Abstract
Vertebrate genome comparisons revealed that there are highly conserved noncoding sequences (HCNSs) among a wide range of species and many of which contain regulatory elements. However, recently emerged sequences conserved in specific lineages have not been well studied. Toward this end, we identified 8,198 primate and 21,128 specific HCNSs as representative ones among mammals from human–marmoset and mouse–rat comparisons, respectively. Derived allele frequency analysis of primate-specific HCNSs showed that these HCNSs were under purifying selection, indicating that they may harbor important functions. We selected the top 1,000 largest HCNSs and compared the lineage-specific HCNS-flanking genes (LHF genes) with ultraconserved element (UCE)-flanking genes. Interestingly, the majority of LHF genes were different from UCE-flanking genes. This lineage-specific set of LHF genes was more enriched in protein-binding function. Conversely, the number of LHF genes that were also shared by UCEs was small but significantly larger than random expectation, and many of these genes were involved in anatomical development as transcriptional regulators, suggesting that certain groups of genes preferentially recruit new HCNSs in addition to old HCNSs that are conserved among vertebrates. This group of LHF genes might be involved in the various levels of lineage-specific evolution among vertebrates, mammals, primates, and rodents. If so, the emergence of HCNSs in and around these two groups of LHF genes developed lineage-specific characteristics. Our results provide new insight into lineage-specific evolution through interactions between HCNSs and their LHF genes.
Keywords: lineage-specific evolution, conserved noncoding sequence, mammals
Introduction
From the inception of molecular evolutionary studies, protein noncoding regions were suspected to be involved in gene regulation (Zuckerkandl and Pauling 1965; Britten and Davidson 1971; King and Wilson 1975). Now it is widely accepted that some noncoding regions play important roles in gene regulation (e.g., Carroll 2005). The functional elements are expected to evolve more slowly than surrounding nonfunctional DNA, as they are under purifying selection (Kimura 1983; Nei 1987). Therefore, sequences that are more highly conserved are likely to be important from the functional point of view. In fact, 5% of the human genome is conserved (Mouse Genome Sequencing Consortium 2002), a considerably higher proportion than that (2%) of the protein coding regions (International Human Genome Sequencing Consortium 2004). Many highly conserved noncoding sequences (HCNSs) among vertebrates have now been identified (Ahituv et al. 2004; Bejerano et al. 2004; Siepel et al. 2005), and some of which are reported to function as distal enhancers for neighboring genes (e.g., Woolfe et al. 2005; Pennacchio et al. 2006).
The regions conserved in only one restricted lineage such as primates, and rodents are considered to be recently emerged HCNSs. These HCNSs may have gained new functions to develop the lineage-specific characteristics after diverging from the ancestral species. However, the commonly accepted strategy for detecting regulatory regions is to identify HCNSs among a wide range of species such as vertebrates. This approach only identifies the regions conserved among diverged species and does not detect sequences conserved in just one particular small lineage. Indeed, such comparisons among vertebrate genomes are known to miss a large number of highly constrained mammalian-specific functional elements despite the fact that these elements are all under similarly intense levels of purifying selection in mammals (Aparicio et al. 2002; Hillier et al. 2004; Cooper et al. 2005).
Challenges and limitations exist for studies seeking to identify the evolution of regulatory regions by detecting changes that were accelerated as a result of lineage-specific positive selection (Pollard et al. 2006; Prabhakar et al. 2006, 2008). These studies focused only on human-specific changes. However, positive selection is not the only possible explanation for these lineage-specific accelerated sequences. Biased gene conversion (BGC) is a neutral mutation process associated with meiotic recombination, which favors a special kind of mutation pattern (Marais 2003). BGC can create strong substitution hotspots, thereby leading to spurious signatures of positive selection (Dreszer et al. 2007; Galtier and Duret 2007; Duret and Galtier 2009; Sumiyama and Saitou 2011). In addition, there are reports that the selective pressure affecting the evolution of regulatory elements in the hominid lineage is significantly relaxed compared with that of the rodent lineage (Kryukov et al. 2005) and that regulatory elements in hominids may be diverging at a neutral rate (Keightley et al. 2005). All these elements point to the difficulty in detecting evidence of positive selection in one lineage.
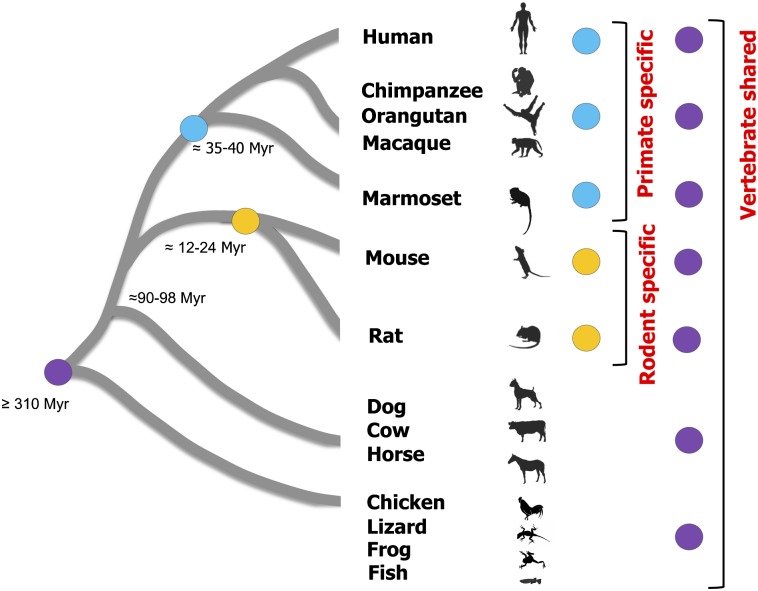
Another challenge for finding the lineage-specific regulatory regions was to identify HCNSs found only in one lineage comprised of very closely related species. The primates is one of the lineages of closely related species compared with the mammals and vertebrates. Sequence comparisons only among primates are likely to capture functional components of the lineage due to shared biological processes (Boffelli et al. 2003). However, to date, this strategy of comparing genomes among closely related species has been applied only to the very limited regions. Furthermore, the goal of this method was to identify all sequences conserved among species at various levels of divergence, such as vertebrates, mammals, and primates but not primate-specific HCNSs. In contrast to the lineage-specific phenotypic changes, the HCNSs which are conserved only in one particular lineage have not been well studied. Thus, to expand our understanding of lineage-specific evolution, we identified HCNSs that were conserved in a particular lineage (either in primates or in rodents) and compared characteristics of the lineage-specific HCNSs with those conserved among mammals and vertebrates. We used human and marmoset genomes for detecting primate-specific HCNSs, whereas mouse and rat genomes were used for detecting rodent-specific HCNSs (fig. 1). The lineage-specific HCNSs identified in this study are expected to provide new insight into how one lineage evolved from a common ancestor.
Fig. 1.—
Phylogenetic relationship of species mainly used in this study. The blue, yellow, and purple circles represent primate-specific, rodent-specific, and vertebrate-shared HCNSs, respectively. The approximate divergence times for ancestral species of each lineage are shown on the tree (Mouse Genome Sequencing Consortium 2002; Hedges and Kumar 2003; Gibbs et al. 2004; She et al. 2006).
Materials and Methods
Genomes Used in This Study
We used a total of 13 vertebrate genomes with over 6X coverage and high quality since alignments containing low-coverage genomes cause misalignment. All genomes were obtained from the UCSC Genome Bioinformatics database (http://genome.ucsc.edu/). They are medaka (Oryzias latipes; oryLat2), stickleback (Gasterosteus aculeatus; gasAcu1), fugu (Takifugu rubripes; fr2), tetradon (Tetraodon nigroviridis; tetNig2), zebrafish (Danio rerio; danRer6), frog (Xenopus tropicalis; xenTro2), lizard (Anolis carolinensis; anoCar1), chicken (Gallus gallus; galGal3), dog (Canis familiaris; canFam2), horse (Equus caballus; equCab1), cow (Bos taurus; bosTau4), rat (Rattus norvegicus; rn4), mouse (Mus musculus; mm9), marmoset (Callithrix jacchus; calJac3), and human (Homo sapiens; hg19). Genomic alignments between human and marmoset and between mouse and rat were also retrieved from the UCSC Genome Bioinformatics database. Genome sequences of rhesus macaque (Macaca mulatta; rheMac2), orangutan (Pongo pygmaeus abelii; ponAbe2), and chimpanzee (Pan troglodytes; panTro3) were also used for extraction of primate-specific HCNSs.
Filtering Repeats and Coding Sequences
Repetitive sequences (chrN_rmsk tables) in the human and mouse genomes were obtained from the UCSC database. All repetitive sequences were excluded from the analysis. Filtering of coding regions was performed based on the annotation (CCDS.20080902) of NCBI CCDS project (http://www.ncbi.nlm.nih.gov/projects/CCDS/) (Pruitt et al. 2009) and the Ensembl human database (http://www.ensembl.org/) (Hubbard et al 2002).
Extraction of Primate-Shared and Rodent-Shared HCNSs
We applied a sliding window analysis to UCSC pairwise noncoding alignments of human–marmoset and mouse–rat (http://genome.ucsc.edu/). We first extracted the repeat-masked noncoding regions and performed sliding window analysis. The window and step size were set to be 100 bp and 25 bp, respectively. When making sliding window sequences, we kept only the sequences that had no gap in a window. To estimate P values for lineage-specific HCNSs, we calculated the divergence of the nongapped noncoding regions between human–marmoset and mouse–rat pairwise alignments (∼10% and ∼14% for autosomes and ∼9% and ∼14% for chromosome X, respectively). We assumed that these average genome divergences are neutral substitution rates and obtained statistical significance of the lineage-specific HCNSs by using a binomial distribution.
Identification of Lineage-Specific HCNSs
Discontiguous MegaBLAST homology search (Zhang et al. 2000) was performed to extract primate-specific HCNSs against the nonprimate vertebrate genomes. Similarly, rodent-specific HCNSs were extracted by performing MegaBLAST search against the nonrodent vertebrate genomes. Parameters for MegaBLAST were discontiguous word template size 16 bp, word matches 12 bp, and mismatch penalty −2. Alignable sequences may be homologous regions. We therefore removed the MegaBLAST hits with ≥30% identity and ≥30 bp in length from primate-shared and rodent-shared HCNSs since the sequences with ≥40% identity may contain functional elements (McGaughey et al. 2008). The homologous sequences among mammals (e.g., human and dog) with ≤30 bp length and ≤30% identity can be found throughout the genome and are assumed to be neutral when assessing average genome identity. However, the homologous sequences among diverged vertebrates (e.g., human and fish) are considered to be functional elements. We removed these alignable sequences among vertebrate genomes (birds, lizard, frog, and fish) from the lineage-specific HCNSs using UCSC multiway alignments. In addition, since there is no closely related species available for rodent lineage, we applied further filtering only for extraction of primate-specific HCNSs and removed the HCNSs that were not found or showed low identity (<98%) in the rhesus macaque, orangutan, and chimpanzee genomes.
