Abstract
The role of adaptation is a fundamental question in molecular evolution. Theory predicts that species with large effective population sizes should undergo a higher rate of adaptive evolution than species with low effective population sizes if adaptation is limited by the supply of mutations. Previous analyses have appeared to support this conjecture because estimates of the proportion of nonsynonymous substitutions fixed by adaptive evolution, α, tend to be higher in species with large Ne. However, α is a function of both the number of advantageous and effectively neutral substitutions, either of which might depend on Ne. Here, we investigate the relationship between Ne and ωa, the rate of adaptive evolution relative to the rate of neutral evolution, using nucleotide polymorphism and divergence data from 13 independent pairs of eukaryotic species. We find a highly significant positive correlation between ωa and Ne. We also find some evidence that the rate of adaptive evolution varies between groups of organisms for a given Ne. The correlation between ωa and Ne does not appear to be an artifact of demographic change or selection on synonymous codon use. Our results suggest that adaptation is to some extent limited by the supply of mutations and that at least some adaptation depends on newly occurring mutations rather than on standing genetic variation. Finally, we show that the proportion of nearly neutral nonadaptive substitutions declines with increasing Ne. The low rate of adaptive evolution and the high proportion of effectively neutral substitution in species with small Ne are expected to combine to make it difficult to detect adaptive molecular evolution in species with small Ne.
Keywords: adaptive evolution, effective population size, eukaryotes
Introduction
Population genetic theory predicts that the effective population size (Ne) of a species should be a major determinant of the rate of adaptive evolution if adaptive evolution is limited by the supply of new mutations. There are two reasons for this. First, the rate of adaptive evolution is expected to be proportional to Nes if , where s is the strength of selection. This is because the fixation probability of a new advantageous mutation is proportional to Nes/N, where N is the census population size, if and s is small (Kimura 1983), and the rate at which new advantageous mutations occur is Nu; hence, the rate of adaptive evolution is expected to be proportional to Nu × Nes/N = uNes. Second, in large populations, a higher proportion of mutations are expected to be effectively selected because a higher proportion are expected to have . Previous analyses have suggested that the proportion of adaptive substitutions (α) is correlated to the effective population size because there is evidence of widespread adaptive amino acid substitutions in species such as Drosophila, house mice, bacteria, and some plant species with large Ne (Bustamante et al. 2002; Smith and Eyre-Walker 2002; Sawyer et al. 2003; Bierne and Eyre-Walker 2004; Charlesworth and Eyre-Walker 2006; Haddrill et al. 2010; Ingvarsson 2010; Slotte et al. 2010; Strasburg et al. 2011), whereas there is little evidence in hominids and other plant species that appear to have small Ne (Chimpanzee Sequencing and Analysis Consortium 2005; Zhang and Li 2005; Boyko et al. 2008; Eyre-Walker and Keightley 2009; Gossmann et al. 2010). There are, however, some exceptions. Maize, for example, has a relatively large effective population size, approaching that of wild house mice, but shows little evidence of adaptive protein evolution (Gossmann et al. 2010), and the yeast Saccharomyces paradoxus, which presumably has a very large Ne, also shows little evidence of adaptive protein evolution (Liti et al. 2009). Furthermore, Drosophila simulans does not appear to have undergone more adaptive evolution than D. melanogaster, even though it is thought to have a larger Ne (Andolfatto et al. 2011).
However, the correlation between α and Ne might be misleading because α depends on the rate of effectively neutral and advantageous substitution, variation in either of which could be caused by Ne (Gossmann et al. 2010), that is, α = Dadaptive/(Dadaptive + Dnonadaptive) where Dadaptive and Dnonadaptive are the rates of adaptive and nonadaptive substitutions, respectively. There is evidence that the proportion of effectively neutral mutations is negatively correlated to Ne across many species (Popadin et al. 2007; Piganeau and Eyre-Walker 2009), so a positive correlation between α and Ne might be entirely explained by variation in the number of effectively neutral substitutions. As a consequence, it has been suggested that ωa, the rate of adaptive substitution relative to the rate of neutral evolution is a more appropriate measure of adaptive evolution for the purpose of comparison between genomic regions or species (Gossmann et al. 2010, see also Bierne and Eyre-Walker 2004; Obbard et al. 2009), that is, ωa = Dadaptive/Dneutral where Dneutral is the substitution rate at sites that evolve neutrally. Contrary to expectation, Gossmann et al. (2010) failed to find any evidence of a correlation between ωa and Ne in plants; but many of the plant species they considered appeared to have low Ne, and there may have been insufficient information from species with larger Ne to reveal a significant positive correlation. In contrast, Strasburg et al. (2011) have recently reported a significant positive correlation between ωa and Ne within sunflowers, including some species that have very large Ne. There are two interpretations of a positive correlation between ωa and Ne in sunflowers. First, the correlation could be due to a higher rate of adaptive substitution, or second, it could be due to an artifact of population size change (Strasburg et al. 2011). It has long been known that approaches to estimate adaptive evolution by methods related to the MK test are sensitive to changes in Ne, if there are slightly deleterious mutations (McDonald and Kreitman 1991; Eyre-Walker 2002; Eyre-Walker and Keightley 2009). For example, if the population has recently expanded, then ωa and α will tend to be overestimated because slightly deleterious mutations, which would have become fixed in the past when the population size was small, no longer segregate as polymorphisms. This bias might be a particular problem in the sunflower data set because each species was contrasted against a common outgroup species, so that each comparison shared much of its divergence with all other comparisons. Therefore, any differences in Ne between the species must have occurred since they split and may have caused a genuine or an artifactual increase in ωa. It is difficult to differentiate between these effects.
In contrast to the pattern in sunflowers, Jensen and Bachtrog (2011) recently estimated the rate adaptive evolution in D. pseudoobscura and D. miranda; they estimated that the two species probably had similar ancestral population sizes but that D. miranda had gone through a recent severe bottleneck. Despite this, the estimate of α along the two lineages was quite similar.
It is also evident that estimates of α or ωa and Ne are not independent because Ne is usually estimated from the neutral diversity, which is also used to estimate α or ωa. Sampling variation will therefore tend to induce a positive correlation between estimates of adaptive evolution and effective population size. This can be dealt with by randomly splitting the neutral sites into two halves, one of which is used to estimate Ne and the other to estimate the rate of adaptive evolution (Piganeau and Eyre-Walker 2009; Stoletzki and Eyre-Walker 2011). This correction is accurate whether or not the sites are linked (Piganeau and Eyre-Walker 2009).
