expression level as measured by protein molecules per cell (Ghaemmaghami et al. 2003) is significantly different among the ten modules (table 1). Substrates in modules Hsp70-Ssa4, Hsp90-Hsp2, and Hsp70-Ssz1 are expressed in the lowest level. Substrates in modules AAA+-Hsp78 and CCT-Cct8 are highly abundant in the cell (fig. 2). Substrates that interact only with the promiscuous Hsp70 chaperones have a higher expression level than substrates within the modules (P = 1.35 × 10−58, using one-sided Kolmogorov–Smirnov). Yeast proteins that are missing from the CSI network have a significantly lower expression level than connected proteins (P = 2.8 × 10−62, using one-sided Kolmogorov–Smirnov). This suggests that those proteins might interact with chaperones but were so far not detected in surveys for chaperone interactors, possibly due to their low expression level. Chaperone expression level shows no significant differences across the ten modules (P = 0.051, using Kruskal–Wallis).
Table 1.
Comparison of Substrate Properties among the Modules
| Variable | As Isa | Random | Correlation with Expression Level in the Networkb | |
| Expression | Expression level | 2.22 × 10−16** | 0.62 | — |
| CAI | 2.38 × 10−06** | 0.37 | 0.54** | |
| Optimal codons | 1.18 × 10−05** | 0.76 | 0.53** | |
| Secondary structure | Alpha helix | 0.0067** | 0.08 | 0.02 |
| Coiled coils | 0.0256** | 0.4 | 0.21** | |
| Beta sheets | 0.0833 | 0.53 | 0.21** | |
| Physiochemical properties | Protein length | 4.13 × 10−09** | 0.94 | −0.17** |
| Hydrophobic amino acids | 0.2177 | 0.23 | 0.18** | |
| Negative amino acids | 0.0008** | 0.56 | 0.08** | |
| Positive amino acids | 0.5682 | 0.72 | −0.06** | |
| Polar amino acids | 0.0081** | 0.83 | −0.31** | |
| Aromaticity index | 0.0017** | 0.43 | −0.04** | |
| Gravy | 0.171 | 0.58 | 0.14** | |
| Amino acid frequencies | Alanine | 6.60 × 10−07** | 0.89 | 0.36** |
| Arginine | 0.3581 | 0.8 | −0.09** | |
| Asparagine | 0.0384* | 0.58 | −0.27** | |
| Aspartate | 4.71 × 10−05** | 0.08 | 0.03 | |
| Cysteine | 0.5354 | 0.23 | −0.09** | |
| Glutamine | 0.0064** | 0.87 | −0.08** | |
| Glutamate | 0.2669 | 0.97 | 0.09** | |
| Glycine | 0.0172** | 0.24 | 0.25** | |
| Histidine | 0.4528 | 0.07 | −0.10** | |
| Isoleucine | 0.0027** | 0.11 | −0.06** | |
| Leucine | 0.0031** | 0.47 | −0.08** | |
| Lysine | 0.4807 | 0.75 | 0.03** | |
| Methionine | 0.3369 | 0.61 | −0.08** | |
| Phenyl-alanine | 0.0012** | 0.48 | −0.04** | |
| Proline | 0.0074** | 0.43 | −0.07** | |
| Serine | 0.0417* | 0.07 | −0.29** | |
| Threonine | 0.4651 | 0.72 | −0.05** | |
| Tryptophan | 0.0612 | 0.48 | 0.03 | |
| Tyrosine | 0.0586 | 0.31 | 0.02 | |
| Valine | 0.0185** | 0.27 | 0.27** | |
| Evolutionary rate | Substitution rate | 2.15 × 10−06** | 0.36 | −0.42** |
| % Identical amino acids | 1.35 × 10−07** | 0.81 | 0.47** | |
| Substitutions per site | 2.58 × 10−07** | 0.75 | −0.46** |
Using Kruskal–Wallis test for equality of median ranks with the null hypothesis, H0: μmodule1 = μmodule2 = … = μmodule10.
Using Spearman rank correlation coefficient.
P value < 0.05.
P value < 0.05 using false discovery rate test for multiple comparisons.
