Abstract
Despite intense investigation for over 25 years, the in vivo structure of plant mitochondrial genomes remains uncertain. Mapping studies and genome sequencing generally produce large circular chromosomes, whereas electrophoretic and microscopic studies typically reveal linear and multibranched molecules. To more fully assess the structure of plant mitochondrial genomes, the complete sequence of the monkeyflower (Mimulus guttatus DC. line IM62) mitochondrial DNA was constructed from a large (35 kb) paired-end shotgun sequencing library to a high depth of coverage (∼30×). The complete genome maps as a 525,671 bp circular molecule and exhibits a fairly conventional set of features including 62 genes (encoding 35 proteins, 24 transfer RNAs, and 3 ribosomal RNAs), 22 introns, 3 large repeats (2.7, 9.6, and 29 kb), and 96 small repeats (40–293 bp). Most paired-end reads (71%) mapped to the consensus sequence at the expected distance and orientation across the entire genome, validating the accuracy of assembly. Another 10% of reads provided clear evidence of alternative genomic conformations due to apparent rearrangements across large repeats. Quantitative assessment of these repeat-spanning read pairs revealed that all large repeat arrangements are present at appreciable frequencies in vivo, although not always in equimolar amounts. The observed stoichiometric differences for some arrangements are inconsistent with a predominant master circular structure for the mitochondrial genome of M. guttatus IM62. Finally, because IM62 contains a cryptic cytoplasmic male sterility (CMS) system, an in silico search for potential CMS genes was undertaken. The three chimeric open reading frames (ORFs) identified in this study, in addition to the previously identified ORFs upstream of the nad6 gene, are the most likely CMS candidate genes in this line.
Keywords: alternative genomic conformations, repeats, rearrangement, homologous recombination, cytoplasmic male sterility
Introduction
To date, the mitochondrial genomes from over 30 species of land plants have been completely sequenced and publicly released (reviewed in Mower et al. 2012). Sequenced genome sizes range from slightly more than 100 kb in the moss Physcomitrella patens (Terasawa et al. 2007) to more than 2.7 Mb in the melon Cucumis melo (Rodriguez-Moreno et al. 2011). Despite the large variation in overall size, plant mitochondrial genomes typically contain a similar gene repertoire: 3 ribosomal RNA (rRNA) genes, an incomplete set of transfer RNA (tRNA) genes, and a variable subset of the same 42 protein-coding genes involved in essential mitochondrial processes, such as electron transport, adenosine triphosphate (ATP) synthesis, and protein translation (summarized in Sloan et al. 2010; Mower et al. 2012). Introns abound in all species sequenced so far, some of which contain additional genes, either endonucleases or maturases, that are essential for proper splicing (reviewed in Bonen 2011). Genomic repeats, both large (>1 kb) and small (<1 kb), are also prevalent in most plants, particularly seed plants (Alverson et al. 2011b).
The physical structure of the plant mitochondrial genome is enigmatic. Across the diversity of plants, from green algae to angiosperms, the genome generally maps as a circular molecule, often termed the “master circle” or “master chromosome” (Lonsdale et al. 1984; Palmer and Shields 1984). The linear chromosomes of maize line CMS-S and some green algae such as Chlamydomonas and Polytomella (Allen et al. 2007; Smith et al. 2010) are clearly exceptions to the circularly mapping arrangements found for most plants. Direct evidence for master chromosomes in plants is generally lacking, although a genome-sized circle was occasionally observed by electron microscopy in the bryophyte Marchantia polymorpha (Oda et al. 1992; Oldenburg and Bendich 1998), and a supercoiled mitochondrial DNA fraction was obtained from Brassica oleracea that was enriched for DNA specific to the smaller of two predicted circular chromosomes (Palmer 1988). Other than these studies, most electrophoretic and microscopic analyses of mitochondrial DNA fail to recover large circular chromosomes. Instead, much of the mitochondrial genome is observed as linear molecules and multibranched conglomerations of subgenome to multigenome size, and when circular molecules are recovered, they are typically much smaller than the expected genome size (Oldenburg and Bendich 1996; Backert and Börner 2000; Manchekar et al. 2006). These findings have led to the idea that the circular map is not an accurate representation of the genome structure in vivo, except perhaps in meristematic tissue to ensure that the genome is faithfully replicated for descendent cells (Backert et al. 1997; Arrieta-Montiel et al. 2001; Sakai et al. 2004; Woloszynska 2010). Nevertheless, circular maps continue to be presented in genome sequencing publications because they are convenient indicators of genome content and sequencing completion. Many authors readily acknowledge that their circular representations may be artifactual, but none have provided convincing evidence to confirm or refute the existence of a master chromosome in plant mitochondria.
In addition to the uncertainties surrounding the in vivo structure of plant mitochondrial genomes, it has also been recognized for some time that the repeated sequences present in these genomes can facilitate genomic rearrangement via homologous recombination (reviewed in Lonsdale et al. 1988; Mackenzie 2007; Maréchal and Brisson 2010). This is indirectly indicated by the near complete scrambling of gene order among closely related species (e.g., Palmer and Herbon 1988; Handa 2003; Ogihara et al. 2005; Alverson et al. 2010) and by high levels of rearrangement even among different varieties of the same species (Allen et al. 2007; Fujii et al. 2010; Darracq et al. 2011; Davila et al. 2011). Larger repeats apparently undergo high-frequency recombination, and the various recombination products appear to be at roughly equal stoichiometry, based on Southern blot analyses (Palmer and Shields 1984; Palmer and Herbon 1986; Stern and Palmer 1986; Folkerts and Hanson 1989; Klein et al. 1994; Siculella et al. 2001; Sloan et al. 2010). The apparent stoichiometric equality of repeat arrangements is attributed to dynamic equilibrium of recombination involving large repeats (Lonsdale et al. 1988; Janska and Woloszynska 1997; Woloszynska 2010). That being said, minor variations in band intensities are sometimes observed in these Southern blot studies, although determining whether the variations reflect real in vivo stoichiometric differences or experimental limitations is challenging (Palmer and Shields 1984). Recently, however, significant stoichiometric differences were shown for a large (3.6 kb) plant mitochondrial repeat shared between two small chromosomes present in Cucumis sativus (Alverson et al. 2011a). The biological significance of this variation is unclear, given that these small chromosomes contain no obvious mitochondrial genes and may not be essential, although a lack of stoichiometry among chromosomes could potentially affect replication rates, mitochondrial gene expression, and further recombination within the genome. Recombination involving smaller repeats is much less frequent; yet, these events are thought to be important for producing substoichiometric molecules which may ultimately generate the highly rearranged genomes found among closely related plants via substoichiometric shifting (reviewed in Mackenzie 2007; Maréchal and Brisson 2010). Recombination around smaller repeats along with recombination-mediated replication (Oldenburg and Bendich 1996; Backert and Börner 2000) could account for the observation of the linear, circular, and complex branching forms of many different sizes (reviewed in Backert et al. 1997).
Although the lack of synteny among plant mitochondrial genomes suggests that, in general, gene order is not important for mitochondrial function, particular rearrangements have been associated with mutant phenotypes and possibly even adaptive benefits (reviewed in Arrieta-Montiel and Mackenzie 2011). The most obvious and widespread phenotype associated with mitochondrial rearrangements in plants is cytoplasmic male sterility (CMS). CMS-causing genes prevent the production of viable pollen; plants that would otherwise be hermaphroditic are rendered female or “male sterile.” Male sterility has long been of interest to plant breeders as male-sterile phenotypes aid in the production of hybrid seed (Kempken and Pring 1999). Because of this application, much of what is known about the genetic basis of CMS comes from studies of economically important species. Each of the CMS-associated genes characterized to date is unique in sequence (even among mitotypes within species). However, they all share a chimeric structure—either the open reading frame (ORF) contains regions of conserved gene sequence or the ORF follows a conserved promoter, usually one associated with an ATP synthase subunit (reviewed in Hanson and Bentolila 2004). Chimerism suggests that these genes arose through recombination between functional mitochondrial genes and unique ORFs (Schnable and Wise 1998). The preponderance of CMS in flowering plants provides strong incentive for understanding the pattern and cause of mitochondrial recombination, particularly in natural populations of wild species.