Materials and Methods
Preparation of Data
Polymorphism data were retrieved from GenBank http://www.ncbi.nlm.nih.gov/Genbank or in case of Arabidopsis thaliana downloaded from http://walnut.usc.edu/2010. A summary of the analyzed data sets is shown in table 1. Phylogenetic trees for the plant and Drosophila species used in our analysis are given in supplementary figures S1 and S2 (Supplementary Material online), respectively (Drosophila 12 Genomes Consortium et al. 2007; Tang et al. 2008; Stevens 2010). Sequences were aligned using ClustalW using default parameter values (Thompson et al. 1994). Coding regions were assigned using protein-coding genomic data coordinates or, if given, derived from the information in the GenBank input files. An outgroup was assigned using the best Blast (Altschul et al. 1990) hit against the outgroup genome or, if included, taken from the GenBank Popset database (http://www.ncbi.nlm.nih.gov/popset). For all analyses, synonymous sites served as the neutral standard. Because some loci had been sampled in more individuals than others and other loci had missing data, we obtained the site frequency spectra (SFS) for each number of chromosomes for each species (e.g., we obtained the SFS for those sites with 4, 5, . . . etc. chromosomes separately). As a consequence, there was usually more than one SFS and its associated divergence data for each species. The estimation of the distribution of fitness effects (DFE), and ωa was done jointly using all available SFS and divergence data for a given species. Summary statistics, such as π, were calculated as weighted averages. The numbers of synonymous and nonsynonymous sites and substitutions were computed using the F3×4 model implemented in PAML (Yang 1997) in which codon frequencies are estimated from the nucleotide frequencies at the three codon positions.
Table 1.
Summary of Data Sets Used for the Analyses
| Species | Outgroup | Loci | Data Set |
| Drosophila melanogaster | Drosophila simulans | 373 | Shapiro et al. (2007) |
| Drosophila miranda | Drosophila affinis | 76 | Haddrill et al. (2010) |
| Drosophila pseudoobscura | Drosophila persimilis | 72 | Haddrill et al. (2010) |
| Homo sapiens | Macaca mulatta | 445 | EGP/PGAa |
| Mus musculus castaneus | Rattus norvegicus | 77 | Halligan et al. (2010) |
| Arabidopsis thaliana | Arabidopsis lyrata | 932 | Nordborg et al. (2005) |
| Capsella grandiflora | Neslia paniculata | 251 | Slotte et al. (2010) |
| Helianthus annuus | Lactuca sativa | 34 | Strasburg et al. (2011) |
| Populus tremula | Populus trichocarpa | 77 | Ingvarsson (2008) |
| Oryza rufipogon | Oryza spp. | 106 | Caicedo et al. (2007) |
| Schiedea globosa | Schiedea adamantis | 23 | Gossmann et al. (2010) |
| Zea mays | Sorghum bicolor | 437 | Wright et al. (2005) |
| Saccharomyces paradoxus | Saccharomyces cerevisiae | 98 | Tsai et al. (2008) |
EGP: http://egp.gs.washington.edu and PGA: http://pga.gs.washington.edu, August 2010.
It is important in this type of analysis to count the numbers of synonymous and nonsynonymous sites correctly and consistently across the divergence and polymorphism data. It is appropriate to use a “mutational opportunity” definition of a site (Bierne and Eyre-Walker 2003) since we are interested in the relative numbers of mutations that can potentially occur at synonymous and nonsynonymous sites. PAML provides estimates of the proportion of sites that are nonsynonymous (and hence also synonymous) from the divergence data, and these were used to calculate the number of nonsynonymous and synonymous sites for the polymorphism data.
Estimation of Ne and ωa
We assumed that synonymous sites were neutral, except when we estimated the strength of selection on synonymous mutations (see below). We estimated Ne from the level of nucleotide diversity, π, at synonymous sites and estimates of the rate of nucleotide mutation per generation, μ, from the literature, since
| (1) |
We estimated the mutation rate per generation in Populus tremula in the following manner. Tuskan et al. (2006) note that sequence divergence in putatively neutral sequences is approximately six times slower in P. tremula than in A. thaliana and that the average generation time for P. tremula is ≈15 years. We therefore estimated the mutation rate per generation in P. tremula by multiplying the mutation rate estimated in A. thaliana from mutation accumulation lines by 15/6 = 1.75 × 10−8.