To make analyses of these lineage-specific HCNSs easier, we extracted the top 1,000 largest HCNSs, as longer sequences were considered to be under stronger constraint. We assumed that the constraints on the HCNSs in the same bin (class of length) were equal. HCNSs were chosen from the first bin to the nth bin until the total number approached 1,000. Additional HCNSs were chosen by random from the (n + 1)-th bin to reach the total HCNS numbers to be 1,000.
SNP Detection
We downloaded human single nucleotide polymorphism (SNP) data from the Hapmap database (http://hapmap.ncbi.nlm.nih.gov/) and mouse SNP data from NCBI dbSNP build 128 (http://www.ncbi.nlm.nih.gov/SNP). We extracted only the SNP with minor allele frequencies of at least 0.01 in one of four populations (YRI, CEU, JPT, and CHB) in humans and one of all strains in mice. The densities of the SNPs in the repeat masked noncoding sequences in the human and mouse genomes were used to estimate the expected SNP numbers in HCNSs, and these were compared with the observed SNP numbers of HCNSs using χ2-analysis.
Derived Allele Frequency Estimation
To estimate derived allele frequency (DAF), we converted the coordinates of primate-specific HCNSs into those of hg18 to obtain allele frequencies in human populations provided by HapMap release 27. We determined the ancestral allele by using chimpanzee alleles defined by UCSC snp126OrthoPt2Pa2Rm2. An SNP locus was discarded whenever the allele of its orthologous chimpanzee locus did not match either human allele. We used 2 × 2 contingency tables to compare DAF distribution for SNPs within primate-specific HCNSs with all nonrepetitive human noncoding genomes.
Gene Ontology Analysis
We looked for significantly enriched gene categories in primate and rodent LHF genes in the Gene Ontology (GO) database (http://www.geneontology.org/) (Ashburner et al. 2000). The assignment of GO terms and the test for statistical enrichment of those terms were performed with GOstat using goa_human and mgi GO gene association database (http://gostat.wehi.edu.au/) (Beissbarth and Speed 2004). The enrichment of InterPro domains (http://www.ebi.ac.uk/interpro/) of human and mouse genes associated with HCNSs was determined by Fisher’s exact test. The correction for multiple comparisons was performed by using the false discovery rate option in GOstat.
Analysis of dN and dS Levels for LHF Genes
We obtained ortholog lists from Ensembl through biomart for human–marmoset, human–rhesus macaque, and mouse–rat pairs (Hubbard et al. 2002) and extracted only the LHF genes (and LHF orthologs) that were located within 1 Mb of HCNSs in all genomes. dN and dS values were also downloaded from Ensembl (Vilella et al 2008). These values were estimated by using codeml in the PAML package (model = 0, NSsites = 0) (Yang 1997). With dN and dS values of one-to-one pair orthologs in Ensembl homolog lists, we calculated the means of dN and dS of LHF genes and all genes in the human and mouse genomes. Statistical analysis (one-sample t-test, two tailed) was conducted using the R package (http://www.r-project.org). For ultraconserved element (UCE)-flanking genes, we used genes that were located within 1 Mb of UCEs in both human and mouse genomes. With these extracted genes, the same procedure was used for estimation of dN and dS for UCE-flanking genes.
Expected Number of Genes Shared by Lineage-Specific HCNSs and UCEs
The expected number of overlapping genes among lineage-specific HCNSs and UCEs was calculated by random sampling simulation. This random sampling weights the chance of choosing a gene by the length of the chromosome where the gene is located. We randomly selected the same number of genes as the primate LHF genes from the human genome, those as the rodent LHF genes from the mouse genome, and those as the noncoding UCE-flanking genes from both human and mouse genomes, in each 10,000 replicates. Using these data sets, we counted the number of shared genes between primate LHF and UCE-flanking genes, rodent LHF and UCE-flanking genes, and primate and rodent LHF genes and obtained the expected numbers for overlapping genes. Chi-squared tests were conducted for observed and expected numbers using the R package.
Results
Determination of Parameters to Extract Lineage-Specific HCNSs
One important parameter when identifying highly conserved sequences among closely related species is the window size used to compare sequences. Although larger windows have more statistical power to detect significantly conserved sequences among closely related species, smaller windows provide better resolution for the analysis. Thus, it is important to set the smallest possible window size that is still large enough to detect conserved sequences. Another important parameter for identifying conserved sequences among closely related species is the threshold for extraction of conserved sequences. Particularly for closely related species, this substitution number must not be too small because the effect of sequencing errors on the determination of significantly conserved sequences may not be negligible. Taking this into consideration, a threshold of 100% identity increases the number of false negative identification of HCNSs and is thus too strict. For these reasons, the window size for sequence comparison among closely related species should be determined by considering the substitution numbers within a given window.
By way of preliminary analysis, we examined which window size was most appropriate for identifying HCNSs in closely related species by assuming a simple model in which the substitution rate within a given window follows a binomial distribution. The window size setting is a more sensitive process in the human and marmoset comparison than that in the mouse and rat comparison because the average genomic divergence between human and marmoset (nongapped noncoding region: ∼10%) is smaller than that between mouse and rat (∼14%). We thus estimated the number of substitutions in HCNSs between human and marmoset. First, we defined that the HCNSs reside in the lowest 5% of the left tail of the distribution and obtained the expected number of substitutions of HCNSs in 50, 100, 150, and 200 bp windows using the average genome identity of nongapped noncoding region as a neutral rate. We then chose to use 100 bp since the length was relatively small but the range of expected substitution numbers in the HCNS was between 0 and 5. This window size also has adequate statistical power for rodents whose genetic divergence was larger than primates. We therefore used the same window length to extract rodent HCNSs for the simplicity of the analysis.
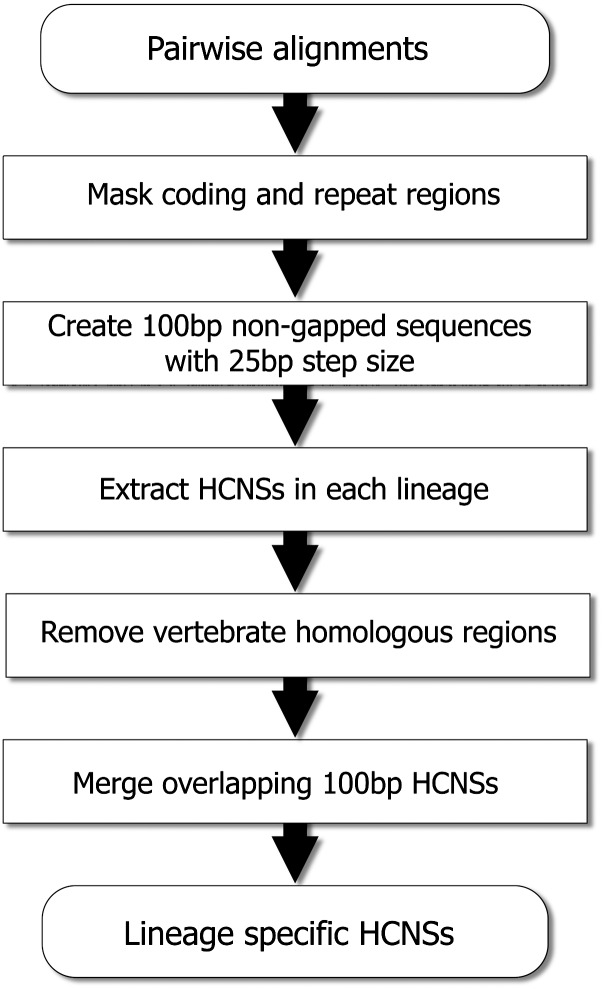
The first step involved extraction of 100 bp primate/rodent shared conserved sequences from human–marmoset/mouse–rat pairwise alignments (2 Gbp and 1.8 Gbp alignments, respectively; fig. 2). The pairwise alignments were obtained from the UCSC Genome Bioinformatics database. We first limited the region used in this analysis to repeat masked noncoding sequences (350 Mbp in human–marmoset and 210 Mbp in mouse–rat pairwise alignments). By using this repeat masked noncoding human–marmoset and mouse–rat pairwise alignments, we created 100-bp sliding windows, which have no gaps from nonrepetitive alignments, with a step size of 25 bp. The total numbers of repeat-masked and non-gapped 100-bp sliding windows in human–marmoset and mouse–rat were 13,618,548 and 8,187,889, respectively.
Fig. 2.—
The procedure of extraction of primate- and rodent-specific HCNSs. Pairwise alignments represent human–marmoset and mouse–rat alignments for extraction of primate- and rodent-specific HCNSs, respectively. After the step “remove vertebrate homologous regions,” another filtering was applied for primate comparison, and the sequences that were not conserved in other primate species (rhesus macaque, orangutan, and chimpanzee) were removed.
From these windows, we extracted HCNSs using empirical conservation cutoffs and obtained only the sequences which have no substitution (0.78% of the total 100-bp fragments), one substitution or fewer (2.2%), and two substitutions or fewer (4.3%) in the human and marmoset pair, and those with no substitution (0.94%), one or fewer (2.5%), and two or fewer (4.5%) in the mouse and rat pair. When we extracted sequences that had three or fewer substitutions, the percentage of the total fragments exceeded 5% in both comparisons. Thus, we determined that a substitution number of two was the appropriate threshold for extraction of HCNSs of primates and rodents (P < 2.1 × 10−3 and P < 1.5 × 10−5, respectively, the binomial model). These numbers determined nucleotide identity thresholds for highly conserved regions in primates and rodents to be ≥98% ([100 − 2]/100). Note that we determined thresholds specifically for chromosome X because the mutation rate on the X chromosome of mammalian genomes is known to be lower than that of autosomes (Miyata et al. 1987; Takahata et al. 1995; Makova and Li 2002). To estimate the appropriate calibration parameter for chromosome X, we examined the number of substitutions in autosome and chromosome X separately in nongapped noncoding pairwise alignments of human–marmoset and mouse–rat pairs. In the human–marmoset comparison, we did not observe any difference in the number of substitution in the HCNSs between autosome and chromosome X and found 2 substitutions in a window (≥98% identity, P < 4.8 × 10−3, the binomial model). However, in the mouse–rat comparison, we observed difference between the two and found only one substitution in a window (≥99% identities, P < 1.2 × 10−5, the binomial model). We calibrated the threshold for chromosome X only for mouse–rat sequences as 99% ([100 − 1]/100) and obtained a total of 590,678 and 356,529 conserved 100-bp sequences from the human–marmoset pair and mouse–rat pair, respectively. These extracted sequences account for less than 2% of the human and mouse genomes. The extraction of conserved sequences is, however, only a starting point for the extraction of lineage-specific HCNSs, which were filtered further in the next step.