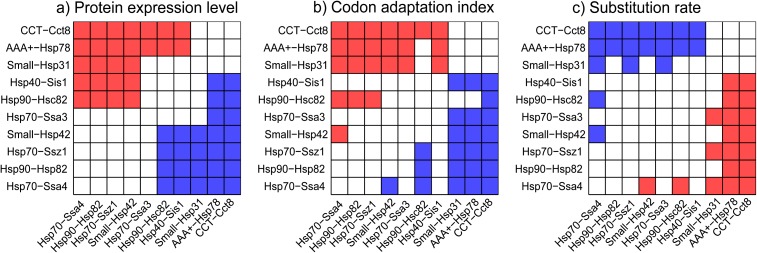
FIG. 2.—
Comparison of expression level (a), codon adaptation index (b), and relative amino acid substitution rates (c) among the modules. A matrix representation of post hoc multiple comparison results (α = 0.05, using Tukey test). Cell aij in the matrix is colored red if the corresponding variable module i > module j, blue if module i < module j, and white if no significant difference between the modules was found.
Protein expression and encoding by preferred codons are known to be positively correlated (Sharp and Li 1987). This correlation is apparent also in the CSI network, where substrate expression level is positively correlated with CAI (table 1). A comparison of codon usage among the modules—measured by the CAI (Sharp and Li 1987)—reveals significant difference across the modules (table 1), with modules Hsp70-Ssa4, Hsp90-Hsp82, and Hsp70-Ssz1 having the lowest CAI values and modules AAA+-Hsp78 and CCT-Cct8 having the highest CAIs (fig. 2). A randomization of protein module classification eliminates the significant CAI differences across the modules (table 1). A pairwise comparison of substrate expression level and CAI between the modules reveals that the correlation between these two properties is apparent at the modules level with highly expressed modules having high CAI values and vice versa (fig. 2).
Substrates in the ten modules vary substantially in their physiochemical properties. The secondary structure of substrates—measured by the proportion of alpha helixes and coiled coils—differs significantly among the modules (table 1). Substrates in module Hsp70-Ssz1 are enriched with coiled coil, whereas substrates in module Small-Hsp31 are enriched with alpha helixes (supplementary fig. 1, Supplementary Material online). No significant difference in the proportion of beta-sheet structures was found among the modules (table 1). The amino acid usage of most hydrophobic amino acids differs significantly between the modules (including Ala, Ile, Leu, Phe, and Val) as well as the usage of the negatively charged amino acid Asp (table 1; supplementary fig. 2, Supplementary Material online). Of the polar amino acids, only Gln usage is significantly different across the modules, with substrates in module Hsp70-Ssz1 encoding the highest Gln content. Phe is the only aromatic amino acid whose content varies across the modules (table 1). Substrates in the modules are significantly different in their aromaticity index with substrates in Small-Hsp31 encoding the lowest content of aromatic amino acids (table 1; supplementary fig. 2, Supplementary Material online). Substrate protein length is significantly different among the modules (table 1). The shortest substrates are found in modules AAA+-Hsp78, Small-Hsp31, and Hsp70-Ssa3 and the longest substrates in module Small-Hsp42 (supplementary fig. 2, Supplementary Material online). Randomizing the module classification of substrates eliminated the significant differences among the modules for all of the substrate properties mentioned above (table 1). Furthermore, none of these protein biochemical properties is correlated with protein expression level within the network (table 1).
No clear enrichment for substrate functional category, cellular localization, chromosomal location (supplementary fig. 1, Supplementary Material online), protein domain (supplementary table 2, Supplementary Material online), or sequence motif (supplementary table 3, Supplementary Material online) was found among the modules.
Module Evolutionary Dynamics
To test the impact of protein interaction with the chaperones on protein evolution, we compared substrate amino acid substitution rate among the modules. Phylogenetic trees were reconstructed from a multiple sequence alignment of S. cerevisiae substrate proteins with their positional ortholog from among 20 sequenced fungal genomes (Wapinski et al. 2007). A comparison of relative amino acid substitution rates among substrates in the ten modules revealed significant differences across the modules (table 1). Randomizing the module classification of substrates eliminates the differences in evolutionary rate among the modules (table 1). Ranking the modules from slow to fast by their relative substrate amino acid substitution rates shows that modules AAA+-Hsp78, CCT-Cct8, and Small-Hsp31 evolve with the slowest rates, whereas modules Hsp40-Sis1, Hsp70-Ssa3, and Hsp70-Ssa4 evolve with the highest rates. Substrates in the fastest module (Hsp70-Ssa3) evolve on average 15.6% faster than substrates in the slowest module (CCT-Cct8). Chaperones in the ten modules evolve in similar evolutionary rates (P = 0.12, using Kruskal–Wallis).