The DFE and ωa, the rate of adaptive substitutions relative to the rate of synonymous substitutions (Gossmann et al. 2010), were estimated using a modified version of the method of Eyre-Walker and Keightley (2009). First, the DFE and demographic parameters of the population are simultaneously estimated from the SFS of nonsynonymous and synonymous sites using the method of Keightley and Eyre-Walker (2007). The DFE is then used to estimate the average fixation probability of mutations at nonsynonymous sites relative to that at neutral sites:
| (2) |
where S = 4Nes, s is the strength of selection, M(S) is the distribution of S as inferred by the method of Keightley and Eyre-Walker (2007) and
| (3) |
is the fixation probability of a new mutation relative to the fixation probability of a neutral mutation (Kimura 1983). The rate of adaptive nonsynonymous substitution relative to the rate of synonymous substitution, ωa, can then be estimated as
| (4) |
where dn and ds are the rates of nonsynonymous and synonymous substitution, respectively. The method of Eyre-Walker and Keightley (2009) does not take into account the fact that some substitutions between species are polymorphisms. This was taken into account in the following manner (Keightley and Eyre-Walker 2012). The Keightley and Eyre-Walker (2007) method estimates the DFE and demographic parameters by generating vectors representing the allele frequency distributions for synonymous and nonsynonymous sites by a transition matrix approach and using these to calculate the likelihood of the observed SFS. Let the density of mutations at i of 2N copies be vn(i) and vs(i) for nonsynonymous and synonymous sites, respectively, and let us assume that we have sampled a single sequence from each species to estimate the divergence. The contribution of polymorphisms to apparent divergence is therefore
| (5) |
for nonsynonymous sites, with an analogous expression for synonymous sites. The factor of two appears because polymorphism in both lineages contributes to apparent divergence, and we assume that the diversity is the same in the two lineages. We can now estimated ωa taking into account the contribution of polymorphism to divergence as
| (6) |
We also estimated ωa using a model in which there was negative selection upon synonymous mutations. We assume that all synonymous mutations are subject to the same strength of selection. Unfortunately, it is not possible to simultaneously estimate the demographic parameters and the strength of selection on synonymous mutations unless one includes information about which codons are preferred by selection (Zeng and Charlesworth 2009), and this is not known for most of the species in our analysis. We therefore infer the strength of selection at synonymous sites from the SFS using the transition matrix approach described in Keightley and Eyre-Walker (2007) assuming a constant population size. The strength of selection at synonymous sites allows us to calculate the probability of fixation of synonymous mutations fs and obtain a corrected estimate of ωa as
| (7) |
It is also necessary to adjust our estimate of Ne to take into account the action of natural selection at synonymous sites. This was performed in one of two ways, depending upon whether our estimate of the mutation rate was a direct estimate from a pedigree or mutation accumulation experiment, as in the Drosophila species, Arabidopsis, Capsella, Populus, and Saccharomyces, or indirectly from phylogenetic analysis, as in Mus, Helianthus, Oryza, Schieda, and Zea. Kimura (1969) showed that the nucleotide diversity at a site subject to recurrent mutation and semidominant selection, of strength s (positive s for advantageous mutations), relative to that at a neutral site is
| (8) |
For those species in which the mutation rate had been estimated directly, we corrected the estimate of Ne obtained from equation (1), by dividing it by H(S), where S is the strength of selection acting at synonymous sites; for those species in which the mutation rate came from a phylogenetic analysis, we corrected for selection at synonymous sites by multiplying the estimates by Q(S)/H(S). Synonymous codon bias was measured using the effective number of codons (ENC; Wright 1990) and ENC taking into account base composition bias (ENC′; Novembre 2002). To investigate whether the proportion of effectively neutral nonsynonymous mutations was correlated to Ne, we calculated a variant on the ψ statistic suggested by Piganeau and Eyre-Walker (2009):
| (9) |
where Pn and Ps are the numbers of nonsynonymous and synonymous polymorphisms, and Ln and Ls are the numbers of nonsynonymous and synonymous sites. ψ is expected to be less biased than Pn/Ps.
Creation of Independent Data Sets
Estimates of ωa and Ne are not independent because they both depend on neutral diversity, so sampling error will tend to induce a positive correlation between Ne and ωa. We avoided this problem by splitting the synonymous site data into two independent sets (which is similar to splitting the data set into odd and even codons as in Smith and Eyre-Walker 2002; Piganeau and Eyre-Walker 2009; Stoletzki and Eyre-Walker 2011) by generating a random multivariate hypergeometric variable as follows:
| (10) |
| (11) |
where Ls is the number of sites and P a vector consisting of the number of nonmutated sites and the site frequency spectrum so that ∑P = Ls. We use Ps1 and Ps2 to compute two corresponding independent variables Ne1 and ωa2. Note that Ne2 and ωa1 could be obtained in a similar manner, however, results were qualitatively comparable and we therefore only show results for Ne1 versus ωa2. The same strategy was used to investigate the relationship between ψ and Ne.
Results
To investigate the correlation between the rate of adaptive evolution and Ne, we compiled data from 13 phylogenetically independent pairs of species (table 1; supplementary figures S1 and S2, Supplementary Material online). We measured the rate of adaptive evolution using the statistic ωa, which is the rate of adaptive substitution at nonsynonymous sites relative to the rate of synonymous substitution, using a method that takes into account the contribution of slightly deleterious mutations to polymorphism and divergence (Eyre-Walker and Keightley 2009; Keightley and Eyre-Walker 2012). We estimated Ne by dividing the synonymous site nucleotide diversity by an estimate of the mutation rate per generation, taken from the literature. We also divided the synonymous sites into two groups when estimating ωa and Ne in order to ensure that the estimates were statistically independent. Estimates of ωa and Ne are given in table 2.
Table 2.
Summary of the Nucleotide Diversity for Silent Sites π, Mutation Rate per Generation μ from the Literature, Estimates of Effective Population Sizes Ne, ωa, ENC, and ENC′ for the 13 Analyzed Species
| Selection on Silent Sites |
||||||||||
| Species | π | μ × 109 | Ne | ωa | N2/N1 | ENC | ENC′ | 4Nes | Nea | ωaa |
| Drosophila melanogaster | 0.019 | 5.8 [1] | 822,351 | 0.03 | 2.31 | 53.56 | 54.42 | −0.0002 | 822,379 | 0.04 |
| Drosophila miranda | 0.008 | 5.8 [1] | 334,502 | −0.00 | 4.95 | 43.27 | 49.27 | −0.0002 | 334,513 | 0.01 |
| Drosophila pseudoobscura | 0.019 | 5.8 [1] | 798,607 | 0.27 | 4.5 | 43.28 | 48.62 | −0.0008 | 798,714 | −0.06 |
| Homo sapiens | 0.001 | 11 [2] | 20,974 | −0.04 | 4.09 | 53.39 | 54.61 | −1.2118 | 26,127 | 0.02 |
| Mus musculus castaneus | 0.008 | 3.4 [3] | 573,567 | 0.18 | 2.79 | 52.95 | 54.51 | −0.4946 | 483,026 | 0.31 |
| Arabidopsis thaliana | 0.007 | 7 [4] | 266,769 | −0.04 | 4.95 | 54.98 | 56.46 | −0.0016 | 266,840 | 0.03 |
| Capsella grandiflora | 0.018 | 7 [4] | 641,262 | 0.06 | 2.8 | 55.08 | 56.11 | −0.0186 | 643,257 | 0.04 |
| Helianthus annuus | 0.024 | 10 [5] | 593,436 | 0.11 | 4.5 | 57.23 | 58.92 | −0.2328 | 548,293 | 0.14 |
| Populus tremula | 0.011 | 17.4 [4,6] | 156,368 | 0.06 | 1.5 | 55.98 | 57.43 | −0.0002 | 156,373 | 0.08 |
| Oryza rufipogon | 0.005 | 10 [7] | 131,083 | −0.07 | 10 | 59.10 | 58.83 | −3.4624 | 28,643 | 0.06 |
| Schiedea globosa | 0.013 | 95 [8,9] | 34,075 | −0.12 | 4.5 | 56.58 | 57.62 | −0.001 | 34,054 | −0.14 |
| Zea mays | 0.019 | 10 [7] | 464,010 | −0.00 | 3.07 | 59.05 | 59.01 | −2.4864 | 168,117 | 0.03 |
| Saccharomyces paradoxus | 0.002 | 0.2 [10] | 256,2065 | −0.02 | 4.5 | 53.31 | 56.85 | −0.0002 | 256,2150 | −0.06 |
Note.—ωa was estimated under a simple demographic model assuming a step change of Ne (k = N2/N1), where the ratio of N2/N1 > 1 and <1 indicates recent population size expansion and contraction, respectively. Estimates of the strength of selection on synonymous sites 4Nes and corresponding corrected estimates of Ne and ωa. The strength of selection s on synonymous mutations was estimated assuming a constant population size. Literature sources for mutation rates: [1] Haag-Liautard et al. (2007); [2] Roach et al. (2010); [3] Keightley and Eyre-Walker (2000); [4] Ossowski et al. (2010); [5] Strasburg and Rieseberg (2008); [6] Tuskan et al. (2006); [7] Swigonová et al. (2004); [8] Filatov and Burke (2004); [9] Wallace et al. (2009); [10] Fay and Benavides (2005).
