Abstract
To sense numerous odorants and chemicals, animals have evolved a large number of olfactory receptor genes (Olfrs) in their genome. In particular, the house mouse has ∼1,100 genes in the Olfr gene family. This makes the mouse a good model organism to study Olfr genes and olfaction-related genes. To date, whether male and female mice possess the same ability in detecting environmental odorants is still unknown. Using the next generation sequencing technology (paired-end mRNA-seq), we detected 1,088 expressed Olfr genes in both male and female olfactory epithelium. We found that not only Olfr genes but also odorant-binding protein (Obp) genes have evolved rapidly in the mouse lineage. Interestingly, Olfr genes tend to express at a higher level in males than in females, whereas the Obp genes clustered on the X chromosome show the opposite trend. These observations may imply a more efficient odorant-transporting system in females, whereas a more active Olfr gene expressing system in males. In addition, we detected the expression of two genes encoding major urinary proteins, which have been proposed to bind and transport pheromones or act as pheromones in mouse urine. This observation suggests a role of main olfactory system (MOS) in pheromone detection, contrary to the view that only accessory olfactory system (AOS) is involved in pheromone detection. This study suggests the sexual differences in detecting environmental odorants in MOS and demonstrates that mRNA-seq provides a powerful tool for detecting genes with low expression levels and with high sequence similarities.
Keywords: mRNA-seq, olfactory epithelium, olfactory receptor, odorant-binding protein, major urinary protein, sexual differentiation
Introduction
There are two olfactory systems in vertebrates: the main olfactory system (MOS) and the accessory olfactory system (AOS). MOS is composed of the main olfactory bulb and the olfactory epithelium (OE), while AOS is composed of the accessory olfactory bulb and the vomeronasal organ (VNO). OE is the main tissue for the expression of olfactory receptors (ORs), while VNO is the tissue for the expression of the vomeronasal receptors (VNRs). The detection of odorants and chemicals by ORs or by VNRs is transmitted to the brain by the main olfactory bulb or the accessory olfactory bulb, respectively. It is commonly thought that in mammals, ORs are responsible for recognizing environmental volatile odorants, such as food odors, whereas VNRs are responsible for detecting chemical cues related to social behaviors, such as pheromones (Touhara 2007). In this study, we examine whether there are sexual differences in the expression profiles of genes expressed in OE, particularly those involved in the odorant detection in mammals.
In the odorant perception process, hydrophobic molecules are transported through the nasal mucus by odorant-binding proteins (OBPs) to ORs. OBPs belong to the lipocalin protein family, which are extracellular transport proteins for carrying small hydrophobic molecules in aqueous solutions (see the review by Godfrey et al. 2004). They can be divided into subclasses based on the tissue specificity and the function in mammals such as OBPs and major urinary proteins (MUPs) (Flower et al. 2000). In mouse, three Obp genes (Obp1a, Obp1b, and Obp2) have been identified and all of them are located on the X chromosome (Bocskei et al. 1992; Briand, Blon, et al. 2004; Briand, Trotier, et al. 2004; Meslin et al. 2011). Obp1a and Obp1b form heterodimers to carry odorant molecules (Pes et al. 1998; Utsumi et al. 1999). In rat, Obp1, Obp1f, Obp2a, Obp2b, and Obp3 were identified (Briand et al. 2000). However, only Obp1 and Obp1f are located in the syntenic region of mouse Obp genes on the X chromosome.
The OR genes (Olfrs) form one of the largest gene families, composing of hundreds of copies in the mammalian genome. The existence of a large number of Olfr genes in a genome may confer the animal the ability to detect numerous kinds of odorants. It appears that the house mouse (Mus musculus), a nocturnal mammal, relies heavily on the sense of smell to perceive the environmental cues as it possesses ∼1,000 functional Olfr genes. The mouse Olfr genes have been classified into at least 241 subfamilies and are mapped to 51 loci distributed on 17 chromosomes with the largest cluster on chromosome 2 (Tegoni et al. 2000).
Sex-specific behaviors due to sex differences in responses to stimuli, such as colors, courtship songs, and chemosensory cues, have been found in animals (Godfrey et al. 2004). The chemosensory cues in animals include pheromone molecules, which are detected specifically by VNRs in mouse (Rubenstein and Lovette 2009). Sex-specific behaviors can be initiated by sex-specific pheromones (Kurtovic et al. 2007; Wyart et al. 2007; Haga et al. 2010) or by sex-differential expression of VNRs in response to the same pheromones (Haga et al. 2010; Roberts et al. 2010). For example, a male-specific pheromone, Darcin (the MUP 20, encoded by Mup20), in the mouse urine only attracts females and stimulates their memory (Herrada and Dulac 1997). As another example, both male and female mice detect exocrine-secreted peptide 1 (ESP1), but only female-specific mating behaviors are stimulated (Roberts et al. 2010), indicating the existence of gender-specific neuronal circuits. In contrast, no evidence of sexual differences in MOS has yet been reported. Thus, it remains unclear whether males and females have the same ability in sensing odorants.
To address the question of sexual differentiation in MOS, an approach with a high resolution and great sensitivity in estimating mRNA levels is required. Previous authors have used oligonucleotide microarrays and single-read mRNA-seq to study the expression of Olfr genes in OE and in matured olfactory sensory neurons (OSNs), respectively (Haga et al. 2010; Magklara et al. 2011). However, the hybridization-based gene expression profiling methods are not ideal for the studies of Olfr genes for two reasons: First, a series of recent expansions of the Olfr gene family have produced many Olfr gene pairs with a very high sequence similarity. Second, Olfr genes are expressed in a “one neuron one receptor” manner (Zhang et al. 2004), leading to extremely low gene expression levels per unit tissue weight. For tissue-specific genes involving high possibilities of cross-hybridization between probes on arrays (Serizawa et al. 2004), a sequencing approach is more suitable for specific quantifications. In addition, the expression profiles of matured OSNs, where Olfr genes are expressed predominantly, do not show the complete expression of Obp genes. Hence, we used the Illumina paired-end mRNA-seq to obtain the transcriptomes of OE in both sexes, which are suitable for studying the expression patterns of Olfr genes and Obp genes.