Although CMS is thought to be extremely common in plants (Laser and Lersten 1972; Kaul 1988; Schnable and Wise 1998; Tiffin et al. 2001), few CMS genes have been characterized genetically in wild plant species, meaning that there are few clues about how often CMS arises in nature and how CMS is affected by evolutionary forces. Although significant strides have been made for understanding CMS systems in some wild plants by comparison with closely related crops or model systems (e.g., Arrieta-Montiel et al. 2001; Darracq et al. 2011), in general, the study of CMS genes in wild species is hindered by a dearth of molecular tools and other genetic resources. The sequence of a CMS gene was recently characterized in an inbred line (hereafter “IM62”) derived from a natural population of Mimulus guttatus (Phrymaceae; Case and Willis 2008), a wild species with no agronomic value. CMS in this line is considered cryptic because all individuals in the wild source population are male fertile, even though they carry the CMS gene (Fishman and Willis 2006; Case and Willis 2008). The lack of male sterility expression results from all individuals also carrying nuclear fertility restoration (Rf) genes (Barr and Fishman 2010), such that the CMS phenotype is only uncovered when crossed to a line lacking the restorer (Fishman and Willis 2006). Male-sterile phenotypes in advanced generation backcrosses of IM62 against a nonrestoring line were associated with the transcription of an unknown ORF upstream from the mitochondrial nad6 gene. Direct evidence confirming CMS induction by this ORF is lacking in M. guttatus, hindered by the limited capacity to manipulate the mitochondrial genome in intact organisms, although this has been done in some species (reviewed in Hanson and Bentolila 2004). Whole-mitochondrial genome sequences can fill in some of the gaps where experimental approaches fall short. Not only will they provide insights into the origin, expression, and evolution of CMS genes but also the effects of CMS on the evolution of the mitochondrial genome.
Mimulus is an emerging model system for evolutionary and ecological genomics (Wu et al. 2008). Its relatively small nuclear genome, short generation time, high fecundity, and ease of propagation facilitate the development and application of genomic tools, whereas its wide distribution in a stunning diversity of habitats broadens its appeal to ecologists and evolutionary biologists. Because of these features, M. guttatus line IM62 was sequenced at the Joint Genome Institute using the whole-genome shotgun sequencing approach. From these data, the nuclear, plastid, and mitochondrial genome sequences were assembled. The sequence, structure, and content of the mitochondrial genome, including in silico evaluation of candidate CMS genes, are described here.
Materials and Methods
Genome Assembly
Paired-end shotgun sequence reads were downloaded from the NCBI Trace Archive repository (ftp://ftp.ncbi.nih.gov/pub/TraceDB/). Initial BlastN searches of the sequence reads revealed that the mitochondrial genome was present at high-copy number (>100×) compared with the nuclear genome (<10×) but lower copy compared with the plastid genome (>1000×). To minimize the accumulation of sizeable nuclear contigs during assembly, all reads were subdivided based on sequence name (defined by the four letter library ID prefix in each name) and library insert size into four independent library subsets: Lib3kA, Lib3kB Lib8k, and Lib35k (supplementary table 1, Supplementary Material online). In each library subset, mitochondrial read coverage should be sufficient for reliable assembly, whereas the very high coverage of plastid reads would cause them to be flagged as repetitive and subsequently masked by the assembler. The four library subsets were independently assembled using PCAP version 06/07/05 (Huang et al. 2003) with modified parameters: 1) the parameter specifying the minimum depth of coverage for repeats was increased from 75 to 200 to prevent mitochondrial reads from being flagged as repetitive; 2) the parameter specifying the overlap percentage identity cutoff was reduced from 4,500 to 3,000 to improve end-joining of contigs; 3) for assembly jobs with >1 million reads, the parameter specifying the number of simultaneous PCAP jobs was increased from 2 to 8. Genome assemblies and read pair mapping patterns were visually inspected using Consed 16.0 (Gordon et al. 1998). In each of the four resulting assemblies, consensus sequences of the mitochondrial contigs were virtually identical. The few discrepancies among assemblies were examined in detail and found to result from assembler miscalls in low-quality regions (near the ends of contigs or at positions of plastid insertions in the mitochondrial genome resulting from intracellular gene transfer).
Genome Finishing
The Lib35k assembly was the most complete and was subsequently used for in silico genome finishing by inspecting the ends of the six mitochondrial contigs for shared overlaps and evidence of repeats (supplementary fig. 1, Supplementary Material online). Three pairs of contig ends overlapped nearly identically by 600–900 bp and were therefore joined. For the remaining contig ends, BlastN searches revealed that they were nearly identical to internal regions of other contigs. These duplicated sequences colocated in pairs, suggesting that each duplicated pair actually defined the ends of a larger repeat. We assumed that these putative large repeats were in fact present in the genome, which closed the remaining sequence gaps. Read pair mapping information viewed in Consed strongly supported all the above finishing work, although it was clear that other arrangements of the genome were also possible.
The intracellular transfer of plastid and nuclear DNA into the mitochondrial genome and mitochondrial DNA into the nuclear genome is a frequent occurrence (Timmis et al. 2004; Mower et al. 2012). Therefore, we took care to avoid assembly errors at regions of shared homology between the different genomes. Because of the low coverage of the nuclear genome, any nuclear-copy reads erroneously assembled into a mitochondrial contig would be at much lower frequency than the mitochondrial reads and should not affect the mitochondrial consensus. Conversely, because of the very high coverage of the plastid genome, any plastid-copy reads that escaped repeat masking might introduce errors into the mitochondrial assembly at sites of plastid integration. Indeed, clusters of polymorphic sites were detected at two regions in the mitochondrial assembly, and both regions showed strong similarity to the plastid genome from Jasminum nudum (GenBank accession number DQ673255). To differentiate between mitochondrion-encoded and plastid-encoded copies, the two haplotypes were reconstructed by examining individual read sequences that link the variable sites within the shared segment to the unique mitochondrial or plastid sequences flanking the shared segment. The reconstructed mitochondrial version was used to correct the mitochondrial consensus sequence.
Genome Assembly Verification
Shotgun sequence reads from the Lib35k library were mapped onto the mitochondrial consensus sequence using BlastN (minimum length of 400 bp and at least 90% sequence identity). Less stringent mapping criteria (200 bp in length and 60% identity) had little effect on the results and no effect on conclusions. When a given read mapped to more than one location, the hit with the highest blast score was taken as the true location. In the case of a tie (i.e., reads that mapped to repeats), the read position could not be distinguished and was mapped to both positions.
Read depth of genomic coverage was measured in a number of ways using the mapped reads and information provided by the paired-end sequencing process, which sequences both ends of clones from a library with a known average insert size. Total read (TR) depth counts all mapped reads. Consistent pair (CP) depth counts only those read pairs that map to the genome in the proper head-to-head orientation and at the expected distance (defined as less than 50% larger or smaller than the average insert size of the library). Inconsistent pair (IP) depth counts those read pairs that map inconsistently; that is, they do not meet the CP criteria. Unpaired read (UR) depth counts those mapped reads whose mate pair does not map to the genome. Coverage was visualized by plotting average depth using a sliding window analysis with a 1,000 bp fixed window and a 100 bp step size.
Genome Annotation
The location of protein coding, rRNA, and tRNA genes were determined using BlastN with known mitochondrial genes from other angiosperms as query sequences. tRNA genes were also predicted using tRNAscan-SE 1.23 (Lowe and Eddy 1997). ORFs >300 bp were located using a custom Perl script. Repeats at least 40 bp in length with fewer than two differences were identified using Vmatch (http://vmatch.de/). Different repeat cutoffs were evaluated, but they had little effect on the frequency or genomic distribution of repeats >50 bp. Sites of RNA editing were predicted using PREP-Mt with a cutoff value of 0.5 (Mower 2009). Sites of plastid integration were identified using a BlastN search with the Jasminum nudiflorum plastid genome as a query and requiring a minimum match of 100 bp, filtering out any hits resulting from homology between plastid and mitochondrial genes. The annotated genome sequence was deposited in GenBank under accession number JN098455.
Quantification of Repeat-Mediated Genomic Rearrangements
Eight different genomic conformations were predicted from the initial finished assembly by assuming that homologous recombination occurs between copies of large repeats in the genome. Using the stringent read mapping criteria described above, read pairs were mapped to all eight alternative conformations. Read pairs were classified depending on whether they mapped consistently to: all eight conformations, some but not all conformations, or none of the conformations. To quantify the abundance of each large repeat arrangement, the number of consistent read pairs that spanned each large repeat in at least one conformation was counted. Because the large repeats are of very different sizes, the total number of spanning pairs is expected to be different for each repeat. To normalize these counts for all three large repeats, a more stringent count was also taken, which required that read pairs map in a fixed window around each repeat copy (from 15 kb to 35 kb to either side of the repeat midpoint). To quantify the abundance of substoichiometric molecules resulting from rearrangement at small repeats, read pairs that were not consistent with any of the eight major genomic conformations were checked for consistency with a putative rearrangement involving a small repeat.