Extraction of Lineage-Specific HCNSs
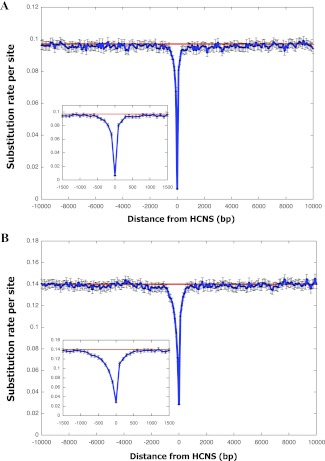
The second step is an extraction of lineage-specific HCNSs. We performed MegaBLAST search against vertebrate genomes with extracted HCNSs (590,678 and 162,304 from primates and rodents, respectively). The primate- and rodent-specific HCNSs are conserved sequences that have emerged after the divergence of these lineages from their ancestors (fig. 1), such that they are found only in primates or rodents. We thus removed all HCNS homologous sequences that were found in nonprimate and nonrodent vertebrates. For extraction of primate-specific HCNSs, an additional filtering criterion was applied, and we limited to the HCNSs that were also conserved in the rhesus macaque, orangutan, and chimpanzee genomes. The remainders were 8,198 and 21,128 for primate- and rodent-specific HCNSs, respectively. For simplicity, we used the top 1,000 largest lineage-specific HCNSs for both rodents and primates when comparing their characteristics. The primate- and rodent-specific HCNSs range in size from 125 to 375 bp and 175 to 425 bp, respectively. Figure 3A and B show the average numbers of substitutions per site (approximated with p-distance) in those 1,000 primate- and rodent-specific HCNSs and their ±10,000 bp flanking regions, respectively. The patterns indicate that only the HCNSs are under the strong constraints, relative to their flanking regions. The ±500 bp flanking regions shown in the insets showed a smaller number of differences compared with those of genome averages. However, the number of substitutions of lineage-specific HCNSs is clearly much lower even when compared with that of ±500 bp flanking regions.
Fig. 3.—
Substitution rates in lineage-specific intergenic HCNSs and their flanking regions. The average substitution number per site within 100-bp window in the range of ±10,000 bp of the top largest 1,000 primate-specific HCNSs (A) and rodent-specific HCNSs (B). The insets show enlarged distributions in the range of ±1,500 bp. The red lines represent average substitution numbers per site of nongapped noncoding regions in the human and mouse genomes, respectively. The error bars are 95% confidence intervals of substitution rate in each window.
SNP Analysis
A SNP is a good indicator for detection of the selective constraint on the sequence in question. We investigated the number of SNPs overlaid on the lineage-specific HCNSs in humans and mice (Sherry et al. 2001; HapMap Consortium 2005) and found that less than 10% of lineage-specific HCNSs had SNPs (MAF < 0.01). The majority of lineage-specific HCNSs have no SNPs and the numbers of SNPs in both primate (SNP density per site: 9.74 × 10−4) and rodent (1.96 × 10−4) lineages were significantly smaller than genome-wide averages (1.6 × 10−3 and 5.0 × 10−4 for human and mouse nonrepetitive genomes, respectively). (P << 0.01 for primate- and rodent-specific HCNSs, Chi-squared test.)
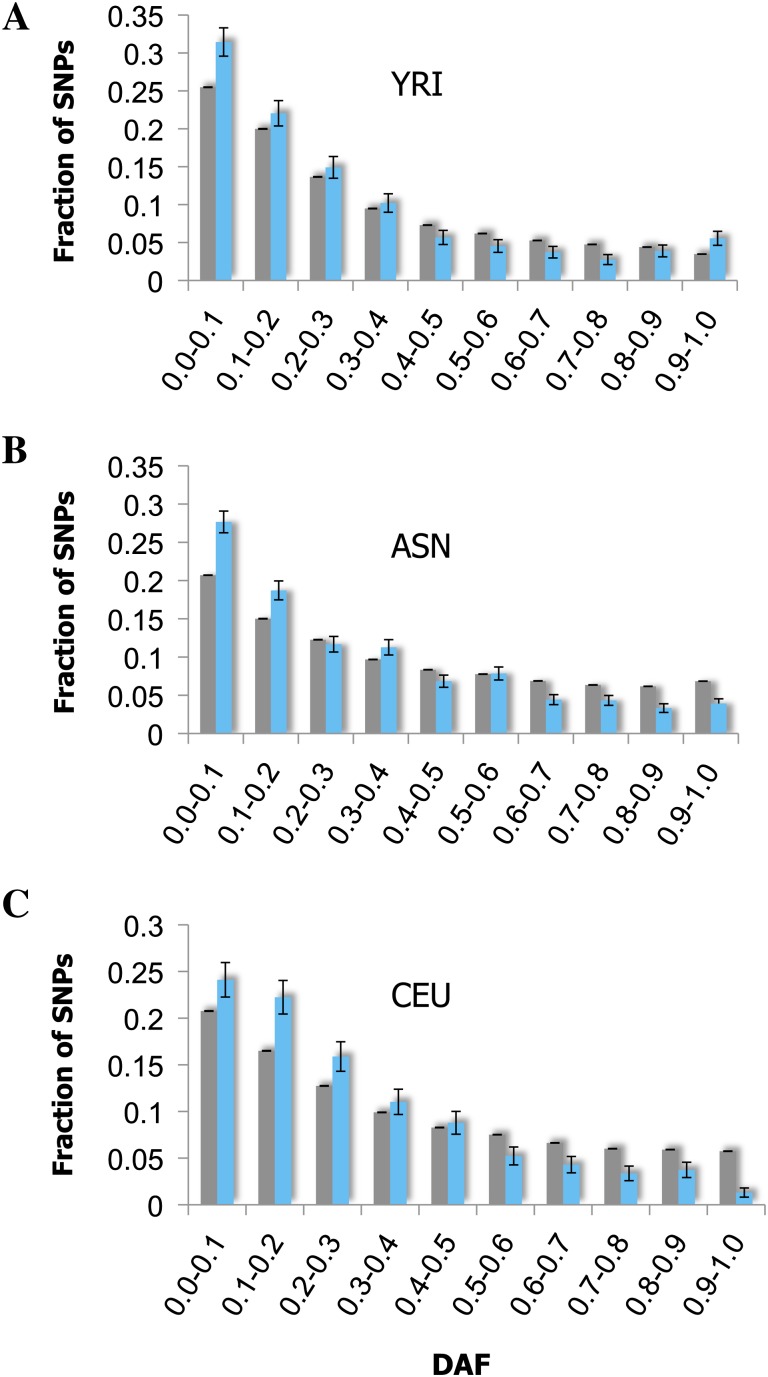
To measure the relative level of purifying selection acting on HCNSs, we analyzed DAF distributions of primate-specific HCNSs and compared these distributions with those of the human genome (fig. 4). Purifying selection is likely the main evolutionary force preventing the vertebrate HCNS from accumulating mutations (Katzman et al. 2007). Quantitatively, the signature of the purifying selection can be observed as a shift in the allele frequency toward ancestral alleles (Drake et al. 2006). We observed the levels of DAF ≤ 0.1 and 0.2 within primate-specific HCNSs in three human populations: Yoruba (YRI), Han Chinese + Japanese (ASN), and American of European Ancestor (CEU). At the level of DAF ≤ 0.1, only the YRI and ASN populations showed a significant excess of rare-derived alleles of SNPs within primate-specific HCNSs compared with the genome average (P < 0.05, Chi-squared test). However, at the level of DAF ≤ 0.2, all populations showed a significant excess of rare allele of the SNPs (P < 0.006, Chi-squared test). This is consistent with previously published results on non-lineage-specific HCNSs (Drake et al. 2006; Ovcharenko 2008, Katzman et al 2007), suggesting that purifying selection is acting on the primate-specific HCNSs.
Fig. 4.—
DAF distribution in primate-specific HCNS. DAF distribution of Yoruba from Nigeria (YRI) (A), Han Chinese from Beijing combined with Japanese from Tokyo (ASN) (B), and American of European ancestry (CEU) (C). Light gray and blue bars represent data for SNPs in the nonrepetitive human genome and SNPs within primate-specific HCNSs. Error bars were estimated using binominal distribution as σ2 = (pq)/n, where p represented the fraction of SNPs in a particular bin, q represented 1 − p, and n represented the total number of SNPs. All primate-specific HCNSs (8,198) were used for this analysis.
General Features of the Lineage-Specific HCNSs
Genic Category
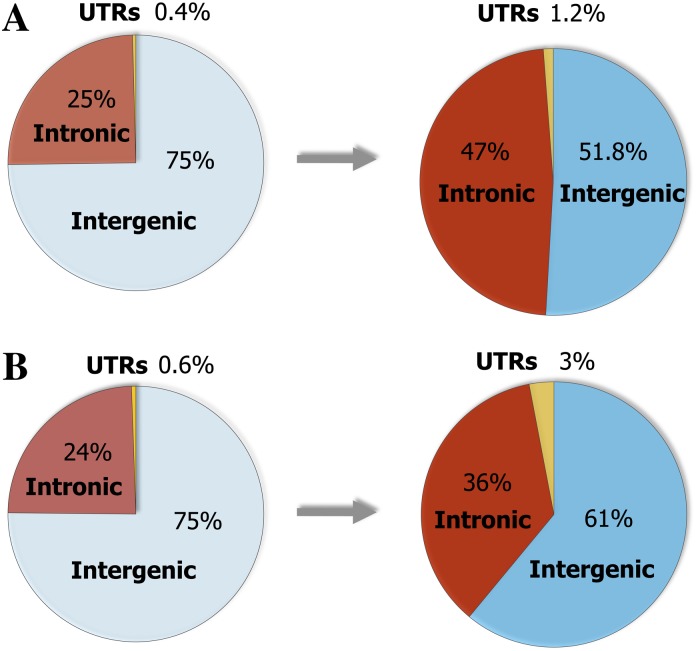
To determine if there are any general trends in the distribution of the lineage-specific HCNSs, we compared three annotation categories (intron, intergenic, and UTR) of the lineage-specific HCNSs. The fractions of lineage-specific HCNSs and their genic categories are shown in figure 5. The fractions of HCNSs residing in UTRs and introns in both primates and rodents were much higher than those of the whole human and mouse genomes, respectively (P < 10−15, Chi-squared test). The most striking difference was found in the UTR category, where the fractions of UTR in the primate and rodent-specific HCNSs were 3 times and 5.5 times higher than those of human and mouse genomes, respectively. This increased fraction of UTRs is consistent with the tendency of HCNSs in vertebrates (e.g., Siepel et al. 2005; Woolfe et al. 2005). However, the fractions of UTR category in lineage-specific HCNSs were lower than those in non-lineage specific vertebrates in UTRs (∼6%). In addition, the fractions of intergenic + UTR and intronic categories differed between primate- and rodent-specific HCNSs (P = 1.04 × 10−4, Chi-squared test).