Materials and Methods
Mice Collection and Total RNA Isolation
All the animals used in this study were processed following the approved protocol of Institutional Animal Care and Use Committee at the National Laboratory Animal Center (NLAC), Taipei, Taiwan.
Pups from brother–sister matings of the BALB/cByJNarl (BALB/c, a common inbred strain), from NLAC were used for this study. To minimize environmental variations between the two sexes, we sacrificed mature pups at 4 weeks of age and isolated their OE tissue before male and female individuals were separated. Only litters with at least three males or three females were selected. The nasal tissue was isolated and preserved in RNALater solution (Ambion) immediately after sacrification. The tissue was allowed for penetration by RNALater solution at 4 °C overnight and then transferred to −20 °C before further isolation of total RNA. OE was dissected from the nasal tissue followed by total RNA isolation, using RNEasy Plus Mini Kit (Qiagen) with an additional on-column DNase treatment recommended by the manufacture (Qiagen). The 15 min DNase treatment was carried out at room temperature by mixing 10 ul DNase and 70 ul RDD buffer and applied to the RNA binding column after the first wash. The RNA quantities and qualities of each individual were analyzed by Nanodrop and BioAnalyzer II (Agilent). If all three samples from the same litter passed the quality control (RNA integrity number [RIN] > 8.0), 10 ug of total RNA from each sample would be pooled to reach final of 30 ug total RNA for sequencing for each sex.
RNA samples from the OE of C57BL/6JNarl (B6, from NLAC, Taiwan) were used to confirm the expression of MUP genes. We collected tissues of livers, spleens, kidneys, testes, ovaries, hearts, and lungs for RNA isolation to confirm the expression of Mup genes. Total RNA was isolated with the same approach described above.
Paired-End mRNA-seq with the Illumina Genome Analyzer IIx
For pair-end mRNA-seq library preparation, we used Illumina mRNA-seq kits. A total of 30 ug total RNA was used for mRNA enrichment by oligo-dT beads followed by cation-catalyzed fragmentation for 4 min at 94 °C. The mRNA fragments were then converted into double stranded cDNA by random priming followed by end repair. The fragmented cDNAs were then ligated to the paired-end adaptors and subjected to size selection. For each pooled RNA sample, three sizes of ∼400 bp, ∼500 bp, and ∼550 bp were selected for the ligated cDNA. The three-gel purified cDNA libraries were then subjected to 15 cycles of polymerase chain reaction (PCR) amplification and purified by Ampure beads (Beckman Agencourt). The absolute concentrations of the libraries were determined by Qubit fluorometry (Invitrogen) and BioAnalyzer High Sensitivity DNA Kit (Agilent). Each size selected mRNA-seq library was loaded in one lane of flow cell and paired-end 2 × 120 nt sequencing was conducted on Illumina Genome Analyzer IIx, totaling three lanes of data per pooled transcriptome. Library preparation and Illumina sequencing was conducted by High Throughput Sequencing Core Facility, Biodiversity Research Center, Academia Sinica, Taiwan.
Analysis of Paired-End Sequences
We trimmed all the paired-end sequencing reads from both ends of each cDNA fragment to 90 bp to reduce sequencing errors. Paired-end sequencing reads were mapped to the genome with TopHat (version 1.2.0) (Qian et al. 2010; Xiong et al. 2010; Chang and Liao 2011), allowing a 100-bp standard deviation for a mean inner distance of 370 bp between paired reads from both ends. Only those pair-end reads mapped to the genome without mismatch were used for subsequent analyses. We first categorized mappable fragments into two groups: “unique” fragments, each of which was mapped to a single position in the genome, and “multiple-hit” fragments, each of which was mapped to more than one position in the genome. To calculate the expression levels, unique fragments were assigned to individual gene first for initial abundance estimation, and the multiple-hit fragments were then redistributed to those genes based on the relative abundances of uniquely mapped fragments. The total mappable fragments increased 2% by including redistributed multiple-hit fragments. The normalized expression levels of genes, measured in fragments per kilobase of exon per million fragments mapped (FPKMs), were calculated using Cufflinks (version 1.0.3) (Trapnell et al. 2009). Total mappable fragments on each chromosome were calculated by SAMtools (Trapnell et al. 2010).
The mouse reference genome was from one of the common lab inbred strains, C57BL/6 (B6), while our transcriptomes were generated from another inbred strain, BALB/c. Single-nucleotide polymorphic sites between the two strains might lead to biased estimation of the expression level of transcripts. We therefore used a data set of 72-bp sequences generated by single-end mRNA-seq to confirm that there was no bias in estimation of expression levels when we mapped BALB/c cDNA sequences to the B6 genome (supplementary data, including table S6, Supplementary Material online).
Reverse Transcription PCR of Mup Genes and Quantitative Reverse Transcription PCR of Obp Genes
A total of 2 ug RNA of each sample was reverse transcribed with MultiScribe Reverse Transcriptase (Life Technologies) into cDNA for both Reverse Transcription PCR (RT-PCR) and Quantitative Reverse Transcription PCR (qRT-PCR) reactions. Total RNA was incubated with RT enzymes at 25 °C for 10 min prior to the RT reaction. RT reactions were performed at 37 °C for 2 h followed by the inactivation of RT enzyme at 85 °C for 10 s. For RT-PCR, 1 μl of 10× diluted cDNA was amplified by Fast Start Taq DNA polymerase (Roche) in a total of 10 ul reaction. For qRT-PCR, 1 μl of 10× diluted cDNA products was quantified with 2× SYBR Green Master Mix (Kapa Biosystems) in a total of 10 ul reaction and performed on LightCycler 480 (Roche).