Corrected for the effect of selection on synonymous sites.
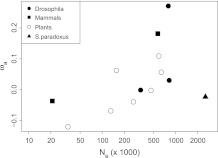
There is a nonsignificant positive correlation between ωa and Ne for the individual data points (Pearson's correlation r = 0.16, P = 0.61; fig. 1). However, there is also a positive correlation between the two variables for all groups for which we have two or more data points (Plants: r = 0.74, P = 0.056; Drosophilidae: r = 0.55, P = 0.63; Mammals: r = 1.00, P not given because there are just two data points), suggesting that differences between taxonomic groups may obscure a significant correlation within the groups. To investigate this further, we performed an analysis of covariance (ANCOVA), grouping organisms as mammals, plants, Drosophila, and fungi. In ANCOVA, a set of parallel lines are fitted to the data, one for each group. This enables a test of whether the common slope of these lines is significantly different from zero, and one can also investigate whether the groups differ in the dependent variable for a given value of the independent variable by testing whether the lines have different intercepts. Using ANCOVA, we find that ωa and Ne are significantly positively correlated (P = 0.017). Furthermore, there is significant variation between the intercepts (P = 0.044). There is also a positive correlation between ωa and log(Ne) (P = 0.018), although the difference between intercepts is no longer significant (P = 0.12). The results therefore suggest that ωa and Ne are positively correlated and that the level of adaptive evolution may vary between groups for a given Ne.
FIG. 1.—
The rates of adaptive evolution (ωa) versus the effective population size (Ne) for 13 species grouped into four phylogenetic sets. Details concerning the analyzed species can be found in table 1.
The correlation between ωa and Ne might be genuine, but it might also have arisen as an artifact, generated by changes in population size. For example, if species with large current Ne tend to have undergone population expansion and/or species with small Ne population size contraction, then a positive correlation between ωa and Ne would be induced because population size expansion leads to an overestimate of ωa and contraction to an underestimate if there are slightly deleterious mutations (Eyre-Walker 2002). We investigated whether changes in population size explain the correlation between ωa and Ne by taking advantage of the fact that the method we used to estimate ωa simultaneously fits a demographic model to the data. In this model, the population experiences a k-fold change in population size t generations in the past. The results of our analysis suggest that the correlation between the estimates of Ne and log(k) are weak and nonsignificant (Pearson: r = −0.41, P = 0.15; ANCOVA: slope P = 0.61) or between log(Ne) and log(k) (Pearson r = −0.15, P = 0.61; ANCOVA: slope P = 0.93); thus, there is no evidence that species with large current Ne have undergone recent expansion and/or that species with small current Ne have undergone recent contraction. We also find little evidence that ωa is correlated to log(k) (Pearson: r = 0.17, P = 0.57; ANCOVA: P = 0.97), implying that the correlation between ωa and Ne is not an artifact of changes in population size. It should be noted, however, that this test is not definitive because MK-based approaches are sensitive to differences in the Ne experienced by the polymorphism and the divergence data (McDonald and Kreitman 1991; Eyre-Walker 2002; Eyre-Walker and Keightley 2009). For example, a species might have experienced an expansion that predates the origin of the polymorphism data but is nevertheless recent in comparison with the overall divergence between the species being considered. In this case, there would be no evidence of expansion in the polymorphism data, but Ne for the polymorphism data would be greater than the average Ne during the divergence of the species. This would artifactually increase the estimate of ωa.
A second explanation for the correlation between ωa and Ne could be selection at synonymous sites. If the effectiveness of selection on synonymous sites increases with Ne, then this predicts a decrease in the level of synonymous divergence relative to polymorphism, leading to overestimation of adaptive nonsynonymous evolution. Although we might expect the effectiveness of selection on synonymous sites to increase with Ne, the evidence is mixed. Selection appears to be more effective on synonymous codon bias in Drosophila simulans than D. melanogaster (Akashi 1996; McVean and Vieira 2001), and Ne is thought to be larger in the former species (Aquadro et al. 1988; Akashi 1996). However, in mammals, selection appears to be more effective on synonymous sites in hominids than rodents (Eory et al. 2010), yet Ne is substantially larger in wild mice than hominids (Eyre-Walker 2002; Halligan et al. 2010). Furthermore, selection on synonymous codon use appears to have little effect on estimates of α in D. pseudoobscura, D. miranda, and D. affinis (Haddrill et al. 2010).
To investigate whether the correlation between ωa and Ne might be due to selection on synonymous sites, we performed two analyses. First, we investigated whether ωa and our estimate of Ne were correlated to codon usage bias, as measured by the ENC and ENC taking into account base composition (ENC′). ωa is negatively correlated to ENC and ENC′, as expected if selection on synonymous codon use was causing an artifactual increase in ωa, but in neither case was the correlation significant (ENC vs. ωa: r = −0.481, P = 0.096; ANCOVA slope: P = 0.40; ENC′ vs. ωa: r = −0.495, P = 0.085; ANCOVA slope: P = 0.430). Furthermore, the correlation between Ne or log(Ne) and ENC or ENC′ are nonsignificant (ENC vs. Ne: r = −0.15, P = 0.61; ANCOVA slope: P = 0.61; ENC′ vs. Ne: r = −0.04, P = 0.89; ANCOVA slope: P = 0.66; ENC vs. log(Ne): r = −0.23, P = 0.44; ANCOVA slope: P = 0.87; ENC′ vs. log(Ne): r = −0.15, P = 0.61; ANCOVA slope: P = 0.86). Hence, there is little evidence that the correlation between ωa and Ne is a consequence codon usage bias.