Fig. 5.—
Fractions of genic categories in whole genomes and lineage-specific HCNSs. The pie charts show percentages of genic categories in the human genome (left) and primate-specific HCNSs (right) (A), in the mouse genome (left) and rodent-specific HCNSs (right) (B). The percentages of UTRs become markedly elevated in the lineage-specific HCNSs. The distribution of genic categories between genomes and lineage-specific HCNSs showed significant difference (P < 10−15, Chi-squared test).
GO Analysis of the Lineage-Specific HCNS-Flanking Genes
The function of genes that are located near lineage-specific HCNSs may provide important information for understanding the lineage-specific evolution. We therefore examined the statistically overrepresented functions of the lineage-specific HCNS-flanking genes (LHF genes). We first obtained the distance between the lineage-specific HCNSs and their LHF genes and found that 96.2% and 97.5% of intergenic HCNSs are located within 1 Mb of the transcription start site of LHF genes in human and mouse, respectively. The longest distance between a target developmental gene and its experimentally verified enhancer is ∼1 Mb; the reported genes are SHH (Lettice et al. 2003; Sagai et al. 2005), SOX9 (Bishop et al. 2000), and SHOX (Sabherwal et al. 2007). These findings indicate that at least some HCNSs may be associated with LHF genes as distal regulatory elements.
Next, we looked for functional categories of LHF genes as defined by the GO database (Beissbarth and Speed 2004) and obtained significantly enriched functions of LHF genes. The top 30 overrepresented gene functions are shown in table 1. In primate and rodent LHF genes, statistically overrepresented functions were developmental process, protein binding, and regulation of transcription. In developmental process, anatomical structure (P = 3.1 × 10−66 and P = 3.1 × 10−66 for primate and rodent LHF genes, respectively) and nervous system development (P = 6.1 × 10−54 and P = 1.9 × 10−13) were enriched. In transcriptional regulation, positive regulation of transcription (P = 7.1 × 10−23 for primate LHF genes) and regulation of transcription (P = 5.0 × 10−7 for rodent LHF genes) were overrepresented. This tendency of overrepresented gene functions is consistent with previous studies of HCNSs among vertebrates (e.g., Bejerano et al. 2004; Siepel et al. 2005; Woolfe et al. 2005). However, both primate- and rodent-specific LHF genes showed significant overrepresentation of protein binding (P = 9.7 × 10−36 and P = 3.3 × 10−16) compared with vertebrate HCNSs (e.g., Bejerano et al. 2004; Siepel et al. 2005; Woolfe et al. 2005). We further examined the LHF genes in the protein-binding category and found that they were enriched in nervous system development (P = 2.93 × 10−37), positive regulation of transcription (P = 8.27 × 10−13), and transcription cofactor binding (P = 2.3 × 10−9), which are also important for developmental process and transcriptional regulation.
Table 1.
Top 30 Overrepresented Functions of LHF Genes
| GO | Gene Function | P Value |
| (A) Primate LHF genes | ||
| GO:0048856 | Anatomical structure development | 3.10 × 10−66 |
| GO:0048731 | System development | 2.10 × 10−64 |
| GO:0007275 | Multicellular organismal development | 2.32 × 10−58 |
| GO:0032502 | Developmental process | 3.18 × 10−55 |
| GO:0007399 | Nervous system development | 6.11 × 10−54 |
| GO:0032501 | Multicellular organismal process | 9.41 × 10−51 |
| GO:0005515 | Protein binding | 9.72 × 10−36 |
| GO:0009653 | Anatomical structure morphogenesis | 2.66 × 10−32 |
| GO:0048869 | Cellular developmental process | 6.39 × 10−29 |
| GO:0030154 | Cell differentiation | 6.39 × 10−29 |
| GO:0048513 | Organ development | 8.93 × 10−29 |
| GO:0007154 | Cell communication | 5.11 × 10−24 |
| GO:0050789 | Regulation of biological process | 2.08 × 10−23 |
| GO:0065007 | Biological regulation | 7.14 × 10−23 |
| GO:0045941 | Positive regulation of transcription | 1.39 × 10−22 |
| GO:0045935 | Positive regulation of nucleobase, nucleoside, nucleotide, and nucleic acid metabolic process | 1.98 × 10−21 |
| GO:0050794 | Regulation of cellular process | 5.97 × 10−21 |
| GO:0031325 | Positive regulation of cellular metabolic process | 1.49 × 10−20 |
| GO:0009893 | Positive regulation of metabolic process | 7.10 × 10−20 |
| GO:0009887 | Organ morphogenesis | 2.53 × 10−19 |
| GO:0007165 | Signal transduction | 8.86 × 10−19 |
| GO:0000902 | Cell morphogenesis | 1.60 × 10−16 |
| GO:0032989 | Cellular structure morphogenesis | 1.60 × 10−16 |
| GO:0022008 | Neurogenesis | 6.43 × 10−16 |
| GO:0008134 | Transcription factor binding | 1.09 × 10−15 |
| GO:0048468 | Cell development | 1.49 × 10−15 |
| GO:0003712 | Transcription cofactor activity | 1.57 × 10−15 |
| GO:0007267 | Cell–cell signaling | 6.28 × 10−15 |
| GO:0048522 | Positive regulation of cellular process | 7.14 × 10−15 |
| GO:0048699 | Generation of neurons | 1.09 × 10−14 |
| (B) Rodent LHF genes | ||
| GO:0005515 | Protein binding | 3.03 × 10−17 |
| GO:0007275 | Multicellular organismal development | 4.43 × 10−15 |
| GO:0048731 | System development | 7.66 × 10−14 |
| GO:0032502 | Developmental process | 1.89 × 10−13 |
| GO:0048856 | Anatomical structure development | 1.92 × 10−13 |
| GO:0007399 | Nervous system development | 1.92 × 10−13 |
| GO:0009653 | Anatomical structure morphogenesis | 3.25 × 10−12 |
| GO:0050789 | Regulation of biological process | 4.09 × 10−12 |
| GO:0065007 | Biological regulation | 2.32 × 10−11 |
| GO:0032990 | Cell part morphogenesis | 1.00 × 10−10 |
| GO:0030030 | Cell projection organization and biogenesis | 1.00 × 10−10 |
| GO:0048858 | Cell projection morphogenesis | 1.00 × 10−10 |
| GO:0009887 | Organ morphogenesis | 1.02 × 10−09 |
| GO:0048666 | Neuron development | 1.02 × 10−09 |
| GO:0050794 | Regulation of cellular process | 1.40 × 10−09 |
| GO:0031175 | Neurite development | 3.86 × 10−09 |
| GO:0007167 | Enzyme-linked receptor protein signaling pathway | 1.18 × 10−08 |
| GO:0048869 | Cellular developmental process | 3.45 × 10−08 |
| GO:0030154 | Cell differentiation | 3.45 × 10−08 |
| GO:0048513 | Organ development | 3.72 × 10−08 |
| GO:0030182 | Neuron differentiation | 4.58 × 10−08 |
| GO:0022610 | Biological adhesion | 4.74 × 10−08 |
| GO:0007155 | Cell adhesion | 4.74 × 10−08 |
| GO:0000904 | Cellular morphogenesis during differentiation | 1.26 × 10−07 |
| GO:0007267 | Cell–cell signaling | 1.68 × 10−07 |
| GO:0005216 | Ion channel activity | 1.81 × 10−07 |
| GO:0016477 | Cell migration | 2.00 × 10−07 |
| GO:0010468 | Regulation of gene expression | 3.90 × 10−07 |
| GO:0022838 | Substrate-specific channel activity | 4.53 × 10−07 |
| GO:0045449 | Regulation of transcription | 4.99 × 10−07 |
Note.—The P value was determined by Fisher’s exact test and corrected with false discovery rate method. Only the gene functions belonging to “Biological process” and “Molecular function” of GO category are shown (Ashburner et al. 2000). A total of 980 and 985 LHF genes that locate within 1 Mbp of HCNSs were used for primate- and rodent-specific HCNSs, respectively.
Selective Constraints on LHF Genes
In order to identify evolutionary constraints on LHF genes, we examined the nonsynonymous (dN) and synonymous (dS) substitution rates in LHF genes. First, dN and dS values for human–marmoset and mouse–rat pairs were obtained from Ensembl (Vilella et al. 2008). Using primate (or rodent) LHF genes that had annotated orthologs within 1 Mbp of the HCNS in human and marmoset (mouse and rat) genomes, we calculated the means of dN and dS of genes in each pair. We then compared dN and dS of LHF genes with those of the genome average. LHF genes are expected to have important functions, and in fact, means of dN/dS ratios in all pairs were significantly smaller than those of genome averages (P < 0.001 in all pairs, one-sample t-test). However, dS values of primate and rodent LHF genes were significantly smaller than those of genome averages as well as dN values (P < 0.001 in human–marmoset and mouse–rodent pairs; table 2), indicating stronger constraint on flanking genes not only at amino acid sequence level but also at nucleotide level.