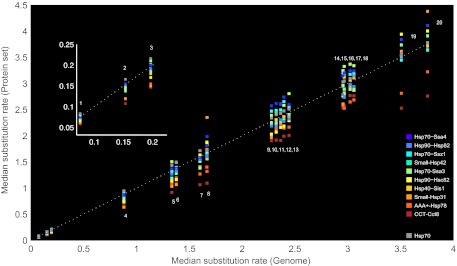
A comparison of module ranking at the species level reveals that module ranking is conserved during evolution (fig. 3). Substrates in the slowest and fastest modules maintain a similar ranking in almost all compared genomes. The conservation of intermediate module ranking varies to a larger extent. Module ranking is mostly diverged in species that are distantly related to yeast such as Debaryomyces hansenii and Candida parapsilosis. The intra-Saccharomyces comparison shows that substrates interacting exclusively with the five ungrouped Hsp70 chaperones evolve at the fastest rates; in more distantly related fungi, these proteins evolve at rates that are comparable to the fastest modules. Species where the module ranking is conserved (e.g., S. paradoxus and S. mikatae) are expected to have a CSI network that is similar to that of yeast (fig. 3).
FIG. 3.—
Evolutionary distances of yeast substrates in the ten modules compared with their positional ortholog in 20 fungal species. The x axis shows the variation of amino acid substitution rates within different fungal genomes in comparison with yeast. The y axis shows the rate variation among proteins in the different modules within the same genome. Module colors correspond to the ranking by substrate expression levels with highly expressed modules in red shades and lowly expressed modules in blue shades. Hsp70 group includes five ungrouped chaperones: Ssb1, Ssa1, Sse1, Ssa2, and Ssb2. Arabic numerals correspond to fungal species: 1) Saccharomyces paradoxus, 2) Saccharomyces mikatae, 3) Saccharomyces bayanus, 4) Saccharomyces castellii, 5) Kluyveromyces Lactis, 6) Ashbya gossypii, 7) Kluyveromyces waltii, 8) Lachancea kluyveri, 9) Candida glabrata, 10) Candida guilliermondii, 11) Candida albicans, 12) Candida tropicalis, 13) Lodderomyces elongosporus, 14) Yarrowia lipolytica, 15) Aspergillus nidulans, 16) Neurospora crassa, 17) Schizosaccharomyces pompe, 18) Schizosaccharomyces japonicus, 19) Debaryomyces hansenii, and 20) Candida parapsilosis.
Amino acid substitution rate and protein expression level are known to be inversely correlated at the genome level (Grantham et al. 1981; Pál et al. 2001, 2006; Krylov et al. 2003; Drummond et al. 2005). This correlation is observed also in the CSI network, where substrate expression level is negatively correlated with evolutionary rate (table 1). A comparison between module ranking by evolutionary rate with that of expression level shows that modules that are highly expressed are also the modules that evolve with the slowest substitution rates. Conversely, substrates in modules have the lowest expression levels and evolve in the highest substitution rates (fig. 2). A comparison of the relative amino acid substitution rates among the ten modules while adjusting for the variability in protein expression level reveals that the effect of expression level could not be rejected (P = 0.56, using analysis of covariance; Plinearity = 2.48 × 10−104; Pslopes homogeneity = 0.27).
Discussion
Chaperones are major hubs within the eukaryotic protein–protein interaction network (Gong et al. 2009). The multiplicity of interacting partners imposes a strong functional constraint on the evolution of hub proteins (Fraser et al. 2002). Moreover, multiple substrates of a certain chaperone evolve under the constraint to interact with that single chaperone. This can explain the similarity in biochemical properties and secondary structure elements among proteins that interact with common chaperones. The differences in substrate physiochemical properties across the modules are probably due to the different structures required for the interaction with the different chaperones.
Notably, the two Hsp70 paralogs Ssb1 and Ssb2 that differ in only two adjacent amino acids (C434V and A435S) were not grouped into the same module, rather each has its independent module. Interaction data of Gong et al. (2009) reveal that they have a different substrate repertoire. Ssb1 interacts with 2,756 (49%) of the substrates in our network; Ssb2 is associated with 1,064 (19%) substrates, and 899 (87%) of them are common with Ssb1 (Gong et al. 2009). The difference in the interaction regime of these two paralogs may be due to the difference in their expression level. Under standard conditions (Ghaemmaghami et al. 2003), Ssb1 is expressed in 170,000 copies in the cell, and Ssb2 is expressed in 104,000 copies. Hence by chance alone, it is more likely that potential Hsp70 substrates will interact more frequently with Ssb1 rather than Ssb2. Substrate specificity in Ssb2 interactions, if exists, is probably determined by chaperone and substrate coexpression or by their specificity to multiprotein complexes (e.g., the RAC complex).