For all large repeat arrangements, we tested for stoichiometric inequality, stoichiometric asymmetry, and sequencing bias using the repeat-spanning read pair counts and Chi-square goodness-of-fit tests. For each large repeat, four different arrangements are possible, and in every case, recombination alternates between two pairs of arrangements. Each coexisting repeat pair can be considered the parental or recombinant forms, depending on the direction of recombination. To test for stoichiometric inequality, the null model assumed that each of the four possible repeat arrangements should be at equal frequencies. Stoichiometric equality would be consistent with similar rates of forward and reverse recombination at each large repeat and more or less equal frequencies of alternate genomic conformations within IM62. We tested for stoichiometric asymmetry by selecting a null model that assumes that coexisting repeat arrangements (i.e., those that coexist in the same master or subgenomic circular chromosome) should be present at equal frequencies. Repeat arrangements should exhibit symmetric stoichiometries if homologous recombination is the only process affecting arrangement abundance, whereas asymmetric stoichiometries could result from additional processes contributing to the amplification or loss of single recombination products. To test whether inequalities or asymmetries could have resulted from sequencing bias rather than recombinational dynamics, we counted read pairs that consistently mapped around six independent single-copy regions far from any repeat, and we assumed that these single-copy regions should exhibit similar frequencies in the absence of sequencing bias. As an additional test for sequencing bias, we assumed that the read counts for each repeat (in all arrangements after correcting for the different repeat sizes) should be equal to each other and to twice the count from single-copy regions. Reduced counts for a particular repeat may indicate sequencing or cloning bias against particular arrangements of that repeat.
Analysis of Candidate Cytoplasmic Male Sterility Genes
Based on previous analyses showing that CMS genes are chimeric (Schnable and Wise 1998; Hanson and Bentolila 2004), a search for chimeric ORFs was conducted. All ORFs at least 150 bp in length were compared with the identified Mimulus mitochondrial genes using BlastN with an e-value cutoff of 1 × 10−3. ORFs containing at least 30 bp of an identified mitochondrial gene were characterized as chimeric, excluding any ORFs that overlap the genomic position of an identified gene. Transmembrane domains in each candidate ORF were predicted using TMHMM Server version 2.0 (http://www.cbs.dtu.dk/services/TMHMM/).
Results
Mitochondrial Genome Assembly and Verification
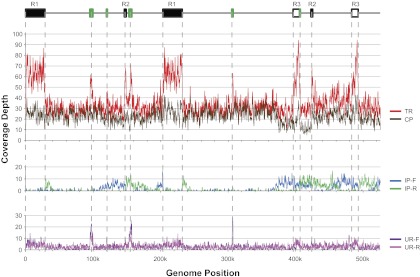
The finished assembly of the mitochondrial genome of M. guttatus IM62 was a single circular chromosome of 525,671 bp. Of the 623,219 paired-end reads in the 35 kb sequencing library, 21,984 mapped to the finished mitochondrial assembly (fig. 1, top panel). Most of these reads (71%) mapped consistently and evenly across the genome (fig. 1, top panel), indicating that the finished assembly is likely correct, the repeats in the consensus sequence are in fact repetitive, and the single-copy regions are present at roughly equal stoichiometry. Another 10% of reads mapped inconsistently to the finished assembly and clustered around the large repeats (fig. 1, middle panel), indicating a multipartite genome structure where additional genomic arrangements could be resulting from high-frequency recombination at the large repeats. Southern blot hybridization of 13 mitochondrial exons against mitochondrial clones from two BAC libraries provided additional evidence for the existence of these different repeat environments (supplementary figs. 2 and 3, Supplementary Material online). The remaining 18% of reads had a mate that did not map to the mitochondrial genome (fig. 1, bottom panel), mostly for trivial reasons (see supplementary text, Supplementary Material online, for further details of assembly verification).
FIG. 1.—
Depth of read coverage across the genome. All reads in the Lib35k sublibrary were mapped to the mitochondrial consensus sequence and read depth was calculated in several ways. Top panel: read depth of TRs (red line) that mapped to the mitochondrial consensus sequence and CPs (brown line) that mapped in the proper orientation and distance. Middle panel: read depth of inconsistent read pairs mapping in a forward (IP-F, green line) or reverse (IP-R, cyan line) orientation. Lower panel: read depth of reads whose mate pair did not map to the mitochondrial genome; these unpaired reads were also split into forward (UR-F, purple line) and reverse (UR-R, pink line) mapping orientations. Above the coverage plots, a linear representation of the genome is given that shows the position of all direct repeats (white boxes), inverted repeats (black boxes), and inserted chloroplast DNA (green boxes) >1 kb in length.
Mitochondrial Genomic Content
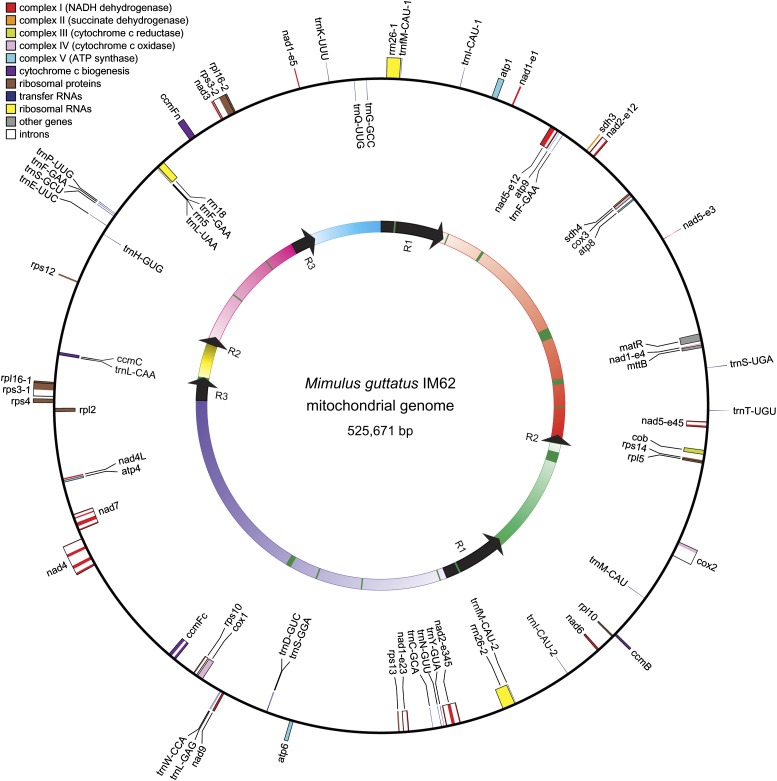
The 525,671 bp mitochondrial consensus sequence for monkeyflower (fig. 2) is an intermediate value among sequenced angiosperms, whose sizes range from 222 kb in Brassica napus (Handa 2003) to 2.7 Mb in Cucumis melo (Rodriguez-Moreno et al. 2011). GC content is 45.1%, which is also typical for flowering plants. Genic regions comprise 7.4% of the genome, including 35 known protein-coding genes (6.1%), 3 rRNAs (1.0%), and 24 tRNAs (0.3%). Intronic regions cover 5–6% of the genome and include 16 cis-spliced introns (4.4%) and 6 trans-spliced introns of uncertain length (∼1%). The remaining 87% of the genome features 3 large repeats >1 kb and 96 small repeats 40–293 bp (9.0%), 16 insertions of plastid DNA >100 bp (3.1%), and a large amount of unannotated DNA (∼75%).
FIG. 2.—
Circular genome map. The outer circle shows sequence and orientation of known protein coding, rRNA, and tRNA genes. Genes are color coded as shown at top left. Genes on the outside and inside of the outer circle are transcribed clockwise and counterclockwise, respectively. The inner circle corresponds to the C1 conformation in figure 3, showing six single-copy regions (gradients of color) separated by the three sets of large repeats R1–R3 (black arrows). Dark green boxes denote chloroplast inserts in the genome.