The primers used in this study are as follows:
Actb, 5′-CCA GTT CGC CAT GGA TGA CGA TAT-3′ and 5′GTC AGG ATA CCT CTC TTG CTC TG-3'. Omp, 5′-GAA GCA GGA TGG TGA GAA GC-3′ and 5′-CGT GTC ATG AGG TTG GTG AG-3′. Mup4, 5′-AGA AGG ACG TGG TCC TGA CA-3′ and 5′-TAA GTT CTG TCC CTT GGA AG-3′. Mup5, F: 5′-GAA AGA CCT GGT ACT GAG AG-3′ and 5′-CTA GCT TCT TCT GCA TGG AC-3′. Gm14743, F: 5′-GGCATTCCAGCTGGAAACCTTAA-3′ and 5′-CTTATGCTGTATCCTCACTT-3′. Obp1b, F: 5′-GGCATTCCAGCTGGAAACCTTAG-3′ and 5′-CTTATGCTGTATCATCACTG-3′. Gm5938, 5′-ATATGCTGTGTCAAGCCACA-3′ and 5′-TAACAGGTCGTAGATCATGAG-3′. Obp1a, 5′-AAGGGAATTCCAGCTGGAAA-3′ and 5′-GGAAGATCATGAGAAGGGGAA-3′. 5430402E10Rik, 5′-GGTGAAGTTCCTGCTAATTGTGA-3′ and 5′-CATCTGGACATGGAATTTGAC-3′. Obp2 (Gm14744), 5′-GGTGAAGTTCCTGCTAATTGCGC-3′ and R: 5′-CATCTGGACATGGAATTTGAT-3′.
Functional Enrichment and Evolutionary Analyses
We looked for functionally enriched gene ontology (GO) terms in sex-biased genes against the rest of expressed genes (FPKMs > 0.05) in OE, using FatiGO (version 3.2) (Li et al. 2009). The significance of differential expression between the two sexes of a given gene was tested by the t-test, using the FPKMs obtained from three cDNA libraries of each sex.
The protein sequence of rat OBP1 was retrieved from the Ensembl database (ENSRNOP00000049161), and that of rat OBP1f was obtained from Briand et al. (Al-Shahrour et al. 2004). The protein sequences, including the hypothetical proteins, of mouse OBPs were retrieved from Ensembl database. Multiple sequence alignment was conducted by ClustalW2 (http://www.ebi.ac.uk/Tools/msa/clustalw2/) and a neighbor-joining tree was constructed using MEGA5 (Tamura et al. 2011).
Results
Transcriptome Data
We used Illumina GA IIx to obtain the OE transcriptomes from one male sample and one female sample separately. Each sample was a pool of mRNA from three littermates of one gender. Three cDNA libraries with insert lengths ranging from 400 to 550 bp were constructed for each sample. In total, we obtained ∼168 million and ∼203 million paired 120-bp reads in the male OE and in the female OE, respectively. From these reads, we obtained 138.9 million mappable-paired fragments for the male OE and 163.6 million for the female OE, corresponding to 83% and 81% mappable rates, respectively. Overall, we detected ∼21,000 genes with FPKM > 0.05 (supplementary table S1, Supplementary Material online). In general, the transcriptomes of the male OE and the female OE were highly correlated (Pearson correlation coefficient, r = 0.94, P< 10−15).
Male-Biased Expression of Olfr Genes in Mice
Among the 1,196 Olfr genes in the mouse genome, we detected 1,088 expressed Olfr genes in both male and female OEs (supplementary table S2, Supplementary Material online). Almost all Olfr genes were found to be very lowly expressed in the transcriptomes (FPKM < 15). The expression levels of Olfr genes were highly correlated between the two sexes (r = 0.96, P < 10−15). Overall, 12 Olfr genes have no detectable expression in the female OE and six have no detectable expression in the male OE. However, the highest expression level of these genes is only 1.82 FPKM and most of them are lower than 0.05 FPKMs (supplementary table S2, Supplementary Material online). Therefore, these “sex-specific” Olfr genes may not contribute to sex differences in function. In addition to functional Olfr genes, we detected the expression of 57 of the 114 Olfr pseudogenes annotated in the mouse genome.
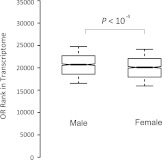
To address the question of whether there are differences in Olfr expression levels between the two sexes, we compared the ranked expression levels of Olfr genes in the two OE transcriptomes. The medians of ranked expression levels of Olfr genes are around 20,000 among all 25,000 genes in both sexes (fig. 1). We made the following interesting observations. First, Olfr genes tend to be expressed at a higher level in males than in females (fig. 1, P < 10−4). Second, all of the 254 Olfr genes that showed sexual preference were expressed at a higher level in the male OE (supplementary table S3, Supplementary Material online). Third, the proportion of male-biased Olfr genes (254/1,088) is significantly higher than the proportion of the male-biased genes in the OE transcriptome (3,884/19,934) (Chi-square = 5.49, P = 0.02). Fourth, although chromosome 2 has the largest Olfr gene cluster, chromosome 7 showed the highest proportion of male-biased Olfr genes (86/228, supplementary table S3, Supplementary Material online). In conclusion, Olfr genes tend to be more strongly expressed in the male OE than in the female OE.
FIG. 1.—
Comparison of ranked expression levels of Olfr genes in the OE transcriptomes of male and female mice. The values of upper quartile, median, and lower quartile are indicated in each box, and the bars outside the box indicate semiquartile ranges. Male Olfr genes tend to express at a higher level (Mann–Whitney U test, P < 10−4).
We further confirmed the sexually biased expression of Olfr genes by examining gene enrichments in GO categories (table 1). We identified a total of 4,300 genes with a significant expression difference between the two sexes. The enrichment analysis shows that these genes involve in biological processes, molecular function, and cellular components (supplementary table S5, Supplementary Material online), particularly in sensory perception of smell, transmembrane receptor activity, and G-protein–coupled receptor activity categories (table 1). Almost 60% of genes in the categories of sensory perception of smell (GO:0007608), transmembrane receptors (GO:0004888) and G-protein–coupled receptors (GO:0004930) show different expression levels between the two sexes. As ORs are G-protein coupled receptors with seven transmembrane domains, the results confirm the differential expression of Olfr genes.
Table 1.
GO Categories Enriched with Sexually Biased Genes
| Functional Categories | GO | Sex-Biased Genes (%) | P |
| Sensory perception of smell | 0007608 | 59.17 | 0.00 |
| Transmembrane receptor activity | 0004888 | 55.87 | 0.00 |
| G-protein–coupled receptor activity | 0004930 | 55.22 | 0.00 |
NOTE.—The proportion of sex-biased genes in each category is shown in percentage (%). For the complete list of GO categories enriched or depleted with sex-biased genes, please see supplementary table S5 (Supplementary Material online).