In the second analysis, we estimated ωa while simultaneously estimating the strength of negative selection on synonymous sites. We also corrected our estimate of the effective population size for the effect of selection on synonymous sites. Estimates of Ne, ωa and the strength of selection on synonymous mutations are given in table 2. The results of this analysis show some evidence of selection on synonymous sites in four species: Oryza rufipogon, Zea mays, human, and mouse. There is independent evidence of selection in Homo sapiens (Iida and Akashi 2000; Hellmann et al. 2003; Chamary et al. 2006; Keightley et al. 2011) and mouse (Chamary and Hurst 2004; Gaffney and Keightley 2005; Keightley et al. 2011) but also in P. tremula (Ingvarsson 2010), D. melanogaster (Zeng and Charlesworth 2009), D. pseudoobscura (Akashi and Schaeffer 1997; Haddrill et al. 2011), and D. miranda (Bartolomé et al. 2005; Haddrill et al. 2011) for which we do not find evidence of selection at synonymous sites. The failure to detect selection on synonymous sites may be due to the strength of the selection being weak, and furthermore, we have assumed a model with constant population size. This was necessary because it is not possible to simultaneously fit a model that allows demographic change and selection on synonymous codon use in the absence of detailed information about codon preferences (Zeng and Charlesworth 2010). Correcting for selection on synonymous sites, we find that the correlation between ωa and Ne is positive but not significant, whereas the correlation between ωa and log(Ne) is positive and significant with ANCOVA (slope P = 0.028, intercept P = 0.032). Although not conclusive, these results suggest that the correlation between ωa and Ne is not due to selection on synonymous codon use.
A third possible explanation for the correlation between ωa and Ne is biased gene conversion (BGC). Like selection upon synonymous codon use, BGC can elevate the ratio of polymorphism to divergence relative to neutral expectations. However, it is less clear that this will affect synonymous sites preferentially.
We might expect that just as the number of adaptive substitutions increases with Ne, the number of effectively neutral substitutions will decline. We estimated the number of effectively neutral substitutions as , and found that is significantly negatively correlated to Ne (r = −0.24, P = 0.43; ANCOVA slope P = 0.05; intercept P = 0.04) and log(Ne) (r = −0.53, P = 0.06; ANCOVA slope P = 0.14; intercept P = 0.23). The slopes of the regression lines, from the ANCOVA, between and Ne are similar in magnitude to those between ωa and Ne (−2.4 × 10−8 vs. 2.5 × 10−8). We also investigated whether aspects of the DFE of deleterious mutations, as estimated from the polymorphism data, are correlated to Ne. We find a significant negative correlation between ψ and Ne with ANCOVA controlling for the nonindependence between these variables (Pearson r = −0.24, P = 0.42; ANCOVA slope P = 0.014; intercepts P = 0.006) and between ψ and log(Ne) with Pearson (Pearson r = −0.64, P = 0.018; ANCOVA slope P = 0.016; intercepts P = 0.041), but correlations between the shape parameter of the DFE and the mean value of Nes and Ne are nonsignificant. The lack of a significant correlation between mean Nes and Ne could be a consequence of the low precision of estimates mean Nes (Keightley and Eyre-Walker 2007).
Discussion
We have presented evidence that the rate of adaptive protein evolution is positively correlated to Ne. We have shown that it is unlikely that this is due to recent demographic changes or selection on synonymous sites. Such a result is not unexpected. If the rate of adaptive evolution is limited by the supply of new mutations, then species with larger Ne are expected to undergo more adaptive evolution than species with small Ne because a greater number of advantageous mutations appear in the population and a higher proportion of these mutations are effectively selected.
The positive correlation between ωa and Ne is consistent with a model in which the rate of adaptive evolution is limited by the supply of new mutations. The correlation seems less consistent with a model in which adaptation comes from standing genetic variation (Pritchard et al. 2010; Pritchard and Rienzo 2010) for two reasons. First, although the level of advantageous, neutral, and slightly deleterious genetic variation is expected to be correlated to Ne, this correlation appears to be weak; levels of diversity, at least in mammalian mitochondrial DNA (mtDNA), are poorly correlated to effective population size (Piganeau and Eyre-Walker 2009). This is probably due to a negative correlation between the rate of mutation per generation and the effective population size (Lynch 2007; Piganeau and Eyre-Walker 2009). Second, the level of diversity of strongly deleterious mutations is expected to be either independent of the effective population size or negatively correlated to it, since species with long generation times, and small effective population size, appear to have higher rates of mutation per generation (Keightley and Eyre-Walker 2000; Piganeau and Eyre-Walker 2009).
We have shown that species with large Ne undergo more adaptive substitutions than species with small Ne. However, this does not necessarily mean that these species adapt faster, though this is likely. This is because the total rate of adaptive evolution is a product of the number of adaptive substitutions and the effects of those substitutions. It is possible that species with large Ne undergo more adaptive substitutions but that these are smaller in magnitude. We have also not considered adaptive evolution outside of protein-coding genes.
The positive correlation between the rate of adaptive evolution and Ne implies that detecting the signature of adaptive evolution using MK approaches is likely to be difficult in species with small Ne because they are expected to have undergone low levels of adaptive evolution. Furthermore, they are likely to have a higher proportion of effectively neutral mutations, which tends to obscure the signature of adaptive evolution. For example, assume that we have two species with the same number of synonymous polymorphisms (20) and substitutions (100) in a sample of genes. Assume that the two species have undergone the same number of adaptive nonsynonymous substitutions (15) but that species A has experienced no neutral mutations, whereas species B has undergone as many effectively neutral nonsynonymous mutations as synonymous mutations. Under the assumption that adaptive mutations contribute little to polymorphism the MK tables for the two species would be as given in table 3. It is evident that adaptive evolution would be detected in species A using a standard MK test (i.e., a χ2 test of independence), but not in species B, because although both species have undergone the same amount of adaptive evolution, this is obscured by the large number of effectively neutral substitutions in species B. The fact that large numbers of effectively neutral substitutions obscure the signature of adaptive evolution means that it will be more difficult to detect adaptive evolution in poorly conserved regions of the genome, such as regulatory sequences.