The 35 protein-coding genes in the Mimulus mitochondrial genome are a subset of the 39 found in Vitis, which appears to represent the ancestral repertoire for core eudicots (table 1). This ancestral eudicot gene count includes the newly identified rpl10 gene found throughout land plants (Mower and Bonen 2009; Kubo and Arimura 2010) but not the rps2 and rps11 genes that were lost early in eudicot history (Adams, Qiu, et al. 2002). A total of 457 sites of RNA editing were predicted to be present in the 35 presumably functional transcripts. Of the four genes missing relative to Vitis, the rps1, rps7, and rps19 genes were lost completely, whereas rpl2 is still present as a frame-shifted pseudogene. BlastP searches using translated Vitis mitochondrial homologs against the annotated set of Mimulus nucleus-encoded proteins identified one or more candidates for RPL2 (mgv1a011898m), RPS1 (mgv1a014033m), RPS7 (mgv11b012968m, mgv1a022477m, and mgv1a024520m), and RPS19 (mgv1a015017m, mgv1a015586m, and mgv1a015638m). In addition to the above-identified protein-coding genes, 143 mitochondrial ORFs at least 300 bp in length were identified in intergenic regions, although none of them are widely conserved among angiosperms. Several ORFs are chimeric, containing one or more fragments of identified mitochondrial genes (see last section of Results). Other ORFs appear to be remnants of degraded nucleus-derived retrotransposons, a common presence in the mitochondrial genomes of plants (Knoop et al. 1996; Kubo et al. 2000; Notsu et al. 2002). The remaining ORFs show little to no similarity to any proteins in GenBank and may not encode functional products.
Table 1.
Mimulus Protein-Coding Gene Content Compared with Selected Angiosperms
| Gene | Mimulus | Nicotiana | Beta | Arabidopsis | Cucurbita | Vitis | Oryza | Triticum |
| 28 genesa | • | • | • | • | • | • | • | • |
| rpl2 | Ψ | • | ○ | • | • | • | • | Ψ |
| rpl10b | • | • | ○ | ○ | • | • | Ψ | ○ |
| rpl16 | • | • | ○ | • | • | • | • | • |
| rps1 | ○ | • | ○ | ○ | • | • | • | • |
| rps2 | ○ | ○ | ○ | ○ | ○ | ○ | • | • |
| rps7 | ○ | ○ | • | • | • | • | • | • |
| rps10 | • | • | ○ | ○ | • | • | ○ | ○ |
| rps11 | ○ | ○ | ○ | ○ | ○ | ○ | Ψ | ○ |
| rps13 | • | • | • | ○ | • | • | • | • |
| rps14 | • | Ψ | ○ | Ψ | Ψ | • | Ψ | Ψ |
| rps19 | ○ | • | ○ | Ψ | • | • | • | Ψ |
| sdh3 | • | • | ○ | ○ | • | • | ○ | ○ |
| sdh4 | • | • | Ψ | Ψ | • | • | ○ | ○ |
| • (present) | 35 | 37 | 30 | 31 | 38 | 39 | 35 | 33 |
| Ψ (pseudo) | 1 | 1 | 1 | 3 | 1 | 0 | 3 | 3 |
| ○ (absent) | 5 | 3 | 10 | 7 | 2 | 2 | 3 | 5 |
The 28 genes include atp[1, 4, 6, 8, 9], ccm[B, C, Fc, Fn], cob, cox[1, 2, 3], matR, mttB, nad[1, 2, 3, 4, 4L, 5, 6, 7, 9], rpl5, and rps[3, 4, 12]. Although Beta vulgaris ccmC is widely reported to be a pseudogene, it is transcribed, edited, and translated (Mower and Palmer 2006; Kitazaki et al. 2009) and is scored as functionally present here. Arabidopsis ccmFn is split into two genes, but the two halves are counted as a single gene here. Although mttB was reported to be a pseudogene in Vitis due to an absence of a conserved start codon (Goremykin et al. 2009), it is probably translated from an alternative start codon as suggested for other species (Sunkel et al. 1994) and is scored as present here.
The rpl10 gene was recently identified in a wide variety of streptophytes (Mower and Bonen 2009; Kubo and Arimura 2010).
The mitochondrial RNA gene complement for Mimulus includes the large subunit, small subunit, and 5S rRNAs found in nearly all land plants so far sequenced (Selaginella moellendorffii lacks a mitochondrion-encoded 5S rRNA; Hecht et al. 2011), as well as 24 tRNAs predicted to recognize all amino acids except alanine, arginine, and valine. All Mimulus tRNAs have homologs in at least one other angiosperm except for a weakly predicted trnT-UGU gene (supplementary fig. 4, Supplementary Material online). This tRNA has no obvious homology to any annotated tRNA currently in GenBank, although it matches unannotated regions in the Nicotiana, Arabidopsis, Vigna, Cucurbita, and Carica mitochondrial genomes. Most Mimulus mitochondrial tRNAs are predicted to carry the same amino acid as their homologs in other plants. However, there are three tRNA-Leu genes (trnL-CAA, trnL-GAG, and trnL-UAA) that are not similar to one another (supplementary fig. 4, Supplementary Material online), and whose closest homologs are plastid-derived tRNAs often found in other angiosperm mitochondrial genomes (Sloan et al. 2010) that carry cysteine (trnC-GCA-cp), isoleucine (trnI-CAU-cp), or proline (trnP-UGG-cp) rather than leucine. There are also three nonidentical copies of trnF-GAA (supplementary fig. 4, Supplementary Material online). Two differ by one nucleotide substitution and a 4 bp indel and are homologous to other angiosperm mitochondrial trnF-GAA genes, whereas the third was inserted as part of a larger plastid integrant. Compared with most other angiosperms, Mimulus has a higher number of mitochondrion-encoded tRNAs, although some may not be functional.
The set of mitochondrial introns within Mimulus includes 16 cis-spliced and 6 trans-spliced group II introns, all of which are homologous to introns in other angiosperms (table 2). Mimulus has lost three introns (cox2-i373, nad7-i676, and rpl2-i917) compared with Vitis, which has retained what appears to be the full complement of 25 introns present in the angiosperm common ancestor. Given the variability of cox2 intron content across angiosperms, the timing of cox2-i373 loss cannot be reliably determined without additional asterid sampling. The nad7-i676 loss is shared between Mimulus and Nicotiana, suggesting a loss in their common ancestor. The loss of rpl2-i917 is unique to Mimulus among sequenced angiosperms, which is curious because rpl2 appears to be a pseudogene in Mimulus. All missing introns were lost precisely from the genome.
Table 2.
Mimulus Intron Content Compared with Selected Angiosperms
| Intron | Mimulus | Nicotiana | Beta | Arabidopsis | Cucurbita | Vitis | Oryza | Triticum |
| 12 cis intronsa | • | • | • | • | • | • | • | • |
| 5 trans intronsb | ⊖ | ⊖ | ⊖ | ⊖ | ⊖ | ⊖ | ⊖ | ⊖ |
| cox2-i373 | ○ | • | • | ○ | ○ | • | • | • |
| cox2-i691 | • | ○ | ○ | • | • | • | ○ | ○ |
| nad1-i728 | ⊖ | ⊖ | ⊖ | • | • | • | ⊖ | ⊖ |
| nad4-i976 | • | • | ○ | • | • | • | • | • |
| nad7-i676 | ○ | ○ | • | • | • | • | • | • |
| rpl2-i917 | ○ | • | × | • | • | • | • | × |
| rps3-i74 | • | • | ○ | • | • | • | • | • |
| rps10-i235 | • | • | × | × | • | • | × | × |
| • (cis-spliced) | 16 | 17 | 14 | 18 | 19 | 20 | 17 | 16 |
| ⊖ (trans-spliced) | 6 | 6 | 6 | 5 | 5 | 5 | 6 | 6 |
| ○ (intron absent) | 3 | 2 | 3 | 1 | 1 | 0 | 1 | 1 |
| × (gene absent) | 0 | 0 | 2 | 1 | 0 | 0 | 1 | 2 |
The 12 cis-spliced introns include ccmFc-i829, nad1-i477, nad2-[i156, i709, i1282], nad4-[i461, i1399], nad5-[i230, i1872], and nad7-[i140, i209, i917].
The five trans-spliced introns include nad1-[i394, i669], nad2-i542, and nad5-[i1455, i1477].
The Mimulus mitochondrial repeat content comprises 3 large and at least 96 small repeats. The three large repeats are each present in two identical copies (based on the absence of sequence polymorphism among reads that map to each repeat). In the finished assembly (fig. 2), repeats R1 (28,763 bp) and R2 (2,742 bp) are in inverted orientations, whereas R3 (9,620 bp) is in direct orientation. Small repeats were found in direct and inverted orientations at roughly equal frequency. Over half of the small repeats were 40–50 bp in length, 31 were 51–100 bp, and 12 were 101–293 bp (supplementary fig. 5, Supplementary Material online).