Female-Biased Expression of X-Linked Obp Genes
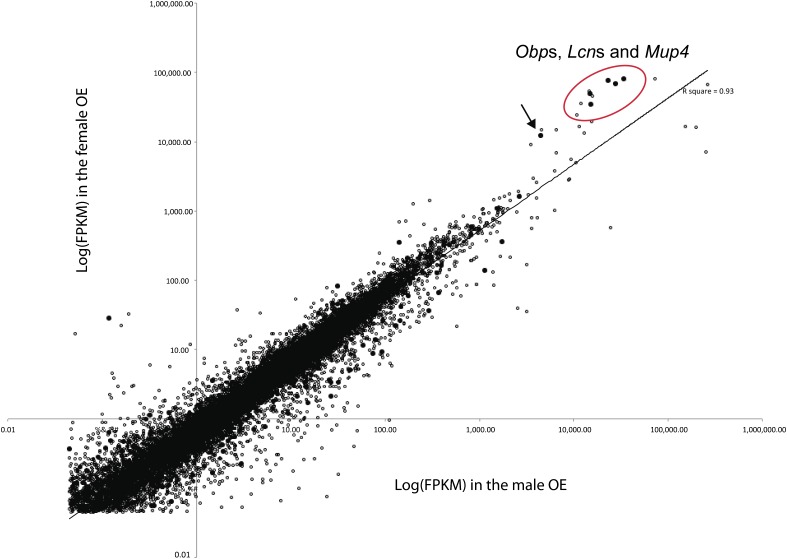
Comparing the paired-end fragments mappable to each chromosome, we found that the X chromosome had 2.8 times more fragments in the female OE than in the male OE (supplementary table S4, Supplementary Material online). A comparison of the expression levels of individual genes on each chromosome revealed that the higher expression of the X chromosome in the female OE was due to five Obp genes expressed abundantly in OE (fig. 2). Removing the FPKMs of these five genes from the transcriptomes resulted in a nonbiased expression pattern of the X-linked genes.
FIG. 2.—
Expression levels of each gene on autosomes and on chromosome X between the two sexes. The FPKM values were logarithmically transformed. The filled dots represent the genes on the X chromosome, whereas the opened dots represent the genes on autosomes. Only genes with detectable expression levels (FPKMs > 0.05) are shown. We found that the higher female mappable fragments of X-linked genes were due to five X-linked Obp genes expressed significantly higher in the female OE (grouped in the red color oval-shaped circle). Highly female-biased genes also included autosomal lipocalin protein genes (Lcn11, Lcn13, and Lcn14) and Mup4. The arrow indicates a highly expressed female-biased pseudogene (ENSMUSG00000082635) located in the X-linked Obp gene cluster.
The five genes are located in a 0.7-Mb region of the X chromosome. Two of them encode subunits of OBP1, Obp1a and Obp1b; the other three genes (Gm14743, Gm14744, and 5430402E10Rik) have unknown functions. The predicted protein sequence of Gm14744 was identical to the OBP2 identified previously, so we denoted this gene as Obp2. We further found that the Gm5938 gene is located within the 0.7-Mb region and shows sequence homology to Obp1a; however, it has very low expression level compared with the other five Obp genes. As the 0.7-Mb region is enriched with Obp genes, we name it the “X-linked Obp gene cluster.”
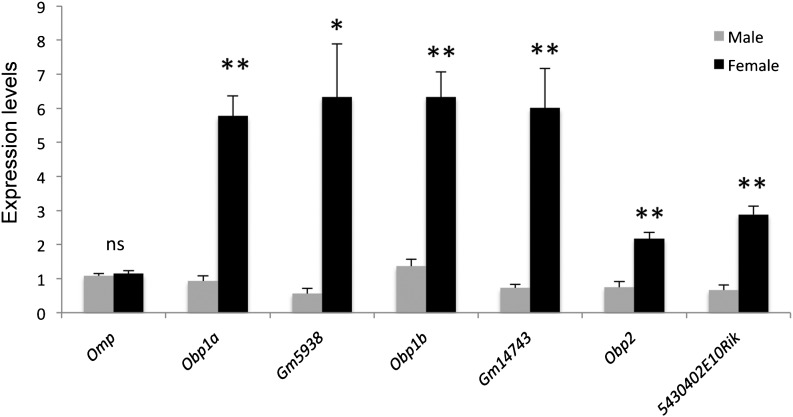
All the genes in the X-linked Obp gene cluster were highly expressed in both sexes and showed a female-biased expression pattern (table 2). We confirmed the female-biased expression patterns of all the six genes in at least three mice by qRT-PCR assay (fig. 3). The nonbiased expression of Gm5938 based on qRT-PCR may be because its expression level is very low, so that it is difficult to detect a difference by qRT-PCR. No female-biased expression was found in the genes located in the flanking regions of this 0.7-Mb region. In particular, probasin (Pbsn), which was proposed to be a member of the Obp gene family with a prostate-specific expression pattern (Utsumi et al. 1999), is located 27-Kb away from the gene cluster (table 2). We concluded that the female-biased expression pattern is specific to the X-linked Obp gene cluster.
Table 2.
Female-Biased Expression Patterns of Genes in the X-Linked Obp Gene Cluster
| Gene Name | Chromosome Location | Chromosome Strand | Male (FPKM) | Female (FPKM) | M/F Ratios | P |
| 0.7-Mb region | ||||||
| Obp1a | 75,330,843–75,336,713 | −1 | 27,620.10 | 69,529.40 | 0.40 | ** |
| Obp1b | 75,432,115–75,437,631 | 1 | 23,272.30 | 75,798.30 | 0.31 | ** |
| Gm14743 | 75,485,800–75,491,260 | −1 | 15,287.80 | 34,579.90 | 0.44 | * |
| Gm5938 | 75,370,805–75,375,742 | 1 | 140.12 | 348.02 | 0.40 | ** |
| Obp2 (Gm14744) | 75,110,096–75,115,372 | 1 | 14,899.80 | 49,584.20 | 0.30 | ** |
| 5430402E10Rik | 75,233,629–75,238,906 | 1 | 34,157.80 | 80,668.10 | 0.42 | ** |
| Flanking genes | ||||||
| Prkx | 75,006,749–75,041,617 | −1 | 27.75 | 14.51 | 1.91 | * |
| Pbsn | 75,083,239–75,098,962 | −1 | 0.21 | 0.40 | 0.52 | ns |
| Prrg1 | 75,715,282–75,829,194 | −1 | 1.95 | 1.48 | 1.32 | ns |
| ENSMUSG00000071735 | 76,511,897–76,564,418 | −1 | 1.97 | 1.43 | 1.38 | ns |
NOTE.—The significance of male (M)-to-female (F) FPKM ratios (M/F ratios) is shown as **P < 0.01, *P < 0.05, and ns: P > 0.05.