Table 3.
Power to Detect Adaptive Changes in Species with Different Effective Population Sizes
| Nonsynonymous Sites |
||||||
| Adaptive | Effectively Neutral | Synonymous Sites | α (%) | ωa (%) | MK Test P Value | |
| Species A (large Ne) | ||||||
| Polymorphisms | n.a. | 0 | 20 | |||
| Substitutions | 15 | 0 | 100 | |||
| 100 | 15 | 0.024 | ||||
| Species B (low Ne) | ||||||
| Polymorphisms | n.a. | 20 | 20 | |||
| Substitutions | 15 | 100 | 100 | |||
| 13 | 15 | 0.685 | ||||
Note.—Comparison between two hypothetical species (A and B) that have the same number of adaptive changes but different effective population sizes illustrated by a difference in the number of effectively neutral nonsynonymous sites. n.a., not applicable.
We have found some evidence that the rate of adaptive evolution varies between groups of organisms for a given Ne. In particular, it is striking that the fungus S. paradoxus has the largest Ne among the species we have considered, but shows no evidence of adaptive evolution. If we remove S. paradoxus from the ANCOVA, we find no evidence that the rate of adaptive evolution differs between groups (ANCOVA intercepts P = 0.47), although ωa is correlated to Ne (ANCOVA slope P = 0.017). It is possible that S. paradoxus has a low rate of adaptive evolution, despite its large Ne, because it is largely asexual (Tsai et al. 2008). Consistent with this, we note that there is a negative correlation between dn/ds and some measure of effective population in a number of nonrecombining genetic systems. In mammalian mtDNA, dn/ds is correlated to body size (Popadin et al. 2007), which is believed to be correlated to Ne, and in both mammals and birds, the largely nonrecombining Y and W chromosomes, which are believed to have lower Ne than the autosomes, have higher dn/ds values (Wyckoff et al. 2002; Berlin and Ellegren 2006). In contrast, we find no evidence of a significant correlation between dn/ds and Ne in our analysis (r = −0.37, P = 0.21; ANCOVA slope P = 0.34). This might be due to our small sample size, but it also may reflect a difference between recombining and nonrecombining loci. In our analysis, we find that the rate of adaptive substitution increases with Ne at a similar rate to the rate at which the effectively neutral substitutions decreases; this leaves the dn/ds uncorrelated to Ne. It might be that rates of adaptive evolution are lower in nonrecombining systems, and hence, the decline in the number of effectively neutral substitutions dominates the relationship between dn/ds and Ne, and species such as S. paradoxus undergo little adaptive evolution.
Supplementary Material
Supplementary figures S1 and S2 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
The authors are grateful to several referees for their comments. T.I.G. was funded by the John Maynard Smith studentship, and P.D.K. acknowledges support from the Wellcome Trust and the Biotechnology and Biological Sciences Research Council.
References
- Akashi H. Molecular evolution between Drosophila melanogaster and D. simulans: reduced codon bias, faster rates of amino acid substitution, and larger proteins in D. melanogaster. Genetics. 1996;144(3):1297–1307. doi: 10.1093/genetics/144.3.1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akashi H, Schaeffer SW. Natural selection and the frequency distributions of “silent” DNA polymorphism in Drosophila. Genetics. 1997;146(1):295–307. doi: 10.1093/genetics/146.1.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Andolfatto P, Wong KM, Bachtrog D. Effective population size and the efficacy of selection on the X chromosomes of two closely related Drosophila species. Genome Biol Evol. 2011;3:114–128. doi: 10.1093/gbe/evq086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aquadro CF, Lado KM, Noon WA. The rosy region of Drosophila melanogaster and Drosophila simulans. I. Contrasting levels of naturally occurring DNA restriction map variation and divergence. Genetics. 1988;119(4):875–888. doi: 10.1093/genetics/119.4.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartolomé C, Maside X, Yi S, Grant AL, Charlesworth B. Patterns of selection on synonymous and nonsynonymous variants in Drosophila miranda. Genetics. 2005;169(3):1495–1507. doi: 10.1534/genetics.104.033068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berlin S, Ellegren H. Fast accumulation of nonsynonymous mutations on the female-specific W chromosome in birds. J Mol Evol. 2006;62(1):66–72. doi: 10.1007/s00239-005-0067-6. [DOI] [PubMed] [Google Scholar]
- Bierne N, Eyre-Walker A. The problem of counting sites in the estimation of the synonymous and nonsynonymous substitution rates: implications for the correlation between the synonymous substitution rate and codon usage bias. Genetics. 2003;165(3):1587–1597. doi: 10.1093/genetics/165.3.1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierne N, Eyre-Walker A. The genomic rate of adaptive amino acid substitution in Drosophila. Mol Biol Evol. 2004;21(7):1350–1360. doi: 10.1093/molbev/msh134. [DOI] [PubMed] [Google Scholar]
- Boyko AR, et al. Assessing the evolutionary impact of amino acid mutations in the human genome. PLoS Genet. 2008;4(5):e1000083. doi: 10.1371/journal.pgen.1000083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustamante CD, et al. The cost of inbreeding in Arabidopsis. Nature. 2002;416(6880):531–534. doi: 10.1038/416531a. [DOI] [PubMed] [Google Scholar]
- Caicedo AL, et al. Genome-wide patterns of nucleotide polymorphism in domesticated rice. PLoS Genet. 2007;3(9):1745–1756. doi: 10.1371/journal.pgen.0030163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamary JV, Hurst LD. Similar rates but different modes of sequence evolution in introns and at exonic silent sites in rodents: evidence for selectively driven codon usage. Mol Biol Evol. 2004;21(6):1014–1023. doi: 10.1093/molbev/msh087. [DOI] [PubMed] [Google Scholar]
- Chamary JV, Parmley JL, Hurst LD. Hearing silence: non-neutral evolution at synonymous sites in mammals. Nat Rev Genet. 2006;7(2):98–108. doi: 10.1038/nrg1770. [DOI] [PubMed] [Google Scholar]