Table 2.
dN and dS Values
| All genesa | Primate LHF genes | Orthologs of rodent LHF genes | |
| (A) dN and dS of LHF genes in the human–marmoset pair | |||
| Number of genes | 15,011 | 462 | 319 |
| Average dN | 0.0407 (0.0494) | 0.0250** (0.0362) | 0.0255** (0.0287) |
| Average dS | 0.1684 (0.1140) | 0.1306** (0.0834) | 0.1740 (0.1279) |
| Average dN/dSb | 0.2495 (0.2599) | 0.1852** (0.2123) | 0.1585** (0.1710) |
| All genesc | Rodent LHF genes | Orthologs of primate LHF genes | |
| (B) dN and dS of LHF genes in the mouse–rat pair | |||
| Number of genes | 16,104 | 517 | 306 |
| Average dN | 0.0408 (0.0509) | 0.0285** (0.0434) | 0.0262** (0.0352) |
| Average dS | 0.2091 (0.0877) | 0.1943* (0.0948) | 0.2002 (0.0815) |
| Average dN/dSb | 0.1910 (0.2360) | 0.1330** (0.1731) | 0.1241** (0.1248) |
| Primates | Rodents | ||
| (C) dN and dS of UCE-flanking genes | |||
| Number of genes | 141 | 122 | |
| Average dN | 0.0245* (0.0378) | 0.0208* (0.0244) | |
| Average dS | 0.1225** (0.0844) | 0.1705** (0.0748) | |
| Average dN/dSb | 0.1857* (0.2381) | 0.1075** (0.1078) | |
Note.—Numbers in parentheses represent standard deviation. Values with asterisks represent significant differences (P < 0.001, one sample t-test) from the genome averages. ** and * represent P < 10−9 and 10−3, respectively.
All genes in the human genome.
dN/dS ratio was calculated only for genes with dS > 0.
All genes in the mouse genome.
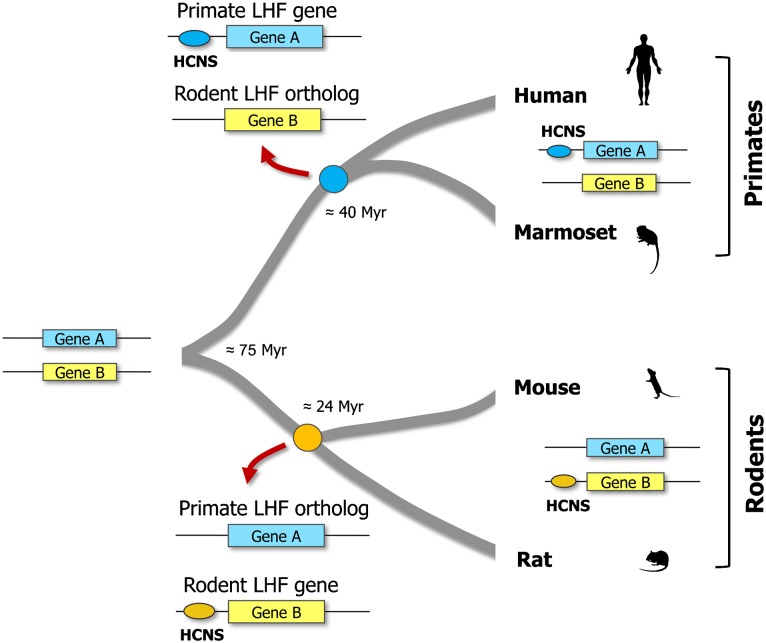
The main advantage of studying HCNSs that are found in only one particular lineage is that we can compare evolutionary constraints on the LHF genes with those of orthologs that have no HCNS in another lineage. To investigate differences in selective constraints on LHF genes and their orthologs that have no lineage-specific HCNS, we compared dN and dS levels of orthologs of primate (rodent) LHF genes with genome averages in rodent (primate) pair. We defined orthologs of primate LHF genes in rodents and those of rodent LHF genes in primates as primate LHF orthologs and rodent LHF orthologs, respectively (see fig. 6). As in the LHF genes, the primate and rodent LHF orthologs had significantly smaller dN/dS ratios as well as dN levels, when compared with those of genome averages in mouse–rat (human–marmoset) pair (P < 0.001, one sample t-test; table 2A and table 2B).
Fig. 6.—
Definition of LHF orthologs. The primate and rodent LHF orthologs are defined as the ortholog of primate LHF gene in rodents (Gene A in rodents), and the ortholog of rodent LHF gene in primates (Gene B in primates). Although primate and rodent LHF genes recruited lineage-specific HCNSs after the divergence of each lineage from the common ancestor, the majority of primate and rodent LHF orthologs did not.
On the other hand, there was no significant difference between dS values of primate and rodent LHF orthologs and those of genome-wide genes (P > 0.05, table 2A and table 2B). This finding indicates that the evolutionary constraint on LHF genes is stronger than the constraint on genes that have no lineage-specific HCNSs. We also analyzed levels of constraints using dN and dS values in genes flanking noncoding UCEs which are an extreme case of HCNSs among vertebrates (Bejerano et al. 2004). All mean values (dN/dS ratio, dN and dS values) of UCE-flanking genes were significantly smaller than genome averages (table 2C). This result further supports the finding lower dS is an important signature of the genes that are associated with HCNSs.
Comparison of Lineage-Specific HCNS-Flanking Genes with Vertebrate HCNS-Flanking Genes
GO analysis of the LHF genes showed that the most statistically overrepresented functions were developmental process and transcriptional regulation, which were quite similar to the overrepresented functions of HCNSs conserved among vertebrates (e.g., Bejerano et al. 2004; Woolfe et al. 2005). This observation raised the question whether the lineage-specific HCNSs are found near the same genes as those of vertebrate HCNSs whose origin are older than primate- and rodent-specific HCNSs.
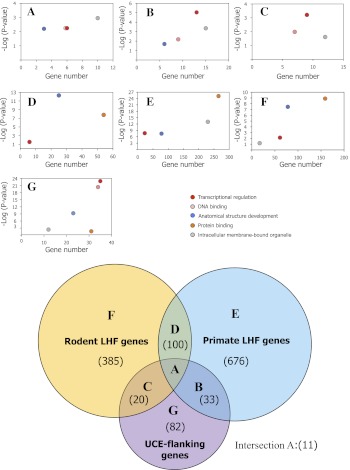
To address this question, we first examined the number of flanking genes that were shared among primate- and rodent-specific HCNSs and noncoding UCEs (fig. 7). The majority of primate- and rodent-specific HCNSs (980 and 985, respectively) had LHF genes within 1 Mbp. They were often clustered near a small subset of genes, and the numbers of LHF genes for primate- and rodent-specific HCNSs were 820 and 516, respectively. This is consistent with previous studies of vertebrate HCNSs including UCEs. Furthermore, a total of 11 LHF genes were shared among lineage-specific HCNS- and UCE-flanking genes (fig. 7A). The number of genes shared by primate-specific HCNSs and UCEs and rodent-specific HCNSs and UCEs were 31 and 41, respectively (fig. 7B and C in the Venn diagram). Interestingly, we found that the numbers of genes shared by lineage-specific HCNSs and UCEs were significantly larger than random expectation (P < 10−4). Overrepresented functions of GO categories for the LHF genes overlapping with UCE-flanking genes were mainly involved in regulation of transcription, DNA binding, anatomical structure development, and intracellular membrane-bound organelle (fig. 7, scatter plots A through C). Note that rodent LHF genes overlapping with UCE-flanking genes were mildly enriched in anatomical structure development (P < 0.056). Many of these genes shared by lineage-specific HCNSs and UCEs are well studied and known to play an important role as transcriptional regulators during vertebrate development.
Fig. 7.—
Comparison of genes among lineage-specific HCNSs and UCEs. Upper 7 panels show the scatter plots of the number of overrepresented gene functions and their P values obtained by GO analysis. The letters A through G in the scatter plots are corresponding to the letters in the Venn diagram, which shows the number of overlapping LHF genes among primate- and rodent-specific HCNSs and UCEs (numbers in parentheses).
Similarly, we examined LHF genes shared by primate- and rodent-specific HCNSs and found that the numbers of overlapping LHF genes were also significantly larger than random expectation (P < 10−4) (fig. 7D in the Venn diagram). In contrast, the proportion of primate and rodent LHF genes that were not shared by any gene set were 82.4% and 74.6%, respectively (fig. 7E and F in the Venn diagram). This finding demonstrates that the majority of the LHF genes are lineage specific. The main overrepresented gene functions were regulation of transcription, protein binding, anatomical structure development, and intracellular membrane-bound organelle (fig. 7 scatter plots D through F). Taking into consideration that the UCE-flanking genes that do not include LHF genes were enriched in both DNA binding and protein binding, whereas genes involved in protein binding were less significant compared with those of DNA binding, the difference between the gene functions of data sets A through F was whether or not DNA binding was overrepresented (fig. 7, scatter plot G).
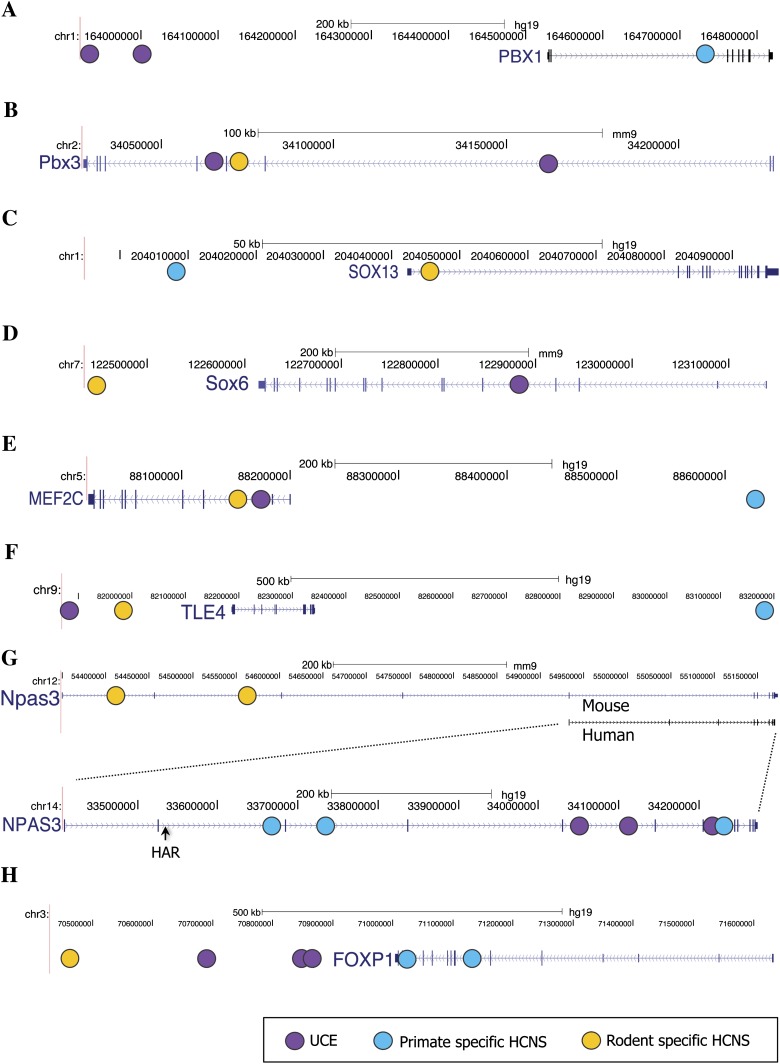
Examples of these overlapping LHF genes are shown in figure 8. Lineage-specific HCNSs as well as UCEs were found in and around the PBX genes. A primate-specific HCNS and UCEs were located within PBX1, and rodent-specific HCNSs and UCEs were located in and around PBX3 (fig. 8A and B). PBX genes act as cofactors for various transcription factors such as HOX genes (e.g., Rauskolb and Wieschaus 1994; Mann 1995, Mann and Chan 1996, Mann and Affolter 1998) and are involved in chromatin modification (e.g., Cirillo et al. 2002; Berkes et al. 2004). The lineage-specific HCNSs were also associated with SOX genes. Both primate- and rodent-specific HCNSs were found in and around SOX13 and SOX6 (Fig. 8C and D), respectively. In addition, a primate-specific HCNS and a rodent-specific HCNS were also located near SOX9 and SOX11, respectively. SOX genes are transcriptional activators that are required for normal development of the central nervous, chondrogenesis, and maintenance of cardiac and skeletal muscle cells (Wegner and Stolt 1995; Wegner 1999).