FIG. 3.—
qRT-PCR confirmed the female-biased expression of X-lined Obp genes of BALB/c mice at 4 weeks of age. The figure shows the expression levels of each X-linked Obp genes and olfactory major protein gene (Omp, a marker gene of matured OSNs) estimated by qRT-PCRs in the two sexes. At least three mice were used in the qRT-PCR assay. The error bars show the standard errors of gene expression levels among all the OE samples tested. The expression levels are significantly different between the two sexes: **P < 0.01, *P < 0.05; the expression levels are not different between the two sexes: ns, P > 0.05.
Rapid Evolution of Mouse Obp Genes
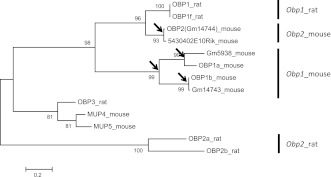
We found that Obp genes clustered on the X chromosome evolved rapidly, while maintaining multiple functional duplicate copies in the mouse genome. A multiple sequence alignment confirmed that the hypothetical proteins Gm5938, Gm14743, and 5430402E10Rik share sequence similarities with OBP1a, OBP1b, and OBP2, respectively (supplementary fig. S1A, Supplementary Material online). A phylogenetic tree constructed from the protein sequences of the OBPs and MUPs expressed in OE revealed that at least four duplication events have occurred in the Obp gene cluster (fig. 4). One of the duplications resulted in the genes for the two Obp1 subunits, Obp1a and Obp1b. The other three duplications occurred independently in the mouse lineage, leading to the existence of three pairs of paralogous Obp genes: 1) Obp1a and Gm5938, 2) Obp1b and Gm14743, and 3) Obp2 and 5430402E10Rik. The branch lengths indicate that the paralog pairs, Obp1b/Gm14743 and Obp2/5430402E10Rik, originated more recently than Obp1a/Gm5938. Only one and two amino acid differences were found between Obp1b/Gm14743 and between Obp2/5430402E10Rik, respectively.
FIG. 4.—
Phylogenetic tree constructed with protein sequences of OBPs and MUPs in mouse and rat. The maximum likelihood method was used with 500 bootstrap replications. The bootstrap values are shown on the figure. The arrows indicate duplication events that occurred within the mouse genome.
In rat, only Obp1 and Obp1f were identified in the syntenic region of the X-linked Obp gene cluster. The syntenic region is flanked by Pbsn and Proline rich Gla (G-carboxyglutamic acid) 1 (Prrg1) in both rat and mouse (supplementary fig. S1B, Supplementary Material online). However, only one duplication event was observed in the syntenic region on the X chromosome of rat, which resulted in Obp1 and Obp1f. Thus, we concluded that the rapid evolution of Obp gene cluster was specific to the mouse lineage.
The three novel genes (Gm14743, Gm5938, and 5430402E10Rik) also maintained female-biased expression patterns based on the NGS data, as did their paralogs. This is likely due to the conservation of the upstream regulatory sequences. The upstream sequences of Obp1b and Gm14743 are conserved up to 5 kb (the number of nucleotide differences per site, π = 0.08), and the upstream sequences of Obp2 and 5430402E10Rik are conserved up to 6 kb (π = 0.10). However, the upstream region of Gm5938 has many insertions and deletions and shows π = 0.43 for about 4 kb compared with that of Obp1a. The loss of conservation in the regulatory region might explain the relatively low expression of Gm5938 in OE. We also detected high expression levels of three Lcn genes, Lcn11, Lcn13, and Lcn14. The three genes are located on chromosome 2 within a cluster of Lcn genes. The abundant expression was only detected in the three genes but not in the others (fig. 2). Interestingly, the three genes also showed a significant higher expression level in the female OE. Although the function of these genes has not been reported in the olfactory systems, Lcn14 was proposed to be an ortholog of OBP2A in human (Mouse Genome Informatics, The Jackson Laboratory). It is possible that there are more proteins that function as OBPs in MOS in order to increase the diversity in transporting various kinds of odorants.
Expression of Mup Genes in OE
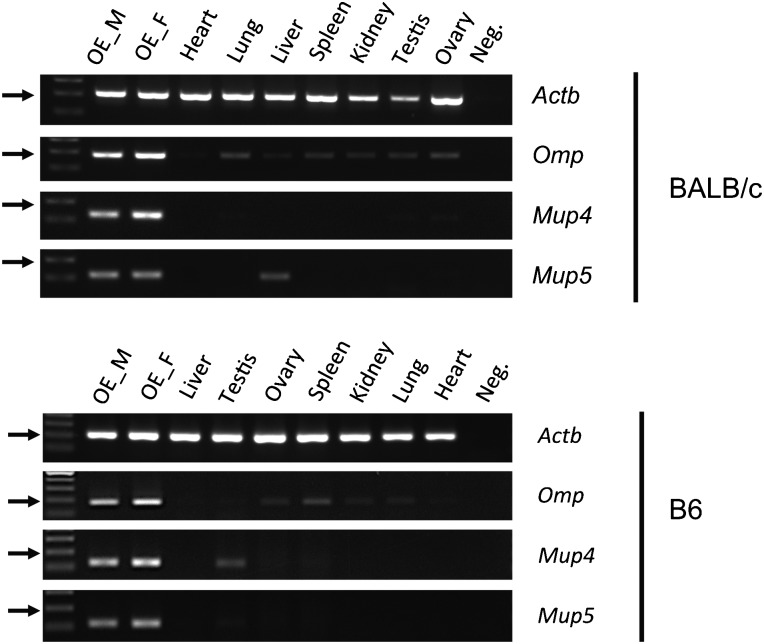
Among Mup genes, including 21 Mup functional genes and the 21 Mup pseudogenes, we detected the expression of Mup4 and Mup5 in OE but not other Mup genes. These two genes are expressed abundantly; they are among the 50 highest expressed genes in OE (fig. 2 and supplementary table S1, Supplementary Material online). The FPKMs of Mup4 are 10,720.6 and 24419.60 in males and in females, respectively. The FPKMs of Mup5 are 6591.54 and 6840.84 in males and in females, respectively. As MUPs were proposed to carry and transport pheromones in urine and/or act as pheromones themselves (Johnson et al. 2000), our result implies a possible role of MOS in pheromone perception. We examined the expression patterns of the two genes in various types of tissue from two laboratory-inbred strains, BALB/c and B6, by RT-PCR. We found that Mup4 and Mup5 were mainly expressed in the OE of both sexes in both strains (fig. 5). In addition, the expression patterns of Mup4 and Mup5 were identical in juvenile (4 weeks old) and adult (6 weeks old) mice (data not shown). Thus, the expression patterns of Mup4 and Mup5 in OE are not sex, age, or strain specific.