Our analysis reveals that highly and lowly expressed proteins interact with different chaperones. Protein amino acid composition and secondary structure are known to impact the rate of protein folding and structural stability (Dobson 2003; Yang et al. 2010). Protein interaction with the chaperones lowers the energetic barrier for protein folding into the functional conformation (Hartl and Hayer-Hartl 2009). Thus, the evolution of protein–chaperone interaction is expected to depend upon the protein propensity to fold spontaneously. Chaperone-mediated folding ensures proper functional conformation, but it costs both time and energy. For example, protein folding by the GroEL/GroES chaperonin system in E. coli takes about 10 s and consumes seven adenosine triphosphate molecules (Horwich et al. 2009). It is therefore probably advantageous to have a subset of proteins that are less dependent upon chaperones for folding. If energetic efficiency is a selective constraint, this subset is likely to be defined by high expression levels and short response time. The spectrum of chaperone interaction with protein substrates can vary. For example, the GroEL/GroES chaperonin system in E. coli interacts with both casual and obligatory substrates. Casual interactors bind to GroEL in vivo but can also gain functional activity independent of GroEL in vitro (Kerner et al. 2005). Casual GroEL substrates have significantly higher expression level than obligatory substrates (Bogumil and Dagan 2010), consistent with the results presented here, which suggest that protein abundance within the cell largely determines the kind and mode of interaction with the chaperones for folding.
Protein expression level is known to be positively correlated with the usage of preferred codons (Sharp and Li 1987) and negatively correlated with evolutionary rate (Grantham et al. 1981; Pál et al. 2001, 2006; Krylov et al. 2003; Drummond et al. 2005). Current theories to explain these correlations evoke either poorly specified network properties of proteins (Fraser et al. 2002) or the specific effects of amino acid misincorporation during protein translation (Drummond et al. 2005; Drummond and Wilke 2008; Warnecke and Hurst 2010). Our results show that dividing the yeast proteins into modules by their chaperone interactions also captures the above correlations. The ten modules are significantly different in terms of each of these three properties, yet the 3-fold correlation prevents naming any one of the three measures as the leading causal effect of substrate–chaperon interactions. The question that remains is how protein interaction with the chaperones is related to protein expression level and codon adaptation. Considering the function of yeast chaperones, the majority of interactions in the CSI network correspond to chaperone-mediated protein folding. We suggest that the correlation between expression level and codon usage stems from the requirement for synchronization between protein translation and folding. Recently, it was shown that codon usage distribution along the protein sequence plays a role in protein translation speed (Cannarozzi et al. 2010; Tuller et al. 2010). Proteins that require chaperones have to be translated at a speed that fits the time required for chaperone recruitment (i.e., chaperone abundance and turnover rate), otherwise the protein will fold spontaneously into the wrong conformation, thereby forming aggregates that hinder the cell viability (Geiler-Samerotte et al. 2011). Proteins that can fold spontaneously into their functional conformation are free from that constraint and can be translated at a higher speed. However, with increasing translation speed, accuracy becomes more important, so that proteins that are translated at high speed should be more conserved (Drummond and Wilke 2008). The involvement of chaperones and folding in the yeast correlations between rates, codon bias, and expression introduces new perspectives on the issue.
Supplementary Material
Supplementary figures 1 and 2 and tables 1–3 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
This work was supported by the German Science Foundation (Deutsche Forschungsgemeinschaft) and a European Research Council grant NETWORKORIGINS to W. Martin. G.L. was supported by US National Library of Medicine grant LM010009-01. Computational support and infrastructure were provided by the Zentrum für Informations- und Medientechnologie (ZIM) at the Heinrich-Heine-University of Duesseldorf. We are thankful to W. Martin, M. J. Lercher, and E. Shakhnovich for their help in refining the manuscript.