Fig. 8.—
Examples of lineage-specific HCNS and UCE distributions. Purple, light blue, and yellow circles represent the position of UCE, primate-, and rodent-specific HCNSs, respectively. Examples of PBX1 (A), Pbx3 (B), SOX13 (C), Sox6 (D), MEF2C (E), TLE4 (F), NPAS3 (G), and FOXP1 (H) are shown in the figure. When LHF genes are of primate-specific HCNSs, the distribution of HCNSs and UCEs are always shown on the human genes. All genes but NPAS3 are highly conserved in vertebrates. For NPAS3, both human and mouse genes are shown since there is no intronic region corresponding to rodent-specific HCNSs in the human gene. As additional information, the human accelerated region (HAR) is shown.
MEF2C is an interesting example of primate and rodent HCNSs and UCE shared LHF genes. The genomic positions of the lineage-specific HCNSs and UCEs for MEF2C are shown in figure 8E. MEF2C belongs to the evolutionarily ancient MADS family of transcription factors which play central roles in the transmission of extracellular signals to the genome and in the activation of the genetic programs that control cell differentiation, proliferation, morphogenesis, survival, and apoptosis of a wide range of cell types (Shore and Sharrocks 1995; Potthoff and Olson 2007). TLE genes also recruited lineage-specific HCNSs. Another LHF gene that was shared among primate- and rodent-specific HCNSs and UCEs is TLE4 (fig. 8F). This is a transcriptional corepressor that binds to a number of transcription factors and inhibits the transcriptional activation mediated by PAX5 and by CTNNB1 and TCF family members in Wnt signaling (Roose et al. 1998; Eberhard et al. 2000; Yaklichkin et al. 2007). An interesting exceptional example of the LHF genes is NPAS3 (fig. 8G). This gene is a brain-enriched transcription factor belonging to the basic helix-loop-helix-PAS superfamily, the members of which carry out diverse functions, including circadian oscillations, neurogenesis, toxin metabolism, hypoxia, and tracheal development (Kamnasaran et al. 2003), shown as HAR in figure 8G. NPAS3 has not only vertebrate HCNSs but also a human-accelerated region (Pollard et al. 2006). Three primate-specific HCNSs, one rodent-specific HCNS, and three UCEs are located in NPAS3. Another example of an LHF gene that is thought to have a critical impact on lineage-specific evolution is FOXP1. Primate-specific HCNSs, a rodent-specific HCNS, and three UCEs were found in and around FOXP1 (fig. 8H). FOXP1 is a member of the FOX family of transcription factor and plays important roles in the regulation of tissue- and cell type-specific gene transcription (e.g., Kaufmann and Knochel 1996; Carlsson and Mahlapuu 2002).
Discussion
In this study, we identified a total of 8,198 primate- and 21,128 rodent-specific HCNSs and found that the lineage-specific HCNSs showed the signature of purifying selection at the SNP level as well as the nucleotide level (figs. 3 and 4). We found that the LHF genes as a whole were enriched in gene functions similar to those UCE-flanking genes (table 2). However, the majority of LHF genes and UCE-flanking genes are independent sets (fig. 7E–G in the Venn diagram). This suggests that a particular group of genes are preferentially associated with lineage-specific HCNSs instead of vertebrate HCNSs (fig. 7, scatter plots E through G). Thus, this group of genes may be regulated through HCNSs in a lineage-specific manner. Similarly, we found that there are UCE-flanking genes that have no lineage-specific HCNSs. These genes are thought to be a core set that may have contributed to the development of fundamental characteristics of vertebrates. On the other hand, we found that the number of LHF genes that were also UCE-flanking genes was significantly larger than random expectation (fig. 7B and C in the Venn diagram). This suggests that certain groups of genes tend to recruit new HCNSs in addition to the vertebrate (old) HCNSs such as UCEs. The genes at the intersection of all lineages were the most extreme example (fig. 7A in the Venn diagram). These genes may have contributed to the evolution of different levels of organisms, for example, vertebrates, primates, and rodents. A particularly noteworthy feature of LHF genes is that even genes that are highly conserved among vertebrates, and which play an important role in the developmental process (e.g., FOXP1 and PBX genes), have lineage-specific HCNSs. This is so because regulatory regions for the conservative genes are also conserved among a wide range of species.
Although the primate- and rodent-specific HCNSs are found only in primates and rodents, respectively (fig. 1), there may be HCNSs that have been lost only in these lineages. Comparisons of ancient vertebrate conserved noncoding elements (aCNEs) which were present in the common ancestor of jawed vertebrates in Hox cluster loci showed that many of aCNEs have diverged beyond recognition in teleost fish (Lee et al. 2008). However, another study of HCNSs among four mammals showed that the loss rate of ultraconserved-like HCNSs in rodents was only 0.086% (Mclean and Bejerano 2008). It is also known that there is a slowdown in substitution rates of UCEs in tetrapods (Stephen et al. 2008). These observations indicate that the loss rate of mammalian and tetrapod HCNSs was smaller than expected. Therefore, if there are HCNSs that have been lost only in primate and rodent lineages, many of them are expected to be derived from aCNEs. The evolutionary process by which new regulatory networks are created may be driven by the addition and loss of HCNSs to genes that play an important role in development.
It is also possible that new lineage-specific HCNSs and old vertebrate or mammalian HCNSs have the same function even if their sequences are not homologous. This suggests that the new-lineage specific HCNSs were created during functional turnover and that gaining the new HCNSs did not contribute to the lineage-specific evolution. In such cases, the gain and loss of HCNSs frequently occur within and around these vertebrate developmental genes and their gene expressions are maintained among a wide range of species. However, it is impossible to know whether or not nonhomologous long sequences have the same function by performing computational analysis alone.
In TF binding sites, it is known that there is turnover, including gain and loss of DNA sequence motifs (Cliften et al. 2003; Kellis et al. 2003; Gasch et al. 2004; Stark et al. 2007) as well as alterations in motif spacing relative to the start of transcription or to other motifs (Ihmels et al. 2005; Tanay et al. 2005). Recently, a number of genome-scale studies using immunoprecipitation were performed to compare TF-binding patterns (Borneman et al. 2007; Tuch et al. 2008; Bradley et al. 2010; Lavoie et al. 2010; Schmidt et al. 2010) or mRNA expression profiles across species (Ihmels et al. 2005; Tanay et al. 2005; Hogues et al. 2008; Field et al. 2009; Wapinski et al. 2010). Many of these studies have identified transcriptional programs that were dramatically rewired over short evolutionary time scales. For instance, Borneman et al. (2007) found that the TF Tec1 binds only 20% of the same target genes in comparisons between Saccharomyces cerevisiae and the closely related S. bayanus and S. mikatae and that this difference is due to the gain and loss of canonical Tec1 cis-regulatory motifs.
Kim et al. (2010) found abundant transcription at neuronal enhancers that are evolutionarily conserved. However, the lineage-specific HCNSs have only a few partial matches with ESTs and known RNA genes. This suggests that the majority of HCNS functions are cis-regulatory elements and in fact many studies reported that vertebrate HCNSs showed enhancer activities (e.g., Poulin et al. 2005; Woolfe et al. 2005; Pennacchio et al. 2006; Lareau et al. 2007; Ni et al. 2007). In addition, we compared lineage-specific HCNSs and CNEs of other known vertebrate HCNSs (Woolfe et al. 2005, 2007) to determine whether there was any difference in the results of comparisons with UCEs. The CNEs are over 1,400 HCNSs that were identified by human–fugu comparison and which include sequences overlapping with UCEs. As expected, we obtained similar results between noncoding UCEs and lineage-specific HCNSs in dN and dS and in shared genes analyses.
We also found that there were differences between dS values of LHF genes and those of genome-wide genes (table 2A and table 2B). It is known that the neutral mutation rate has regional biases in mammalian genomes (Mouse Genome sequencing Consortium 2002; Hardison et al. 2003), and low dS genes tend to have similar GO functional categories such as transcriptional regulation and development (Chuang and Li 2004). However, the low mutation rate of LHF genes does not affect substitution numbers in HCNSs because there is no correlation between dS values and distances between HCNSs and their LHF genes (Pearson’s r = 0.04 to 0.05). This raises the question as to why there is correlation between HCNSs and low dS genes.
One possible explanation is that a lower dS gene may be constrained at nucleotide level when it affects splicing and/or mRNA stability (Chamary et al. 2006). To investigate this, we examined the number of splicing variants in LHF genes. Both the primate and rodent LHF genes have significantly higher numbers of splicing variants than genome averages (one sample t-test, P < 2.50 × 10−2 and P < 1.87 × 10−7, respectively). However, LHF orthologs also showed relatively higher number of splicing variants (P < 3.0 × 10−2 and P < 2.8 × 10−2, respectively). This does not explain the difference in dS values between LHF genes and the LHF orthologs. A second possibility is the change in chromatin structure. Genes that display a strong constraint at synonymous sites are preferentially located in closed regions of the genome because they require tight transcriptional regulation (Prendergast et al. 2007). Moreover, this strong constraint on LHF genes at the nucleotide level suggests that many regulatory proteins may bind to the genes and interact with HCNSs for tight regulation of the gene expression.