FIG. 5.—
RT-PCR confirmed the expression of Mup4 and Mup5 in OE. Expression of Omp, Mup4, and Mup5, in various tissues of the two inbred strains. The β-actin gene (Actb) was used as the positive control. OE_M indicates the male OE, and OE_F indicates the female OE. The expression patterns of Mup4 and Mup5 suggest that these two genes are mainly expressed in OE.
Discussion
To obtain a deep coverage of lowly expressed genes, particularly Olfr genes, three cDNA libraries with different insert sizes were constructed for each RNA sample. We found that the expression levels of a given gene estimated from the three cDNA libraries were highly correlated (supplementary fig. S2, Supplementary Material online). Furthermore, the ranked expression levels of Olfr genes estimated from three cDNA libraries were not significantly different, except for the library with the shortest fragment sizes in the male OE (supplementary fig. S3, Supplementary Material online). Toung et al. (Beynon and Hurst 2003; Roberts et al. 2010) suggested that a minimum of 500 million reads is required to estimate FPKMs accurately for a human B cell transcriptome. In this study, we found no significant difference between the FPKMs of Olfr genes estimated from a single cDNA library and those estimated from the pool of three libraries (supplementary fig. S4, Supplementary Material online). Therefore, it seems that a sample of ∼80 million 120-bp paired-end reads can provide unbiased estimation of gene expression levels for the mouse OE transcriptome.
Previous studies have used array-based and sequencing-based approaches to estimate the expression levels of Olfr genes in OE or matured OSNs (Toung et al. 2011). Conducting single-end mRNA-seq, Magklara et al. (2011) showed that the expression of Olfr genes ranged from 0 to 46 RPKMs in matured OSNs (only 19 Olfrs had RPKMs > 15, whereas 717 Olfrs had RPKMs >1). Our study shows similar results in the expression levels of Olfr genes with Magklara et al. (2011), that is, Olfr genes are lowly expressed, consistent with the “one neuron one receptor” hypothesis
Overall, our study has detected more expressed Olfr genes than the previous studies and has identified Olfr genes and novel duplicate Obp genes that are expressed differentially between the two sexes. Thus, mRNA-seq, particularly paired-end technology, provides a high resolution in estimating expression levels of genes from highly duplicated gene families, such as Olfr and Obp genes.
Sexual Differences in the Olfactory Systems
Sexual dimorphisms have been found in the olfactory systems of mouse and fly. In mice, sexual dimorphisms were found in AOS, where pheromones are perceived by VNRs (Zhang et al. 2004; Clowney et al. 2011; Magklara et al. 2011). In flies, olfaction is mediated by OSNs in sensilla of the third antennal segment and the maxillary palps. Each OSN usually expresses one (sometimes two) of the 62 Olfr genes, and OBPs are secreted in the perilymph by support cells in the sensilla. Unlike vertebrates, ORs and OBPs in flies are responsible for sensing both odorants and pheromones. Although Zhou et al. (Kurtovic et al. 2007; Wyart et al. 2007; Haga et al. 2010) showed sexual dimorphic expression patterns of Obp genes in flies, the dimorphic patterns varied among genes. That is, some of the Obp genes were expressed at a higher level in males and some were expressed at a higher level in females. In contrast, in our study all of the Obp genes and the other Lcn genes showed a consistently higher expression level in female mice at 4 weeks of age, whereas Olfr genes tend to have a higher expression level in males. The high expression of Obp genes in both sexes indicates an important function of OBP in OE. However, it requires experiments to test whether the differential expression in Obp genes lead to differences in the ability of smelling odorant between the two sexes. These observations raise an interesting question as to whether sexual dimorphism exists in other inbred lab strains or in the wild house mice, and what the advantage of evolving dimorphic expression patterns in MOS is. Further research is required to address these questions.
Rapid Evolution of Obp Genes
We found that several duplication events have produced multiple copies of Obp genes in the X-lined Obp gene cluster in the house mouse, increasing the potential in transporting more kinds of odorants. We noted that these genes have evolved rapidly, similar to the rapid evolution of Lcn genes observed in other species (Zhou et al. 2009).
Several models have been proposed for the evolution of duplicate genes (Stopkova et al. 2010; Meslin et al. 2011). One model proposes that duplicate genes tend to reduce their expression levels in order to achieve dosage balances, that is, the same amount of proteins would be produced before and after the duplication (Lynch and Force 2000; Shiao et al. 2008; Kaessmann et al. 2009), especially for genes involved in the protein complexes (Qian et al. 2010). In mouse, OBP1 is a heterodimer protein formed by two subunits, OBP1a and OBP1b. We found that each subunit has a duplicate copy in the mouse genome and both are expressed. According to the above model, the duplicate genes, Gm5938 (Obp1a-dup) and Gm14743 (Obp1b-dup), and the parental genes, Obp1a and Obp1b, should all reduce their expression levels to rebalance the dosage of OBP1. However, Obp1a, Obp1b, and Obp1b-dup were expressed at high levels, whereas Obp1a-dup was expressed at an extremely low level. It is possible that Obp1a-dup is undergoing pseudogenization. For this possibility, we note the great sequence divergence between Obp1a-dup and Obp1a.