References
- Akaike H. A new look at the statistical model identification. IEEE Trans Automat Contr. 1974;19:716–723. [Google Scholar]
- Ashburner M, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogumil D, Dagan T. Chaperonin-dependent accelerated substitution rates in prokaryotes. Genome Biol Evol. 2010;2:602–608. doi: 10.1093/gbe/evq044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannarozzi G, et al. A role for codon order in translation dynamics. Cell. 2010;141:355–367. doi: 10.1016/j.cell.2010.02.036. [DOI] [PubMed] [Google Scholar]
- Cherry JM, et al. Genetic and physical maps of Saccharomyces cerevisiae. Nature. 1997;387:67–73. [PMC free article] [PubMed] [Google Scholar]
- Conz C, et al. Functional characterization of the atypical Hsp70 subunit of yeast ribosome-associated complex. J Biol Chem. 2007;282:33977–33984. doi: 10.1074/jbc.M706737200. [DOI] [PubMed] [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D. ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics. 2011;27:1164–1175. doi: 10.1093/bioinformatics/btr088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobson CM. Protein folding and misfolding. Nature. 2003;426:884–890. doi: 10.1038/nature02261. [DOI] [PubMed] [Google Scholar]
- Drummond DA, Bloom JD, Adami C, Wilke CO, Arnold FH. Why highly expressed proteins evolve slowly. Proc Natl Acad Sci U S A. 2005;102:14338–14343. doi: 10.1073/pnas.0504070102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond DA, Wilke CO. Mistranslation-induced protein misfolding as a dominant constraint on coding-sequence evolution. Cell. 2008;134:341–352. doi: 10.1016/j.cell.2008.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis RJ. Proteins as molecular chaperones. Nature. 1987;328:378–379. doi: 10.1038/328378a0. [DOI] [PubMed] [Google Scholar]
- Esser C, et al. A genome phylogeny for mitochondria among alpha-proteobacteria and a predominantly eubacterial ancestry of yeast nuclear genes. Mol Biol Evol. 2004;21:1643–1660. doi: 10.1093/molbev/msh160. [DOI] [PubMed] [Google Scholar]
- Fares MA, Ruiz-Gonzalez MX, Moya A, Elena SF, Barrio E. GroEL buffers against deleterious mutations. Nature. 2002;417:398. doi: 10.1038/417398a. [DOI] [PubMed] [Google Scholar]
- Fraser HB, Hirsh AE, Steinmetz LM, Scharfe C, Feldman MW. Evolutionary rate in the protein interaction network. Science. 2002;296:750–752. doi: 10.1126/science.1068696. [DOI] [PubMed] [Google Scholar]
- Gautschi M, et al. RAC, a stable ribosome-associated complex in yeast formed by the DnaK-DnaJ homologs Ssz1p and zuotin. Proc Natl Acad Sci U S A. 2001;98:3762–3767. doi: 10.1073/pnas.071057198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiler-Samerotte KA, et al. Misfolded proteins impose a dosage-dependent fitness cost and trigger a cytosolic unfolded protein response in yeast. Proc Natl Acad Sci U S A. 2011;108:680–685. doi: 10.1073/pnas.1017570108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaemmaghami S, et al. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- Gong Y, et al. An atlas of chaperone-protein interactions in Saccharomyces cerevisiae: implications to protein folding pathways in the cell. Mol Syst Biol. 2009;5:275. doi: 10.1038/msb.2009.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grantham R, Gautier C, Gouy M, Jacobzone M, Mercier R. Codon catalog usage is a genome strategy modulated for gene expressivity. Nucleic Acids Res. 1981;9:43–74. doi: 10.1093/nar/9.1.213-b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Gupta RS. Evolution of the chaperonin families (Hsp60, Hsp10 and Tcp-1) of proteins and the origin of eukaryotic cells. Mol Microbiol. 1995;15:1–11. doi: 10.1111/j.1365-2958.1995.tb02216.x. [DOI] [PubMed] [Google Scholar]
- Hartl FU, Hayer-Hartl M. Converging concepts of protein folding in vitro and in vivo. Nat Struct Mol Biol. 2009;16:574–581. doi: 10.1038/nsmb.1591. [DOI] [PubMed] [Google Scholar]
- Horwich AL, Apetri AC, Fenton WA. The GroEL/GroES cis cavity as a passive anti-aggregation device. FEBS Lett. 2009;583:2654–2662. doi: 10.1016/j.febslet.2009.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh WK, et al. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- Jones DT. Protein secondary structure prediction based on position-specific scoring matrices. J Mol Biol. 1999;292:195–202. doi: 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]