We carefully chose the species used in this study by considering their coverage and quality. Nevertheless, there are some limitations in these analyses of lineage-specific HCNSs due to the small number of genomes of closely related species. We were able to use only two species genomes for the rodent lineage. We found small differences in genic categories overlapping primate- and rodent-specific HCNSs (fig. 5A and B). The number of rodent-specific HCNSs overlapping intergenic and UTR was significantly larger than the comparable number in primates (P < 10−4, Chi-squared test). We found another small difference in overrepresented gene functions. However, we cannot determine whether these differences are derived from lineage-specific characteristics, from the different number of species used for extraction of primate and rodent HCNSs or from annotation problems. Further studies of lineage-specific HCNSs are necessary to obtain clear pictures of lineage-specific evolution.
In spite of the small differences, similar tendencies were found in the constraints on the primate and rodent LHF genes and their functions. Our analyses of LHF genes imply that the lineage-specific HCNSs were created in and around two categories of genes. In the first category, lineage-specific HCNSs are created near protein coding genes that had no HCNSs before. This expands the set of LHF genes that differ from those of ancestral (mammalian and vertebrate) HCNSs. These genes are more enriched in protein binding, many of which are involved in nervous development, compared with ancestral HCNS flanking genes, suggesting that the lineage-specific evolution may be driven by changes in the regulation of protein interaction during nervous system development. In the second category, lineage-specific HCNSs are newly added to particular groups of genes that already have vertebrate HCNSs. One of the major gene groups codes transcriptional regulators involved in anatomical development and may be involved in the various levels of lineage-specific evolution such as vertebrates, mammals, primates, and rodents. Many of the lineage-specific HCNSs and vertebrate HCNSs are likely to be associated with different gene sets. The lineage-specific evolution through HCNSs thus occurred by obtaining both new HCNS and LHF gene sets that differ from the “core” sets of vertebrate HCNS and its associated gene.
Acknowledgments
We thank the Genome Sequencing Center at WUSTL for providing marmoset genome sequences; Drs Jaume Bertranpetit, Arcadi Navarro, Hafid Laayouni, Kenta Sumiyama, and Kirill Kryukov for useful discussion. M.T. received Research Fellowship of the Japan Society for the Promotion of Science for Young Scientists. This study was supported partly by Grant-in-Aid for scientific research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan to N.S. and by SOKENDAI Short-term Study-abroad Program to M.T.
References
- Ahituv N, Rubin EM, Nobrega MA. Exploiting human–fish genome comparisons for deciphering gene regulation. Hum Mol Genet. 2004;13:R261–R266. doi: 10.1093/hmg/ddh229. [DOI] [PubMed] [Google Scholar]
- Aparicio S, et al. Whole-genome shotgun assembly and analysis of the genome of Fugu rubripes. Science. 2002;297:1301–1310. doi: 10.1126/science.1072104. [DOI] [PubMed] [Google Scholar]
- Ashburner M, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beissbarth T, Speed TP. GOstat: find statistically overrepresented Gene Ontologies within a group of genes. Bioinformatics. 2004;20:1464–1465. doi: 10.1093/bioinformatics/bth088. [DOI] [PubMed] [Google Scholar]
- Bejerano G, et al. Ultraconserved elements in the human genome. Science. 2004;304:1321–1325. doi: 10.1126/science.1098119. [DOI] [PubMed] [Google Scholar]
- Berkes CA, et al. Pbx marks genes for activation by MyoD indicating a role for a homeodomain protein in establishing myogenic potential. Mol Cell. 2004;14:465–477. doi: 10.1016/s1097-2765(04)00260-6. [DOI] [PubMed] [Google Scholar]
- Bishop CE, et al. A transgenic insertion upstream of sox9 is associated with dominant XX sex reversal in the mouse. Nat Genet. 2000;26:490–494. doi: 10.1038/82652. [DOI] [PubMed] [Google Scholar]
- Boffelli D, et al. Phylogenetic shadowing of primate sequences to find functional regions of the human genome. Science. 2003;299:1391–1394. doi: 10.1126/science.1081331. [DOI] [PubMed] [Google Scholar]
- Borneman AR, et al. Divergence of transcription factor binding sites across related yeast species. Science. 2007;317:815–819. doi: 10.1126/science.1140748. [DOI] [PubMed] [Google Scholar]
- Bradley RK, et al. Binding site turnover produces pervasive quantitative changes in transcription factor binding between closely related Drosophila species. PLoS Biol. 2010;8:e1000343. doi: 10.1371/journal.pbio.1000343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britten RJ, Davidson EH. Repetitive and non-repetitive DNA sequences and a speculation on the origins of evolutionary novelty. Q Rev Biol. 1971;46:111–138. doi: 10.1086/406830. [DOI] [PubMed] [Google Scholar]
- Carlsson P, Mahlapuu M. Forkhead transcription factors: key players in development and metabolism. Dev Biol. 2002;250:1–23. doi: 10.1006/dbio.2002.0780. [DOI] [PubMed] [Google Scholar]
- Carroll SB. Evolution at two levels: on genes and form. PLoS Biol. 2005;3:e245. doi: 10.1371/journal.pbio.0030245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamary JV, Parmley JL, Hurst LD. Hearing silence: non-neutral evolution at synonymous sites in mammals. Nat Rev Genet. 2006;7:98–108. doi: 10.1038/nrg1770. [DOI] [PubMed] [Google Scholar]
- Chuang JH, Li H. Functional bias and spatial organization of genes in mutational hot and cold regions in the human genome. PLoS Biol. 2004;2:E29. doi: 10.1371/journal.pbio.0020029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirillo LA, et al. Opening of compacted chromatin by early developmental transcription factors HNF3 (FoxA) and GATA-4. Mol Cell. 2002;9:279–289. doi: 10.1016/s1097-2765(02)00459-8. [DOI] [PubMed] [Google Scholar]
- Cliften P, et al. Finding functional features in Saccharomyces genomes by phylogenetic footprinting. Science. 2003;301:71–76. doi: 10.1126/science.1084337. [DOI] [PubMed] [Google Scholar]
- Cooper GM, et al. Distribution and intensity of constraint in mammalian genomic sequence. Genome Res. 2005;15:901–913. doi: 10.1101/gr.3577405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake JA, et al. Conserved noncoding sequences are selectively constrained and not mutation cold spots. Nat Genet. 2006;38:223–227. doi: 10.1038/ng1710. [DOI] [PubMed] [Google Scholar]
- Dreszer TR, Wall GD, Haussler D, Pollard KS. Biased clustered substitutions in the human genome: the footprints of male driven biased gene conversion. Genome Res. 2007;17:1420–1430. doi: 10.1101/gr.6395807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duret L, Galtier N. Comment on “human-specific gain of function in a developmental enhancer.”. Science. 2009;323:714c. doi: 10.1126/science.1165848. [DOI] [PubMed] [Google Scholar]
- Eberhard D, Jiménez G, Heavey B, Busslinger M. Transcriptional repression by Pax5 (BSAP) through interaction with corepressors of the Groucho family. EMBO J. 2000;19:2292–2203. doi: 10.1093/emboj/19.10.2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field Y, et al. Gene expression divergence in yeast is coupled to evolution of DNA-encoded nucleosome organization. Nat Genet. 2009;41:438–445. doi: 10.1038/ng.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galtier N, Duret L. Adaptation or biased gene conversion? Extending the null hypothesis of molecular evolution. Trends Genet. 2007;23:273–277. doi: 10.1016/j.tig.2007.03.011. [DOI] [PubMed] [Google Scholar]
- Gasch AP, et al. Conservation and evolution of cis-regulatory systems in ascomycete fungi. PLoS Biol. 2004;2:e398. doi: 10.1371/journal.pbio.0020398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs RA, et al. Genome sequence of the Brown Norway rat yields insights into mammalian evolution. Nature. 2004;428:493–521. doi: 10.1038/nature02426. [DOI] [PubMed] [Google Scholar]
- HapMapConsortium. A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardison RC, et al. Covariation in frequencies of substitution, deletion, transposition, and recombination during eutherian evolution. Genome Res. 2003;13:13–26. doi: 10.1101/gr.844103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedges SB, Kumar S. Genomic clocks and evolutionary timescales. Trends Genet. 2003;19:200–206. doi: 10.1016/S0168-9525(03)00053-2. [DOI] [PubMed] [Google Scholar]
- Hillier LW, et al. Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature. 2004;432:695–716. doi: 10.1038/nature03154. [DOI] [PubMed] [Google Scholar]
- Hogues H, et al. Transcription factor substitution during the evolution of fungal ribosome regulation. Mol Cell. 2008;29:552–562. doi: 10.1016/j.molcel.2008.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard T, et al. The Ensembl genome database project. Nucleic Acids Res. 2002;32:D468–D470. [Google Scholar]
- Ihmels J, et al. Rewiring of the yeast transcriptional network through the evolution of motif usage. Science. 2005;309:938–940. doi: 10.1126/science.1113833. [DOI] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium. Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–945. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- Kamnasaran D, Muir WJ, Ferguson-Smith MA, Cox DW. Disruption of the neuronal PAS3 gene in a family affected with schizophrenia. Med Genet. 2003;40:325–332. doi: 10.1136/jmg.40.5.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzman S, et al. Human genome ultraconserved elements are ultraselected. Science. 2007;317:915. doi: 10.1126/science.1142430. [DOI] [PubMed] [Google Scholar]
- Kaufmann E, Knochel W. Five years on the wings of fork head. Mech Dev. 1996;57:3–20. doi: 10.1016/0925-4773(96)00539-4. [DOI] [PubMed] [Google Scholar]
- Keightley PD, Lercher MJ, Eyre-Walker A. Evidence for widespread degradation of gene control regions in hominid genomes. PLoS Biol. 2005;3:e42. doi: 10.1371/journal.pbio.0030042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellis M, Patterson N, Endrizzi M, Birren B, Lander ES. Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature. 2003;423:241–254. doi: 10.1038/nature01644. [DOI] [PubMed] [Google Scholar]