A pseudogene, Gm14750 (ENSMUSG00000082635), in the X-linked Obp gene cluster was highly expressed in OE (male FPKM = 4,463, female FPKM = 12,172). The pseudogene resides between Obp2 and 5430402E10Rik (Obp2-duplicate) on the X chromosome and was determined as a pseudogene because of a prematured stop codon. Interestingly, even though not sharing sequence homology with the Obp genes in the cluster, Gm14750 showed a female-biased expression pattern as well (fig. 2). Although pseudogenes are not functional in most cases, it has been proposed that they may play a role in regulating their paralogous partners (Papp et al. 2003; Yang et al. 2003; Chang and Liao 2011) and a role in pathological defense in bacteria (Lim et al. 2004). The expression of Gm14750 indicates that all of the genes in the X-linked Obp gene cluster share the same regulation mechanism.
A Potential Role of MOS in Pheromone Perception
We found that the expression of Mup4 and Mup5 is not strain, age, or sex specific in OE. MUPs were found in urine and a number of secretary glands, such as lachrymal, mammary, and salivary glands (Zhou et al. 2009). In liver, MUPs are produced and secreted into serum, which is followed by a rapid excretion into urine. It was proposed that male urine has about 3 times more MUPs than female urine (Shaw et al. 1983; Shahan et al. 1987; Stopka et al. 2007). One of the function of MUPs is to bind to low molecular weight volatile pheromones and affect their transport and release of pheromones in VNO (Stopka et al. 2007). Another function is that they act as pheromones and regulate sex-specific social behaviors (Beynon and Hurst 2003). Furthermore, MUPs were only found in the urine of matured individuals. The expression of Mup4 and Mup5 has been identified in the nasal tissue in mouse (Roberts et al. 2010), and we found them to be expressed in the OE of juvenile and adult mice. The abundant expression of Mup4 and Mup5 based on our data indicate that the two genes may play an important role in MOS.
One possible explanation of the expression of two Mups in OE is that these two proteins bind to odorant molecules in nasal and act as OBPs, as suggested by Utsumi et al. (1999). We compared the protein sequences of MUP4 and MUP5 with other OBPs in mouse and found that the protein sequences of MUPs have highly diverged from OBPs (fig. 4). Thus, MUP4 and MUP5 are very likely to have been functionally differentiated from the OBPs. Furthermore, the activation of accessory olfactory bulbs was observed when OE was stimulated by certain odorants, and it was abolished in response to urine when OE underwent a lesion treatment (Utsumi et al. 1999). The above evidence indicates a role of MOS in pheromone detection.
Supplementary Material
Supplementary figures S1–S4 and tables S1–S6 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
Thanks to Jiun-Jie Chen, Chi-Wei Chang, and Yi-Wei Huang for providing experimental assistance that has contributed to the data reported in this paper. Dr Michael Baum at Boston University provided inspiring discussions for the study. This work was supported by Academia Sinica and National Science Council (NSC) of Taiwan (NSC 99-2628-B-001-009-MY3, NSC 99-2311-B-400-003-MY2, NSC 100-2319-B-400-001, and NSC 98-2311-B-492-001-MY3).
References
- Al-Shahrour F, Diaz-Uriarte R, Dopazo J. FatiGO: a web tool for finding significant associations of Gene Ontology terms with groups of genes. Bioinformatics. 2004;20:578–580. doi: 10.1093/bioinformatics/btg455. [DOI] [PubMed] [Google Scholar]
- Beynon RJ, Hurst JL. Multiple roles of major urinary proteins in the house mouse, Mus domesticus. Biochem Soc Trans. 2003;31:142–146. doi: 10.1042/bst0310142. [DOI] [PubMed] [Google Scholar]
- Bocskei Z, et al. Pheromone binding to two rodent urinary proteins revealed by X-ray crystallography. Nature. 1992;360:186–188. doi: 10.1038/360186a0. [DOI] [PubMed] [Google Scholar]
- Briand L, Blon F, Trotier D, Pernollet JC. Natural ligands of hamster aphrodisin. Chem Senses. 2004;29:425–430. doi: 10.1093/chemse/bjh044. [DOI] [PubMed] [Google Scholar]
- Briand L, Trotier D, Pernollet JC. Aphrodisin, an aphrodisiac lipocalin secreted in hamster vaginal secretions. Peptides. 2004;25:1545–1552. doi: 10.1016/j.peptides.2003.10.026. [DOI] [PubMed] [Google Scholar]
- Briand L, et al. Ligand-binding properties and structural characterization of a novel rat odorant-binding protein variant. Eur J Biochem. 2000;267:3079–3089. doi: 10.1046/j.1432-1033.2000.01340.x. [DOI] [PubMed] [Google Scholar]
- Chang AY, Liao BY. DNA methylation rebalances gene dosage after mammalian gene duplications. Mol Biol Evol. 2011;29:133–144. doi: 10.1093/molbev/msr174. [DOI] [PubMed] [Google Scholar]
- Clowney EJ, et al. High-throughput mapping of the promoters of the mouse olfactory receptor genes reveals a new type of mammalian promoter and provides insight into olfactory receptor gene regulation. Genome Res. 2011;21:1249–1259. doi: 10.1101/gr.120162.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flower DR, North AC, Sansom CE. The lipocalin protein family: structural and sequence overview. Biochim Biophys Acta. 2000;1482:9–24. doi: 10.1016/s0167-4838(00)00148-5. [DOI] [PubMed] [Google Scholar]
- Godfrey PA, Malnic B, Buck LB. The mouse olfactory receptor gene family. Proc Natl Acad Sci U S A. 2004;101:2156–2161. doi: 10.1073/pnas.0308051100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haga S, et al. The male mouse pheromone ESP1 enhances female sexual receptive behaviour through a specific vomeronasal receptor. Nature. 2010;466:118–122. doi: 10.1038/nature09142. [DOI] [PubMed] [Google Scholar]
- Herrada G, Dulac C. A novel family of putative pheromone receptors in mammals with a topographically organized and sexually dimorphic distribution. Cell. 1997;90:763–773. doi: 10.1016/s0092-8674(00)80536-x. [DOI] [PubMed] [Google Scholar]