- Kampinga HH, Craig EA. The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat Rev Mol Cell Biol. 2010;11:579–592. doi: 10.1038/nrm2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Kuma KI, Toh H, Miyata T. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005;33:511–518. doi: 10.1093/nar/gki198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerner MJ, et al. Proteome-wide analysis of chaperonin-dependent protein folding in Escherichia coli. Cell. 2005;122:209–220. doi: 10.1016/j.cell.2005.05.028. [DOI] [PubMed] [Google Scholar]
- Krylov DM, Wolf YI, Rogozin IB, Koonin EV. Gene loss, protein sequence divergence, gene dispensability, expression level, and interactivity are correlated in eukaryotic evolution. Genome Res. 2003;13:2229–2235. doi: 10.1101/gr.1589103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman MEJ. Finding community structure in networks using the eigenvectors of matrices. Phys Rev E. 2006;74:036104. doi: 10.1103/PhysRevE.74.036104. [DOI] [PubMed] [Google Scholar]
- Pál C, Papp B, Hurst LD. Highly expressed genes in yeast evolve slowly. Genetics. 2001;158:927–931. doi: 10.1093/genetics/158.2.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pál C, Papp B, Lercher MJ. An integrated view of protein evolution. Nat Rev Genet. 2006;7:337–348. doi: 10.1038/nrg1838. [DOI] [PubMed] [Google Scholar]
- Queitsch C, Sangster TA, Lindquist S. Hsp90 as a capacitor of phenotypic variation. Nature. 2002;417:618–623. doi: 10.1038/nature749. [DOI] [PubMed] [Google Scholar]
- Rospert S, et al. Identification and functional analysis of chaperonin 10, the groES homolog from yeast mitochondria. Proc Natl Acad Sci U S A. 1993;90:10967–10971. doi: 10.1073/pnas.90.23.10967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford SL. Between genotype and phenotype: protein chaperones and evolvability. Nat Rev Genet. 2003;4:264–274. doi: 10.1038/nrg1041. [DOI] [PubMed] [Google Scholar]
- Sharp PM, Li WH. The codon adaptation index—a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 1987;15:1281–1295. doi: 10.1093/nar/15.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shorter J, Lindquist S. Hsp104, Hsp70 and Hsp40 interplay regulates formation, growth and elimination of Sup35 prions. EMBO J. 2008;27:2712–2724. doi: 10.1038/emboj.2008.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark C, et al. Biogrid: a general repository for interaction datasets. Nucleic Acids Res. 2006;34:535–539. doi: 10.1093/nar/gkj109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D, et al. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 2011;39:561–568. doi: 10.1093/nar/gkq973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokuriki N, Tawfik DS. Chaperonin overexpression promotes genetic variation and enzyme evolution. Nature. 2009;459:668–673. doi: 10.1038/nature08009. [DOI] [PubMed] [Google Scholar]
- Tuller T, et al. An evolutionarily conserved mechanism for controlling the efficiency of protein translation. Cell. 2010;141:344–354. doi: 10.1016/j.cell.2010.03.031. [DOI] [PubMed] [Google Scholar]
- Voos W, Röttgers K. Molecular chaperones as essential mediators of mitochondrial biogenesis. Biochim Biophys Acta. 2003;1592:51–62. doi: 10.1016/s0167-4889(02)00264-1. [DOI] [PubMed] [Google Scholar]
- Wapinski I, Pfeffer A, Friedman N, Regev A. Natural history and evolutionary principles of gene duplication in fungi. Nature. 2007;449:54–61. doi: 10.1038/nature06107. [DOI] [PubMed] [Google Scholar]
- Warnecke T, Hurst LD. GroEL dependency affects codon usage—support for a critical role of misfolding in gene evolution. Mol Syst Biol. 2010;6:340. doi: 10.1038/msb.2009.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JR, Zhuang SM, Zhang J. Impact of translational error-induced and error-free misfolding on the rate of protein evolution. Mol Syst Biol. 2010;6:421. doi: 10.1038/msb.2010.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JC, Agashe VR, Siegers K, Hartl FU. Pathways of chaperone-mediated protein folding in the cytosol. Nat Rev Mol Cell Biol. 2004;5:781–791. doi: 10.1038/nrm1492. [DOI] [PubMed] [Google Scholar]