- Charlesworth J, Eyre-Walker A. The rate of adaptive evolution in enteric bacteria. Mol Biol Evol. 2006;23(7):1348–1356. doi: 10.1093/molbev/msk025. [DOI] [PubMed] [Google Scholar]
- Chimpanzee Sequencing and Analysis Consortium. Initial sequence of the chimpanzee genome and comparison with the human genome. Nature. 2005;437(7055):69–87. doi: 10.1038/nature04072. [DOI] [PubMed] [Google Scholar]
- Drosophila 12 Genomes Consortium, et al. Evolution of genes and genomes on the Drosophila phylogeny. Nature. 2007;450(7167):203–218. doi: 10.1038/nature06341. [DOI] [PubMed] [Google Scholar]
- Eory L, Halligan DL, Keightley PD. Distributions of selectively constrained sites and deleterious mutation rates in the hominid and murid genomes. Mol Biol Evol. 2010;27(1):177–192. doi: 10.1093/molbev/msp219. [DOI] [PubMed] [Google Scholar]
- Eyre-Walker A. Changing effective population size and the McDonald-Kreitman test. Genetics. 2002;162(4):2017–2024. doi: 10.1093/genetics/162.4.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyre-Walker A, Keightley PD. Estimating the rate of adaptive molecular evolution in the presence of slightly deleterious mutations and population size change. Mol Biol Evol. 2009;26(9):2097–2108. doi: 10.1093/molbev/msp119. [DOI] [PubMed] [Google Scholar]
- Fay JC, Benavides JA. Evidence for domesticated and wild populations of Saccharomyces cerevisiae. PLoS Genet. 2005;1(1):66–71. doi: 10.1371/journal.pgen.0010005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filatov DA, Burke S. DNA diversity in Hawaiian endemic plant Schiedea globosa. Heredity. 2004;92(5):452–458. doi: 10.1038/sj.hdy.6800440. [DOI] [PubMed] [Google Scholar]
- Gaffney DJ, Keightley PD. The scale of mutational variation in the murid genome. Genome Res. 2005;15(8):1086–1094. doi: 10.1101/gr.3895005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gossmann TI, et al. Genome wide analyses reveal little evidence for adaptive evolution in many plant species. Mol Biol Evol. 2010;27(8):1822–1832. doi: 10.1093/molbev/msq079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haag-Liautard C, et al. Direct estimation of per nucleotide and genomic deleterious mutation rates in Drosophila. Nature. 2007;445(7123):82–85. doi: 10.1038/nature05388. [DOI] [PubMed] [Google Scholar]
- Haddrill PR, Loewe L, Charlesworth B. Estimating the parameters of selection on nonsynonymous mutations in Drosophila pseudoobscura and D. miranda. Genetics. 2010;185(4):1381–1396. doi: 10.1534/genetics.110.117614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haddrill PR, Zeng K, Charlesworth B. Determinants of synonymous and nonsynonymous variability in three species of Drosophila. Mol Biol Evol. 2011;28(5):1731–1743. doi: 10.1093/molbev/msq354. [DOI] [PubMed] [Google Scholar]
- Halligan DL, Oliver F, Eyre-Walker A, Harr B, Keightley PD. Evidence for pervasive adaptive protein evolution in wild mice. PLoS Genet. 2010;6(1):e1000825. doi: 10.1371/journal.pgen.1000825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellmann I, Ebersberger I, Ptak SE, Pääbo S, Przeworski M. A neutral explanation for the correlation of diversity with recombination rates in humans. Am J Hum Genet. 2003;72(6):1527–1535. doi: 10.1086/375657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iida K, Akashi H. A test of translational selection at ‘silent’ sites in the human genome: base composition comparisons in alternatively spliced genes. Gene. 2000;261(1):93–105. doi: 10.1016/s0378-1119(00)00482-0. [DOI] [PubMed] [Google Scholar]
- Ingvarsson PK. Multilocus patterns of nucleotide polymorphism and the demographic history of Populus tremula. Genetics. 2008;180(1):329–340. doi: 10.1534/genetics.108.090431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingvarsson PK. Natural selection on synonymous and nonsynonymous mutations shapes patterns of polymorphism in Populus tremula. Mol Biol Evol. 2010;27(3):650–660. doi: 10.1093/molbev/msp255. [DOI] [PubMed] [Google Scholar]
- Jensen JD, Bachtrog D. Characterizing the influence of effective population size on the rate of adaptation: Gillespie’s Darwin domain. Genome Biol Evol. 2011;3:687–701. doi: 10.1093/gbe/evr063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keightley PD, Eöry L, Halligan DL, Kirkpatrick M. Inference of mutation parameters and selective constraint in mammalian coding sequences by approximate Bayesian computation. Genetics. 2011;187(4):1153–1161. doi: 10.1534/genetics.110.124073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keightley PD, Eyre-Walker A. Deleterious mutations and the evolution of sex. Science. 2000;290(5490):331–333. doi: 10.1126/science.290.5490.331. [DOI] [PubMed] [Google Scholar]
- Keightley PD, Eyre-Walker A. Joint inference of the distribution of fitness effects of deleterious mutations and population demography based on nucleotide polymorphism frequencies. Genetics. 2007;177(4):2251–2261. doi: 10.1534/genetics.107.080663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keightley PD, Eyre-Walker A. Estimating the rate of adaptive molecular evolution when the evolutionary divergence between species is small. J Mol Evol. 2012;74:61–68. doi: 10.1007/s00239-012-9488-1. [DOI] [PubMed] [Google Scholar]
- Kimura M. The number of heterozygous nucleotide sites maintained in a finite population due to steady flux of mutations. Genetics. 1969;61(4):893–903. doi: 10.1093/genetics/61.4.893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M. The neutral theory of molecular evolution. New York: Cambridge University Press; 1983. [Google Scholar]
- Liti G, et al. Population genomics of domestic and wild yeasts. Nature. 2009;458(7236):337–341. doi: 10.1038/nature07743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M. The origins of genome architecture. Vol. 98. Sunderland (MA): Sinauer Associates; 2007. [Google Scholar]
- McDonald JH, Kreitman M. Adaptive protein evolution at the Adh locus in Drosophila. Nature. 1991;351(6328):652–654. doi: 10.1038/351652a0. [DOI] [PubMed] [Google Scholar]