Evidence of Stoichiometric Inequality and Asymmetry for Large Repeat Arrangements
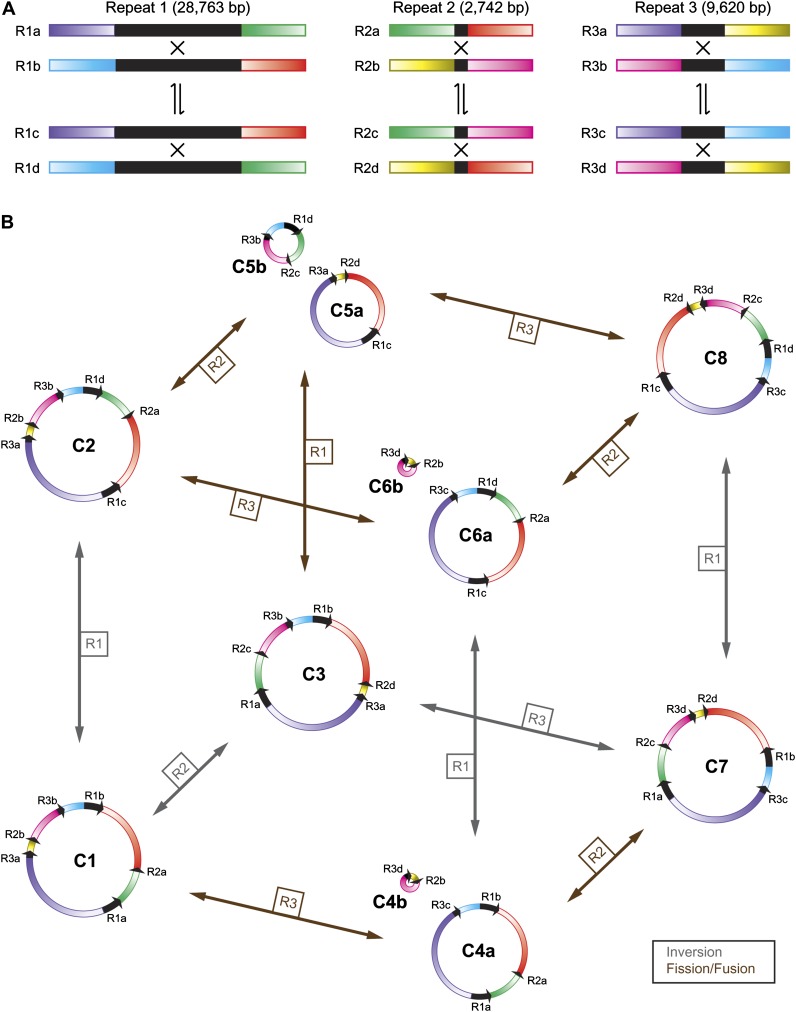
Our assembly validation procedure (fig. 1; supplementary text, Supplementary Material online) indicated that the large repeats exist in several alternative arrangements resulting from homologous recombination (fig. 3A), a well-known phenomenon for plant mitochondrial genomes. Starting from the initial circular assembly (labeled conformation C1) and assuming homologous recombination across each large repeat, seven additional genomic conformations (C2–C8) can be predicted (fig. 3B). In this master circular model of multipartite genome structure, all eight conformations contain exactly the same genomic information; the only differences are the order and orientation of the nonrepetitive and repetitive segments and, in some cases, the number of chromosomes.
FIG. 3.—
Alternative repeat arrangements and mitochondrial genomic conformations. (A) Each of the three large repeats (R1, R2, and R3) is shown in all four possible arrangements (a, b, c, and d). Coexisting arrangements found within particular genomic conformations are paired together as recombination products (a + b and c + d). (B) Eight complete conformations (C1–C8) are possible for the Mimulus mitochondrial genome as the result of homologous recombination between one of the three large repeat pairs (direction of black arrows on genome maps indicates orientation between repeat copies). Recombination between inverted repeats leads to inversions of genomic segments in different conformations (gray arrows), whereas recombination between direct repeats causes genomic fission or fusion events (brown arrows). The repeat facilitating a particular recombination event is labeled on each arrow. The six single-copy genomic regions are shown in gradients of color; the same color for each single-copy region was used in all conformations.
To examine the stoichiometric equality of these alternative repeat arrangements, we compared the number of consistent read pairs from the 35 kb library that span each repeat (table 3, “All” column). The four environments for R1 are not significantly different from equality (X2 = 1.64; df = 3; P = 0.64). In contrast, the four R2 environments are significantly unequal in frequency (X2 = 66.4; df = 3; P = 3 × 10−14), as are the four R3 environments (X2 = 48.7; df = 3; P = 1 × 10−10). Because of the small size of the yellow single-copy region and the adjacent R2 and R3 repeats in all eight conformations (fig. 3), we also evaluated the stoichiometry of the four possible environments around this combined segment and again found stoichiometric inequality (X2 = 30.6; df = 3; P = 1 × 10−6). Interestingly, across repeat regions, all the most abundant environments are compatible with the C4 and C6 conformations (table 3).
Table 3.
Read Pair Counts for Alternative Repeat Conformations
| Spanning Read Pairs |
||||
| Region ID | Region Environmenta | All | Fixed Distance | Compatible Conformations |
| A. Repeat R1 (28,763 bp) | ||||
| R1a | Violet-R1-Green | 72 | 58 | C1, C3, C4, C7 |
| R1b | Cyan-R1-Red | 59 | 53 | C1, C3, C4, C7 |
| R1c | Violet-R1-Red | 67 | 58 | C2, C5, C6, C8 |
| R1d | Cyan-R1-Green | 72 | 61 | C2, C5, C6, C8 |
| 230 | All (×2) | |||
| B. Repeat R2 (2,742 bp) | ||||
| R2a | Green-R2-Red | 283 | 54 | C1, C2, C4, C6 |
| R2b | Yellow-R2-Pink | 356 | 81 | C1, C2, C4, C6 |
| R2c | Green-R2-Pink | 202 | 33 | C3, C5, C7, C8 |
| R2d | Yellow-R2-Red | 196 | 37 | C3, C5, C7, C8 |
| 205 | All (×2) | |||
| C. Repeat R3 (9,620 bp) | ||||
| R3a | Violet-R3-Yellow | 130 | 27 | C1, C2, C3, C5 |
| R3b | Pink-R3-Cyan | 241 | 65 | C1, C2, C3, C5 |
| R3c | Violet-R3-Cyan | 217 | 52 | C4, C6, C7, C8 |
| R3d | Pink-R3-Yellow | 265 | 72 | C4, C6, C7, C8 |
| 216 | All (×2) | |||
| D. Segment R2 + R3 (29,250 bp) | ||||
| R3a + R2b | Violet-R3-Yellow-R2-Pink | 22 | 19 | C1, C2 |
| R3a + R2d | Violet-R3-Yellow-R2-Red | 16 | 14 | C3, C5 |
| R3d + R2b | Pink-R3-Yellow-R2-Pink | 54 | 48 | C4, C6 |
| R3d + R2d | Pink-R3-Yellow-R2-Red | 23 | 19 | C7, C8 |
| 100 | All | |||
| E. Single-copy regionsb | ||||
| SC1 | Red (40 kb) | 517 | 76 | All |
| SC2 | Red (110 kb) | 555 | 109 | All |
| SC3 | Green (190 kb) | 604 | 115 | All |
| SC4 | Violet (275 kb) | 529 | 91 | All |
| SC5 | Violet (355 kb) | 565 | 110 | All |
| SC6 | Pink (460 kb) | 646 | 136 | All |
These spanning read pair counts were also used to evaluate whether the various recombination products were present in symmetrical stoichiometry (i.e., coexisting repeat arrangements have equal stoichiometry with each other but not necessarily with the other pair of coexisting arrangements). Stoichiometric symmetry should result from homologous recombination in the absence of processes that amplify or reduce the frequency of single products. For R1, this assumption appears to be valid; there is no significant difference in the frequencies for R1a and R1b from their average of 65.5, nor for R1c and R1d from their average of 69.5 (X2 = 1.5; df = 3; P = 0.69). For R2, although significant differences exist among all four environments as shown in the stoichiometric equality test above, there is only weak evidence against stoichiometric symmetry (X2 = 8.4; df = 3; P = 0.038). This result is likely due to the moderate (1.3-fold) disagreement between the frequencies of coexisting arrangements R2a and R2b because there is virtually no frequency difference between coexisting arrangements R3c and R2d. For R3, however, the evidence against stoichiometric symmetry is strong (X2 = 38; df = 3; P = 3 × 10−8), due to the large (1.9-fold) frequency difference between R3a and R3b, coupled with the moderate (1.2-fold) difference between R3c and R3d.