- Johnson MA, Hernandez I, Wei Y, Greenberg N. Isolation and characterization of mouse probasin: an androgen-regulated protein specifically expressed in the differentiated prostate. Prostate. 2000;43:255–262. doi: 10.1002/1097-0045(20000601)43:4<255::aid-pros4>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- Kaessmann H, Vinckenbosch N, Long M. RNA-based gene duplication: mechanistic and evolutionary insights. Nat Rev Genet. 2009;10:19–31. doi: 10.1038/nrg2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtovic A, Widmer A, Dickson BJ. A single class of olfactory neurons mediates behavioural responses to a Drosophila sex pheromone. Nature. 2007;446:542–546. doi: 10.1038/nature05672. [DOI] [PubMed] [Google Scholar]
- Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim MM, et al. Enhanced partner preference in a promiscuous species by manipulating the expression of a single gene. Nature. 2004;429:754–757. doi: 10.1038/nature02539. [DOI] [PubMed] [Google Scholar]
- Lynch M, Force A. The probability of duplicate gene preservation by subfunctionalization. Genetics. 2000;154:459–473. doi: 10.1093/genetics/154.1.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magklara A, et al. An epigenetic signature for monoallelic olfactory receptor expression. Cell. 2011;145:555–570. doi: 10.1016/j.cell.2011.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meslin C, et al. The evolutionary history of the SAL1 gene family in eutherian mammals. BMC Evol Biol. 2011;11:148. doi: 10.1186/1471-2148-11-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papp B, Pal C, Hurst LD. Dosage sensitivity and the evolution of gene families in yeast. Nature. 2003;424:194–197. doi: 10.1038/nature01771. [DOI] [PubMed] [Google Scholar]
- Pes D, et al. Cloning and expression of odorant-binding proteins Ia and Ib from mouse nasal tissue. Gene. 1998;212:49–55. doi: 10.1016/s0378-1119(98)00131-0. [DOI] [PubMed] [Google Scholar]
- Qian W, Liao BY, Chang AY, Zhang J. Maintenance of duplicate genes and their functional redundancy by reduced expression. Trends Genet. 2010;26:425–430. doi: 10.1016/j.tig.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts SA, et al. Darcin: a male pheromone that stimulates female memory and sexual attraction to an individual male's odour. BMC Biol. 2010;8:75. doi: 10.1186/1741-7007-8-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubenstein DR, Lovette IJ. Reproductive skew and selection on female ornamentation in social species. Nature. 2009;462:786–789. doi: 10.1038/nature08614. [DOI] [PubMed] [Google Scholar]
- Serizawa S, Miyamichi K, Sakano H. One neuron-one receptor rule in the mouse olfactory system. Trends Genet. 2004;20:648–653. doi: 10.1016/j.tig.2004.09.006. [DOI] [PubMed] [Google Scholar]
- Shahan K, Gilmartin M, Derman E. Nucleotide sequences of liver, lachrymal, and submaxillary gland mouse major urinary protein mRNAs: mosaic structure and construction of panels of gene-specific synthetic oligonucleotide probes. Mol Cell Biol. 1987;7:1938–1946. doi: 10.1128/mcb.7.5.1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw PH, Held WA, Hastie ND. The gene family for major urinary proteins: expression in several secretory tissues of the mouse. Cell. 1983;32:755–761. doi: 10.1016/0092-8674(83)90061-2. [DOI] [PubMed] [Google Scholar]
- Shiao MS, Liao BY, Long M, Yu HT. Adaptive evolution of the insulin two-gene system in mouse. Genetics. 2008;178:1683–1691. doi: 10.1534/genetics.108.087023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stopka P, Janotova K, Heyrovsky D. The advertisement role of major urinary proteins in mice. Physiol Behav. 2007;91:667–670. doi: 10.1016/j.physbeh.2007.03.030. [DOI] [PubMed] [Google Scholar]
- Stopkova R, et al. Novel OBP genes similar to hamster Aphrodisin in the bank vole, Myodes glareolus. BMC Genomics. 2010;11:45. doi: 10.1186/1471-2164-11-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tegoni M, et al. Mammalian odorant binding proteins. Biochim Biophys Acta. 2000;1482:229–240. doi: 10.1016/s0167-4838(00)00167-9. [DOI] [PubMed] [Google Scholar]
- Touhara K. Molecular biology of peptide pheromone production and reception in mice. Adv Genet. 2007;59:147–171. doi: 10.1016/S0065-2660(07)59006-1. [DOI] [PubMed] [Google Scholar]
- Toung JM, Morley M, Li M, Cheung VG. RNA-sequence analysis of human B-cells. Genome Res. 2011;21:991–998. doi: 10.1101/gr.116335.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utsumi M, et al. Expression of major urinary protein genes in the nasal glands associated with general olfaction. J Neurobiol. 1999;39:227–236. doi: 10.1002/(sici)1097-4695(199905)39:2<227::aid-neu7>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- Wyart C, et al. Smelling a single component of male sweat alters levels of cortisol in women. J Neurosci. 2007;27:1261–1265. doi: 10.1523/JNEUROSCI.4430-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y, et al. RNA sequencing shows no dosage compensation of the active X-chromosome. Nat Genet. 2010;42:1043–1047. doi: 10.1038/ng.711. [DOI] [PubMed] [Google Scholar]
- Yang J, Lusk R, Li WH. Organismal complexity, protein complexity, and gene duplicability. Proc Natl Acad Sci U S A. 2003;100:15661–15665. doi: 10.1073/pnas.2536672100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, et al. High-throughput microarray detection of olfactory receptor gene expression in the mouse. Proc Natl Acad Sci U S A. 2004;101:14168–14173. doi: 10.1073/pnas.0405350101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou S, Stone EA, Mackay TF, Anholt RR. Plasticity of the chemoreceptor repertoire in Drosophila melanogaster. PLoS Genet. 2009;5:e1000681. doi: 10.1371/journal.pgen.1000681. [DOI] [PMC free article] [PubMed] [Google Scholar]