- McVean GA, Vieira J. Inferring parameters of mutation, selection and demography from patterns of synonymous site evolution in Drosophila. Genetics. 2001;157(1):245–257. doi: 10.1093/genetics/157.1.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordborg M, et al. The pattern of polymorphism in Arabidopsis thaliana. PLoS Biol. 2005;3(7):e196. doi: 10.1371/journal.pbio.0030196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novembre JA. Accounting for background nucleotide composition when measuring codon usage bias. Mol Biol Evol. 2002;19(8):1390–1394. doi: 10.1093/oxfordjournals.molbev.a004201. [DOI] [PubMed] [Google Scholar]
- Obbard DJ, Welch JJ, Kim KW, Jiggins FM. Quantifying adaptive evolution in the Drosophila immune system. PLoS Genet. 2009;5(10):e1000698. doi: 10.1371/journal.pgen.1000698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossowski S, et al. The rate and molecular spectrum of spontaneous mutations in Arabidopsis thaliana. Science. 2010;327(5961):92–94. doi: 10.1126/science.1180677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piganeau G, Eyre-Walker A. Evidence for variation in the effective population size of animal mitochondrial DNA. PLoS One. 2009;4(2):e4396. doi: 10.1371/journal.pone.0004396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popadin K, Polishchuk LV, Mamirova L, Knorre D, Gunbin K. Accumulation of slightly deleterious mutations in mitochondrial protein-coding genes of large versus small mammals. Proc Natl Acad Sci U S A. 2007;104(33):13390–13395. doi: 10.1073/pnas.0701256104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard JK, Pickrell JK, Coop G. The genetics of human adaptation: hard sweeps, soft sweeps, and polygenic adaptation. Curr Biol. 2010;20(4):R208–R215. doi: 10.1016/j.cub.2009.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard JK, Rienzo AD. Adaptation—not by sweeps alone. Nat Rev Genet. 2010;11(10):665–667. doi: 10.1038/nrg2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roach JC, et al. Analysis of genetic inheritance in a family quartet by whole-genome sequencing. Science. 2010;328(5978):636–639. doi: 10.1126/science.1186802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyer SA, Kulathinal RJ, Bustamante CD, Hartl DL. Bayesian analysis suggests that most amino acid replacements in Drosophila are driven by positive selection. J Mol Evol. 2003;57(Suppl 1):S154–S164. doi: 10.1007/s00239-003-0022-3. [DOI] [PubMed] [Google Scholar]
- Shapiro JA, et al. Adaptive genic evolution in the Drosophila genomes. Proc Natl Acad Sci U S A. 2007;104(7):2271–2276. doi: 10.1073/pnas.0610385104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotte T, Foxe JP, Hazzouri KM, Wright SI. Genome-wide evidence for efficient positive and purifying selection in Capsella grandiflora, a plant species with a large effective population size. Mol Biol Evol. 2010;27(8):1813–1821. doi: 10.1093/molbev/msq062. [DOI] [PubMed] [Google Scholar]
- Smith NGC, Eyre-Walker A. Adaptive protein evolution in Drosophila. Nature. 2002;415(6875):1022–1024. doi: 10.1038/4151022a. [DOI] [PubMed] [Google Scholar]
- Stevens PF. Angiosperm phylogeny website. 2010. Version 9. [cited 2011 October]. Available from: http://www.mobot.org/mobot/research/apweb/ [Google Scholar]
- Stoletzki N, Eyre-Walker A. Estimation of the neutrality index. Mol Biol Evol. 2011;28(1):63–70. doi: 10.1093/molbev/msq249. [DOI] [PubMed] [Google Scholar]
- Strasburg JL, Rieseberg LH. Molecular demographic history of the annual sunflowers Helianthus annuus and H. petiolaris—large effective population sizes and rates of long-term gene flow. Evolution. 2008;62(8):1936–1950. doi: 10.1111/j.1558-5646.2008.00415.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasburg JL, et al. Effective population size is positively correlated with levels of adaptive divergence among annual sunflowers. Mol Biol Evol. 2011;28:1569–1580. doi: 10.1093/molbev/msq270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swigonová Z, et al. Close split of sorghum and maize genome progenitors. Genome Res. 2004;14(10A):1916–1923. doi: 10.1101/gr.2332504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H, et al. Synteny and collinearity in plant genomes. Science. 2008;320(5875):486–488. doi: 10.1126/science.1153917. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22(22):4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai IJ, Bensasson D, Burt A, Koufopanou V. Population genomics of the wild yeast Saccharomyces paradoxus: quantifying the life cycle. Proc Natl Acad Sci U S A. 2008;105(12):4957–4962. doi: 10.1073/pnas.0707314105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuskan GA, et al. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray) Science. 2006;313(5793):1596–1604. doi: 10.1126/science.1128691. [DOI] [PubMed] [Google Scholar]
- Wallace LE, Weller SG, Wagner WL, Sakai AK, Nepokroeff M. Phylogeographic patterns and demographic history of Schiedea globosa (Caryophyllaceae) on the Hawaiian Islands. Am J Bot. 2009;96(5):958–967. doi: 10.3732/ajb.0800243. [DOI] [PubMed] [Google Scholar]
- Wright F. The ‘effective number of codons’ used in a gene. Gene. 1990;87(1):23–29. doi: 10.1016/0378-1119(90)90491-9. [DOI] [PubMed] [Google Scholar]
- Wright SI, et al. The effects of artificial selection on the maize genome. Science. 2005;308(5726):1310–1314. doi: 10.1126/science.1107891. [DOI] [PubMed] [Google Scholar]
- Wyckoff GJ, Li J, Wu CI. Molecular evolution of functional genes on the mammalian Y chromosome. Mol Biol Evol. 2002;19(9):1633–1636. doi: 10.1093/oxfordjournals.molbev.a004226. [DOI] [PubMed] [Google Scholar]
- Yang Z. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput Appl Biosci. 1997;13(5):555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- Zeng K, Charlesworth B. Estimating selection intensity on synonymous codon usage in a nonequilibrium population. Genetics. 2009;183(2):651–662. doi: 10.1534/genetics.109.101782. , 1SI–23SI. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng K, Charlesworth B. Studying patterns of recent evolution at synonymous sites and intronic sites in Drosophila melanogaster. J Mol Evol. 2010;70(1):116–128. doi: 10.1007/s00239-009-9314-6. [DOI] [PubMed] [Google Scholar]
- Zhang L, Li WH. Human SNPs reveal no evidence of frequent positive selection. Mol Biol Evol. 2005;22(12):2504–2507. doi: 10.1093/molbev/msi240. [DOI] [PubMed] [Google Scholar]