Variation in Repeat Stoichiometry Is Not due to Cloning or Sequencing Bias
Although significant differences in stoichiometry were found for some repeat environments, we tested whether this pattern could result from some sort of sequencing bias (as opposed to in vivo stoichiometric differences). Sequencing bias could reflect cloning bias during the construction or maintenance of the libraries or from systematic bias of the sequencing platform. The overall stability of TR coverage across the single-copy regions of the genome (evenness of the red line across fig. 1) suggests that sequencing bias is minimal. However, our statistical test for sequencing bias showed that the 1.25-fold variation in read pair counts among single-copy regions (table 3E) is significant (X2 = 20.5; df = 5; P = 0.001). These tallies and the TR coverage in figure 1 suggest a slight excess of reads from the pink single-copy region that lies between R2 and R3 at roughly 420–480 kb in conformation C1.
This 1.25-fold variation among environments can be considered a threshold for detecting stoichiometric differences that cannot be attributed to sequencing bias. At this threshold, the differences in frequency among the four R2 or R3 environments are still significantly greater than expected under stoichiometric equality as they show 1.8- to 2-fold variation in read pair counts among environments, respectively (table 3B and 3C), and a 3.3-fold range for the combined R2 + R3 segment (table 3D). With respect to stoichiometric symmetry, R3 is still significantly asymmetric because of the 1.9-fold variation that exists between R3a and R3b (table 3C). However, the weakly significant result for stoichiometric asymmetry at R2 is less reliable because the frequency difference between R2a and R2b is only 1.26-fold (table 3B), so potential sequencing bias effects cannot be excluded.
Sequencing bias was also examined by comparing the length-adjusted number of spanning read pairs for each large repeat and single-copy region (table 3; “Fixed distance” column). If the stoichiometric differences for R2 and R3 environments are due to pervasive sequencing bias against particular environments rather than real in vivo differences, then the total number of spanning read pairs for all R2 or R3 environments should be lower than for the R1 environments or for twice the number at the single-copy regions. However, this is not the case (X2 = 1.4; df = 3; P = 0.68). In addition, there is no major drop in TR coverage in the single-copy regions adjacent to R2 and R3 compared with other single-copy regions (red line in fig. 1). Thus, there does not appear to be a major reduction in the overall representation of R2 and R3 in the sequence data. Instead, the more pronounced variation in read pair counts among R2 and R3 environments likely reflects true in vivo preferences for particular environments.
Genomic Basis for CMS in M. guttatus IM62
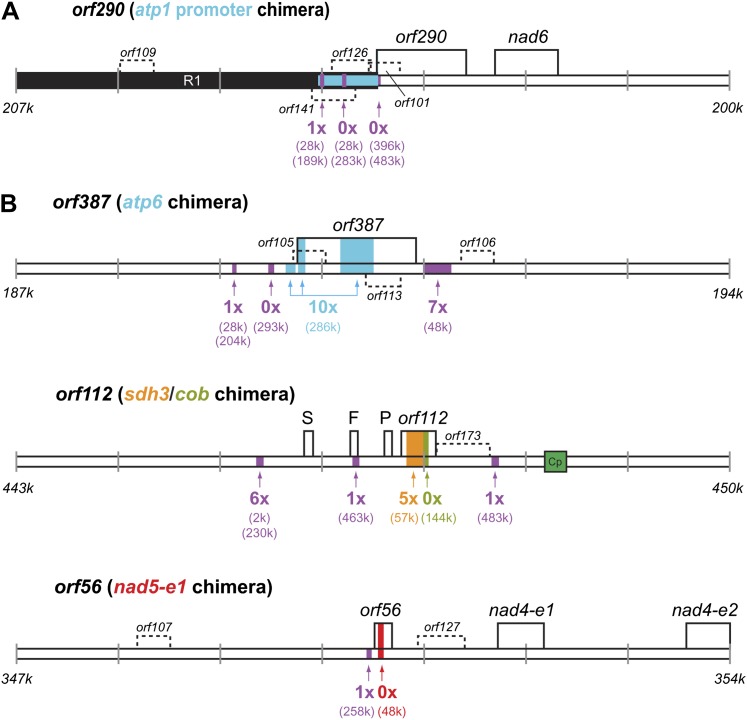
A previous study on the molecular basis of cryptic CMS in M. guttatus IM62 found that the region upstream from the nad6 gene was associated with the CMS phenotype (Case and Willis 2008). It was suggested that one or more ORFs upstream of nad6 (fig. 4A) might be cotranscribed with nad6 and cause CMS in the absence of a fertility restorer allele. A particularly strong candidate for CMS is orf290; it is immediately downstream from a second copy of the atp1 promoter, which is at the end of the largest inverted repeat that is immediately adjacent to orf290, and in fact, the first 8 bp of orf290 and atp1 are identical (fig. 4A). Association with an ATP synthase subunit is important because nearly all the CMS-associated genes known in plants involve proximity to or inclusion of an ATP synthase gene or promoter (Hanson and Bentolila 2004). It is also strongly predicted to encode a protein with a transmembrane domain, which is another common feature of CMS proteins (Hanson and Bentolila 2004).
FIG. 4.—
Candidate CMS genes. (A) Genomic map surrounding the ORFs upstream of the nad6 gene that were previously shown to be associated with the CMS phenotype (Case and Willis 2008). The atp1 promoter region is shown in blue. The beginning is estimated to be 1736 bp upstream of the start of nad6 (Case and Willis 2008). (B) Genomic map surrounding the three ORFs (orf387, orf112, and orf56) identified in this study that may be alternative or additional CMS genes due to their chimeric nature. Small repeats that generate the chimeric portions of the ORFs are color coded according to the genome map in figure 2. Other small repeats are shown on the map in purple. Below each small repeat, the number of read pairs that indicate substoichiometric rearrangements with the other repeat copy is shown, and the genomic position of the other repeat copy (or copies) is given in parentheses. The large repeat R1 is shown in black. A plastid insertion site is shown in green. Additional ORFs not considered candidate CMS genes are shown as shorter boxes with hatched borders.
As an independent strategy to identify additional or alternative candidate CMS genes in the Mimulus mitochondrial genome, an in silico search for chimeric genes was performed. Nine ORFs at least 150 bp in length were identified that contain a >30 bp fragment of a known mitochondrial gene (table 4). Three of these ORFs (orf387, orf112, and orf56) contain the largest fragments of one or more mitochondrial protein-coding genes (fig. 4B), and the fragments are in the same reading frame as the full-length gene copies from which the fragments were presumably derived (table 4). Thus, these ORFs have the potential to cause CMS by directly competing with their functional protein counterparts and disrupting bioenergetic complexes. All three are predicted to encode one or more transmembrane domains, and orf387 is a particularly likely additional candidate because it includes portions of atp6 (table 4; fig. 4B). The six remaining ORFs contain smaller fragments of known genes that are from the complementary DNA strand and/or from ribosomal RNA genes that are not normally translated (table 4), and none is predicted to contain a transmembrane domain. These six ORFs are less likely to be CMS-causing genes unless they act at the RNA level or in a more indirect manner to disrupt mitochondrial function.
Table 4.
Chimeric Mitochondrial Genes
| Chimeric ORF | Gene fragment (nt position in ORF)a | Fragment size (nt) |
| orf387 | atp6 (8–78), atp6 (423–748) | 71, 326 |
| orf112 | sdh3 (64–233), cob (229–280) | 170, 52 |
| orf56 | nad5–e1 (34–88) | 55 |
| orf85 | rrnL (1–44 rc) | 44 |
| orf60 | cox1 (49–92 rc) | 44 |
| orf123 | rrnS (42–78) | 37 |
| orf75 | rrnS (179–212 rc) | 34 |
| orf49 | rrnL (5–37) | 33 |
| orf99 | rrnS (9–38 rc) | 30 |
Gene fragments in bold are in proper orientation and reading frame. rc = reverse complement.
Interestingly, each of the four strongest candidate CMS regions contains small repeats that may facilitate genomic rearrangement (fig. 4). Evaluation of read pair information provides evidence of low-level recombination between orf387 and atp6 and between orf112 and sdh3 (fig. 4), suggesting that recombinant DNA molecules are present at a substoichiometric level in M. guttatus IM62. If these ORFs are CMS genes, rearrangements at these small repeats may be responsible for regulating CMS expression via substoichiometric shifting (McCauley and Olson 2008).