- Kim TK, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M. The neutral theory of molecular evolution. Cambridge: Cambridge University Press; 1983. [Google Scholar]
- King MC, Wilson AC. Evolution at two levels in humans and chimpanzees. Science. 1975;188:107–116. doi: 10.1126/science.1090005. [DOI] [PubMed] [Google Scholar]
- Kryukov GV, Schmidt S, Sunyaev S. Small fitness effect of mutations in highly conserved non-coding regions. Hum Mol Genet. 2005;14:2221–2229. doi: 10.1093/hmg/ddi226. [DOI] [PubMed] [Google Scholar]
- Lareau LF, Inada M, Green RE, Wengrod JC, Brenner SE. Unproductive splicing of SR genes associated with highly conserved and ultraconserved DNA elements. Nature. 2007;446:926–929. doi: 10.1038/nature05676. [DOI] [PubMed] [Google Scholar]
- Lavoie H, et al. Evolutionary tinkering with conserved components of a transcriptional regulatory network. PLoS Biol. 2010;8:e1000329. doi: 10.1371/journal.pbio.1000329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lettice LA, et al. A long-range Shh enhancer regulates expression in the developing limb and fin and is associated with preaxial polydactyly. Hum Mol Genet. 2003;12:1725–1735. doi: 10.1093/hmg/ddg180. [DOI] [PubMed] [Google Scholar]
- Lee AP, Kerk SY, Tan YY, Brenner S, Venkatesh B. Ancient vertebrate conserved noncoding elements have been evolving rapidly in teleost fishes. Mol Biol Evol. 2008;28:1205–1215. doi: 10.1093/molbev/msq304. [DOI] [PubMed] [Google Scholar]
- Makova KD, Li WH. Strong male-driven evolution of DNA sequences in humans and apes. Nature. 2002;416:624–626. doi: 10.1038/416624a. [DOI] [PubMed] [Google Scholar]
- Mann RS. The specificity of homeotic gene function. BioEssays. 1995;17:855–863. doi: 10.1002/bies.950171007. [DOI] [PubMed] [Google Scholar]
- Mann RS, Affolter M. Hox proteins meet more partners. Curr Opin Genet Dev. 1998;8:423–429. doi: 10.1016/s0959-437x(98)80113-5. [DOI] [PubMed] [Google Scholar]
- Mann RS, Chan SK. Extra specificity from extradenticle: the partnership between HOX and PBX/EXD homeodomain proteins. Trends Genet. 1996;12:258–262. doi: 10.1016/0168-9525(96)10026-3. [DOI] [PubMed] [Google Scholar]
- Marais G. Biased gene conversion: implications for genome and sex evolution. Trends Genet. 2003;19:330–338. doi: 10.1016/S0168-9525(03)00116-1. [DOI] [PubMed] [Google Scholar]
- McGaughey DM, et al. Metrics of sequence constraint overlook regulatory sequences in an exhaustive analysis at phox2b. Genome Res. 2008;18:252–260. doi: 10.1101/gr.6929408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean C, Bejerano G. Dispensability of mammalian DNA. Genome Res. 2008;18:1743–1751. doi: 10.1101/gr.080184.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyata T, Hayashida H, Kuma K, Mitsuyasu K, Yasunaga T. Male-driven molecular evolution: a model and nucleotide sequence analysis. Cold Spring Harb Symp Quant Biol. 1987;52:863–867. doi: 10.1101/sqb.1987.052.01.094. [DOI] [PubMed] [Google Scholar]
- Mouse Genome Sequencing Consortium. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- Nei M. Molecular evolutionary genetics. New York: Columbia University Press; 1987. [Google Scholar]
- Ni JZ, et al. Ultraconserved elements are associated with homeostatic control of splicing regulators by alternative splicing and nonsense-mediated decay. Genes Dev. 2007;21:708–718. doi: 10.1101/gad.1525507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovcharenko I. Widespread ultraconservation divergence in primates. Mol Biol Evol. 2008;25:1668–1676. doi: 10.1093/molbev/msn116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennacchio LA, et al. In vivo enhancer analysis of human conserved non-coding sequences. Nature. 2006;444:499–502. doi: 10.1038/nature05295. [DOI] [PubMed] [Google Scholar]
- Pollard KS, et al. An RNA gene expressed during cortical development evolved rapidly in humans. Nature. 2006;433:167–172. doi: 10.1038/nature05113. [DOI] [PubMed] [Google Scholar]
- Potthoff MJ, Olson EN. MEF2: a central regulator of diverse developmental programs. Development. 2007;134:4131–4140. doi: 10.1242/dev.008367. [DOI] [PubMed] [Google Scholar]
- Poulin F, et al. In vivo characterization of a vertebrate ultraconserved enhancer. Genomics. 2005;85:774–781. doi: 10.1016/j.ygeno.2005.03.003. [DOI] [PubMed] [Google Scholar]
- Prabhakar A, Noonan JP, Pääbo S, Rubin EM. Accelerated evolution of conserved noncoding sequences in humans. Science. 2006;314:786. doi: 10.1126/science.1130738. [DOI] [PubMed] [Google Scholar]
- Prabhakar S, et al. Human-specific gain of function in a developmental enhancer. Science. 2008;321:1346–1350. doi: 10.1126/science.1159974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prendergast JG, et al. Chromatin structure and evolution in the human genome. BMC Evol Biol. 2007;7:72. doi: 10.1186/1471-2148-7-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt KD, et al. The consensus coding sequence (CCDS) project: identifying a common protein-coding gene set for the human and mouse genomes. Genome Res. 2009;19:1316–1323. doi: 10.1101/gr.080531.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauskolb C, Wieschaus E. Coordinate regulation of downstream genes by extradenticle and the homeotic selector proteins. EMBO J. 1994;13:3561–3569. doi: 10.1002/j.1460-2075.1994.tb06663.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roose J, et al. The Xenopus Wnt effector XTcf-3 interacts with Groucho-related transcriptional repressors. Nature. 1998;395:608–612. doi: 10.1038/26989. [DOI] [PubMed] [Google Scholar]
- Sabherwal N, et al. Long-range conserved non-coding SHOX sequences regulate expression in developing chicken limb and are associated with short stature phenotypes in human patients. Hum Mol Genet. 2007;16:210–222. doi: 10.1093/hmg/ddl470. [DOI] [PubMed] [Google Scholar]
- Sagai T, Hosoya M, Mizushina Y, Tamura M, Shiroishi T. Elimination of a long-range cis-regulatory module causes complete loss of limb-specific Shh expression and truncation of the mouse limb. Development. 2005;132:797–803. doi: 10.1242/dev.01613. [DOI] [PubMed] [Google Scholar]
- Schmidt D, et al. Five-vertebrate ChIP-seq reveals the evolutionary dynamics of transcription factor binding. Science. 2010;328:1036–1040. doi: 10.1126/science.1186176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- She X, et al. A preliminary comparative analysis of primate segmental duplications shows elevated substitution rates and a great-ape expansion of intrachromosomal duplications. Genome Res. 2006;16:576–583. doi: 10.1101/gr.4949406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherry ST, et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shore P, Sharrocks AD. The MADS-box family of transcription factors. Eur J Biochem. 1995;229:1–13. doi: 10.1111/j.1432-1033.1995.tb20430.x. [DOI] [PubMed] [Google Scholar]
- Siepel A, et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark A, et al. Discovery of functional elements in 12 Drosophila genomes using evolutionary signatures. Nature. 2007;450:219–232. doi: 10.1038/nature06340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephen S, Pheasant M, Makunin IV, Mattick JS. Large-scale appearance of ultraconserved elements in tetrapod genomes and slowdown of the molecular clock. Mol Biol Evol. 2008;25:402–408. doi: 10.1093/molbev/msm268. [DOI] [PubMed] [Google Scholar]
- Sumiyama K, Saitou N. Loss-of–function mutation in a repressor module of human-specifically activated enhancer HACNS1. Mol Biol Evol. 2011;28:3005–3007. doi: 10.1093/molbev/msr231. [DOI] [PubMed] [Google Scholar]
- Takahata N, Satta Y, Klein J. Divergence time and population size in the lineage leading to modern humans. Theor Popul Biol. 1995;48:198–221. doi: 10.1006/tpbi.1995.1026. [DOI] [PubMed] [Google Scholar]
- Tanay A, Regev A, Shamir R. Conservation and evolvability in regulatory networks: the evolution of ribosomal regulation in yeast. Proc Natl Acad Sci U S A. 2005;102:7203–7208. doi: 10.1073/pnas.0502521102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuch BB, Galgoczy DJ, Hernday AD, Li H, Johnson AD. The evolution of combinatorial gene regulation in fungi. PLoS Biol. 2008;6:e38. doi: 10.1371/journal.pbio.0060038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilella AJ, et al. EnsemblCompara GeneTrees: analysis of complete, duplication aware phylogenetic trees in vertebrates. Genome Res. 2008;19:327–335. doi: 10.1101/gr.073585.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wapinski I, et al. Gene duplication and the evolution of ribosomal protein gene regulation in yeast. Proc Natl Acad Sci U S A. 2010;107:5505–5510. doi: 10.1073/pnas.0911905107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegner M. From head to toes: the multiple facets of Sox proteins. Nucleic Acids Res. 1999;27:1409–1420. doi: 10.1093/nar/27.6.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegner M, Stolt CC. From stem cells to neurons and glia: a Soxist’s view of neural development. Trends Neurosci. 1995;28:583–588. doi: 10.1016/j.tins.2005.08.008. [DOI] [PubMed] [Google Scholar]
- Woolfe A, et al. Highly conserved non-coding sequences are associated with vertebrate development. PLoS Biol. 2005;3:e7. doi: 10.1371/journal.pbio.0030007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolfe A, et al. CONDOR: a database resource of developmentally-associated conserved non-coding elements. BMC Dev Biol. 2007;7:100. doi: 10.1186/1471-213X-7-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaklichkin S, Steiner AB, Lu Q, Kessler DS. FoxD3 and Grg4 physically interact to repress transcription and induce mesoderm in Xenopus. J Biol Chem. 2007;282:2548–2557. doi: 10.1074/jbc.M607412200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. “PAML: a program package for phylogenetic analysis by maximum likelihood.”. Comput Appl Biosci. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Schwartz S, Wagner L, Miller W. A greedy algorithm for aligning DNA sequences. J Comput Biol. 2000;7:203–214. doi: 10.1089/10665270050081478. [DOI] [PubMed] [Google Scholar]
- Zuckerkandl E, Pauling L. Evolutionary divergence and convergence in proteins. In: Bryson V, Vogel HJ, editors. Evolving genes and proteins. New York: Academic Press; 1965. pp. 97–166. [Google Scholar]