In Northern hybridizations using total RNA from male-sterile and male-fertile full sibs (Case and Willis 2008), atp6, nad6, and cob probes all showed transcript heteromorphism among sibs (sdh3 and nad5 were not tested). However, only atp6 and nad6 heteromorphism was sterility associated in advanced generation backcrosses, although it is possible that the cob portion of orf112 was too small for reliable hybridization. Confirmation of either or all these as active CMS genes awaits additional evidence, such as accumulation of predicted proteins or change in sterility expression with alterations to these ORFs.
Discussion
This study provides comprehensive detail on the sequence, in vivo structure, and genetic content of the mitochondrial genome of M. guttatus IM62, a hermaphroditic wild plant that nonetheless carries a cytoplasmic male sterility system, including one or more mitochondrial sterility genes and one or more nuclear restorer genes.
Repeat Activity and Stoichiometric Variation
The first major result of this work is that the different recombinational environments of the large repeats are all abundant but not always in precisely equal or symmetric stoichiometries (table 3). By using deep paired-end sequencing from large insert libraries, we were able to detect and statistically verify small (∼2-fold) shifts in stoichiometry. The reliability of our computational approach using read pair counts to estimate stoichiometric abundance was experimentally confirmed in Cucumis sativus, in which the 9:1 stoichiometric variation between the 3.6 kb repeat environments was calculated using read pair counts and corroborated by Southern blot analysis (Alverson et al. 2011a). This approach provides a useful tool to address a question that was previously difficult (perhaps impossible) to resolve using more traditional approaches. Southern blotting techniques are not truly quantitative; quantitative polymerase chain reaction, which relies on amplifying products at nearly 100% efficiency, would not be reliable at the distances required to span large repeats, thus limiting its utility for quantifying each unique repeat environment.
This study is among the first to quantitatively and statistically assess the frequency of all large repeat environments in a plant mitochondrial genome (see also Alverson et al. 2011a). Early mapping studies used Southern blot mapping data to show that large plant mitochondrial repeats are recombinogenic (Lonsdale et al. 1984; Palmer and Shields 1984), and many subsequent studies have detected or directly sequenced the alternative environments (e.g., Klein et al. 1994; Ogihara et al. 2005; Sugiyama et al. 2005). Fewer studies have examined the relative frequency of the different environments (e.g., Palmer and Shields 1984; Klein et al. 1994; Sloan et al. 2010), but the consensus view is that they are in dynamic equilibrium due to frequent and reversible homologous recombination between the large repeat arrangements (Lonsdale et al. 1988; Woloszynska 2010). Because these previous studies typically relied on semiquantitative assessments of Southern blot intensities, it would have been easy to overlook the subtle (but significant) level of variation uncovered here, which is 2-fold or less between the most and least abundant arrangements for any one repeat and roughly 3-fold for the combined R2 + R3 segment (table 3). Re-evaluation of the reported stoichiometric equality in other species is likely to uncover additional cases of subtle stoichiometric variation, especially for those species in which minor variation is apparent from Southern blot data.
The In Vivo Structure of the Mitochondrial Genome
In this study, we have presented clear evidence of high-frequency rearrangement at large repeats. Additionally, we showed that many small repeats are also active, albeit at a much lower frequency (supplementary text, Supplementary Material online). Altogether, of the 9,001 pairs of reads that mapped to the mitochondrial genome, >99% can be mapped in a consistent fashion to either the initial assembly or some alternative arrangement derived from recombination involving a large or small repeat. The remaining <1% of read pairs may indicate some novel arrangement formed by homologous recombination at an unidentified small repeat, by illegitimate recombination, or after transfer to the nucleus. Alternatively, they may simply reflect a low level of sequence chimeras or handling errors. Regardless, these results show that very little mitochondrial DNA exists in some unidentified substoichiometric arrangement in vivo in Mimulus. However, they do not tell us about the structure of the genome as a whole. Do mitochondrial genomes truly exist as a collection of master and subgenomic circles, as shown in fig. 3, perhaps in a state so fragile that the circles cannot be recovered intact during electrophoretic or microscopy studies (the so-called “broken circles” theory)? Or are they a collection of linear and multibranching molecules, each with random genomic endpoints resulting in a circularly permuted (and thus circularly mapping) genome?
Our statistical tests of stoichiometric symmetry of the large repeat arrangements were designed to distinguish between circular and linear molecules. If the genome exists primarily as a collection of large circular molecules, we can make two clear predictions about stoichiometric symmetry. First, repeat arrangements that are physically linked in the same master or subgenomic circle must have equal stoichiometry. Second, repeat arrangements that lie on separate subgenomic circles (such as R2a vs. R2b and R3c vs. R3d in conformations C4 and C6) may have unequal stoichiometry if the subgenomic circles experience differential amplification and/or degradation, although there should be consistency in the direction and magnitude of the stoichiometric differences among the repeats on each subgenomic circle (e.g., if C4B is more abundant than C4A, then repeat arrangements R2b and R3d should both exhibit a higher abundance relative to their coexisting arrangements R2a and R3c, respectively). In contrast, if the genome exists primarily as a collection of large linear molecules, then coexisting repeat arrangements will not necessarily be physically linked, allowing them to exhibit asymmetric and uncorrelated stoichiometries due to independent gains or losses of particular linear chromosomal fragments.
Our strongest evidence against the large circular chromosome model for IM62 can be seen in the striking asymmetry of coexisting repeat arrangements R3a and R3b. The 2-fold lower frequency of R3a relative to R3b is unlikely to have been caused by sequencing bias and instead probably reflects real in vivo stoichiometric differences. Because this large difference in abundance is incompatible with the constraint of physical linkage on conformations C1, C2, and C3, it may point to unequal copies of the separate subgenomic circles in conformation C5. However, arrangements R1d and R2c are also separated from their respective coexisting repeats R1c and R2d in conformation C5; yet, neither of these coexisting pairs exhibit a 2-fold difference in abundance. Thus, none of the circular conformations in figure 3B can account for the stoichiometric differences between R3a and R3b. The anomalously low frequency of R3a relative to the other R3 arrangements suggests an independent repression of R3, which could only occur if R3a is physically unlinked from R3b and not circularized with R1c and R2d.
The stoichiometric asymmetry of the R2a and R2b arrangements is also inconsistent with the large circular model, but this case may have other explanations. The frequency difference between R2 arrangements was smaller than at R3 and small enough that sequencing bias could not be ruled out. That being said, we find it striking that the most abundant arrangements for repeats R2, R3, and R2 + R3 were all associated with the C4 and C6 conformations and also all involved the pink and yellow single-copy regions. The C4 and C6 conformations predict an identical 84 kb subgenomic circle comprising R2, R3, and the pink and yellow regions. It is possible that the increased abundance of R2b vs. R2a, R3d vs. R3c, R3d + R2b vs. the other combined segments, and the pink single-copy region (SC1) vs. other single-copy regions in table 3 reflects disproportional amplification of this particular subgenomic circle relative to the rest of the genome.
Our results clearly indicate dynamic recombinational stoichiometry and variable conformational structure in the mitochondrial genome of IM62 in vivo. Given the asymmetry of some repeat arrangements, we argue that the master and subgenomic circles shown in figure 3B are not the predominant conformations of the mitochondrial genome. We do not know whether the observed mitochondrial genomic variability occurs at the scale of different individuals in a population or within individual plants because the DNA used for whole-genome shotgun sequencing was prepared from multiple plants. However, the plants were grown from seeds produced by self-pollination of a single highly inbred line, suggesting that there should be minimal variation among individuals. Furthermore, many studies have observed multiple genomic conformations within individual plants (reviewed in Kmiec et al. 2006; McCauley and Olson 2008; Woloszynska 2010), indicating that the different large repeat environments are present at appreciable levels within a single plant. Thus, it is unlikely that the variation we detect is due to the averaging of minor to major stoichiometric differences among individual plants. Rather, the inequality we find is probably due to slight deviations from dynamic equilibrium within an individual.
Future work is necessary to evaluate genomic conformations between different tissue types. It remains possible that large circular chromosomes may exist in specific tissues or at a low level throughout the plant. For instance, it is possible that master circle conformations may be the predominant form in meristematic tissue to ensure faithful replication and propagation of the mitochondrial genome (Backert et al. 1997; Arrieta-Montiel et al. 2001; Sakai et al. 2004; Woloszynska 2010). But, based on our evidence, most mitochondrial DNA in whole individuals cannot exist in a large circular conformation in Mimulus.
Verification of the Assembled Genome
In our experience, plant mitochondrial genomes rarely assemble into a single contig in genome sequencing projects. This is typically due to the inability of assembly software to correctly position multiple copies of large repeats and the confounding influence of alternative genomic environments in which these repeats are found. Given these difficulties and the uncertainty regarding the in vivo structure of plant mitochondrial genomes, it is surprising that most complete genome reports provide few details on the methods used to close gaps in the assembly and little to no evidence to support the accuracy of their finished assembly. Without these details, it is impossible to know whether a genome was assembled correctly.
For the Mimulus genome reported here, we conducted four independent assemblies using different library insert sizes, all of which produced essentially identical consensus sequences, thus providing verification of the assembly at the single-nucleotide level and at larger genomic scales. Mapping of the read pairs from the 35 kb insert library (fig. 1) provided unambiguous support for the methods used to close gaps including the contig end-joins and inferred repeats (supplementary fig. 1, Supplementary Material online) and confirmed that the entire consensus sequence is supported by read pair data. The high quality of the sequencing data used here, which was generated by paired-end Sanger sequencing of clonal libraries with large and variable insert sizes, ensured that the assembly would also be of high quality. However, as sequencing projects shift to using next generation sequencing technologies, which typically use smaller library insert sizes and produce shorter and less accurate reads, it will become ever more important to provide evidence of assembly accuracy. We hope that our comprehensive verification procedure will serve as a model for future plant mitochondrial genome projects.
Genome Evolution: tRNAs, Ribosomal Proteins, and Chimeric ORFs
In most respects, the genetic content of the Mimulus mitochondrial genome is fairly typical of other angiosperms. However, the four novel tRNAs predicted to carry threonine or leucine are unusual because tRNAs for these amino acids are rare to absent in sequenced mitochondrial genomes from other angiosperms (summarized in Sloan et al. 2010). The trnT-GGU gene is similar (∼80%) to unannotated regions in at least five other angiosperms, suggesting a previously unrecognized tRNA of potential functionality in many species. The origins and evolutionary implications of the three nonhomologous trnL genes are also curious. Presumably, these three tRNAs independently shifted their anticodon from nonleucine amino acids to leucine, but it is not known if they are functional or why they became necessary in Mimulus. Further comparative analysis is required from other asterids (Lamiales, in particular) to determine the timing and functional significance of these apparent anticodon changes.
For the four ribosomal proteins lost from the Mimulus mitochondrial genome, the availability of the assembled nuclear genome allowed us to search for their functional replacements. The putative nuclear RPL2 and RPS1 proteins are most similar (>50% identity) to several mitochondrial homologs from other plants available in GenBank, suggesting that the Mimulus nuclear genes were derived by direct functional transfer of the mitochondrial genes to the nuclear genome. In contrast, Mimulus nuclear RPS7 and RPS19, which were identified by strongest similarity to Vitis mitochondrial homologs, are in fact much more similar (>90% identity) to plastid or cytosolic ribosomal proteins in GenBank. In other words, we did not find any evidence for a mitochondrial gene or mitochondrion-derived nuclear gene encoding RPS7 or RPS19 in Mimulus. It is possible that mitochondrion-derived nuclear genes are absent from the assembly (i.e., they lie in the currently unassembled regions of the nuclear genome). However, the presence of multiple nuclear genes encoding plastid or cytosolic ribosomal proteins for RPS7 and RPS19 suggests that one or more of these products may be retargeted to the mitochondrion to functionally replace the lost mitochondrial version, as also observed for several other ribosomal proteins in other plants (Adams, Daley, et al. 2002; Mollier et al. 2002; Mower and Bonen 2009; Kubo and Arimura 2010).
Finally, we were surprised to find four ORFs exhibiting strong features of known CMS-causing genes, including a chimeric structure (two containing part of an ATP synthase subunit) and the presence of predicted transmembrane domains. Work is underway to determine whether any or all these ORFs are expressed and functional, either as CMS genes or otherwise, because direct association with the male-sterile phenotype is currently limited to transcription of orf290 (Case and Willis 2008). Although segregation of male sterility in controlled crosses was consistent with a single nuclear restorer locus (Fishman and Willis 2006), Rf in M. guttatus IM62 actually involves two tightly linked nuclear loci (Barr and Fishman 2010). Multiple Rf loci do not necessarily imply that there are multiple CMS genes in the IM62 mitochondrial genome, although it is possible. These two Rf loci do not act epistatically because a single dominant allele at either locus was sufficient to restore male fertility in CMS lines in M. guttatus (Barr and Fishman 2010). Analysis of mitochondrial gene expression with alternate Rf genotypes may reveal whether each locus alters nad6 or atp6 transcription profiles in addition to restoring male fertility.
All four candidate CMS genes contained multiple small repeats, although not all were obviously actively recombining in the tissues used to create the IM62 genomic libraries. Historical activity at these repeats may have created the chimeric genes in the first place and/or placed them in locations within the genome that favored their transcription. Studies in crop systems suggest that CMS expression may be regulated by current activity at small repeats that alter or relocate a CMS gene prior to transcription (reviewed in McCauley and Olson 2008) and that nuclear loci may be responsible for the timing or cues for rearrangement (Arrieta-Montiel et al. 2009). It is also possible that it is the substoichiometric recombination products (rather than the chimeric ORFs themselves) that cause CMS. Whether these substoichiometric molecules are variably present among individual mitochondria, among cells or tissue types, or among different individuals cannot be determined from read pair data alone. If they were, then the expression of CMS in natural populations would vary irrespective of their genotype at the nuclear restorer loci. Indeed, Barr and Fishman (2010) found a very small number of M. guttatus IM62 individuals that lacked both restorer alleles but were still male fertile. This may indicate a role for stoichiometry or recombination in regulating CMS expression, although several attempts to document such an effect have been inconclusive. Markers for alternate forms of orf290 suggest that many wild accessions of M. guttatus do contain a mixture of mitotypes where orf290 is either complete or truncated (Floro 2011; Case AL, unpublished data). Further characterization of all candidate CMS genes and their recombination products is necessary to unambiguously identify the source of cryptic CMS in M. guttatus IM62. Comparative analyses with other M. guttatus mitochondrial genomes that do not harbor cryptic CMS will be important for understanding the consequences of a history of CMS on mitochondrial genome structure and evolution.
Supplementary Material
Supplementary text, figures 1–5, and table 1 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
The authors thank Ken Wolfe for providing lab resources and assistance with the genome assembly, the Irish Centre for High-End Computing for providing computational facilities and support, the U.S. Department of Energy Joint Genome Institute (especially Dan Rokhsar and Kerrie Barry) for primary genome sequencing and support, Anna Blenda and the Clemson University Genomics Institute for clone library hybridizations, and Dan Sloan for providing insightful comments on the manuscript. This work was supported by a postdoctoral fellowship from the Irish Research Council for Science, Engineering and Technology (J.P.M.), a Clark Fellowship in Comparative Genomics from Duke University (A.L.C.), start-up funds from the University of Nebraska-Lincoln (J.P.M.) and from Kent State University (A.L.C.), and the National Science Foundation (IOS-1027529 and MCB-1125386 to J.P.M.; EF-0328636 and EF-0723814 to J.H.W.). The work conducted by the Joint Genome Institute is supported by the Office of Science of the U.S. Department of Energy under Contract No. DE-AC02-05CH11231. Data deposition: Mimulus raw shotgun sequence reads are available from the NCBI Trace archives at ftp://ftp.ncbi.nih.gov/pub/TraceDB/mimulus_guttatus/. The assembled mitochondrial genome sequence was deposited in GenBank under accession # JN098455.
References
- Adams KL, Daley DO, Whelan J, Palmer JD. Genes for two mitochondrial ribosomal proteins in flowering plants are derived from their chloroplast or cytosolic counterparts. Plant Cell. 2002;14:931–943. doi: 10.1105/tpc.010483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams KL, Qiu YL, Stoutemyer M, Palmer JD. Punctuated evolution of mitochondrial gene content: high and variable rates of mitochondrial gene loss and transfer to the nucleus during angiosperm evolution. Proc Natl Acad Sci U S A. 2002;99:9905–9912. doi: 10.1073/pnas.042694899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen JO, et al. Comparisons among two fertile and three male-sterile mitochondrial genomes of maize. Genetics. 2007;177:1173–1192. doi: 10.1534/genetics.107.073312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alverson AJ, et al. Insights into the evolution of mitochondrial genome size from complete sequences of Citrullus lanatus and Cucurbita pepo (Cucurbitaceae) Mol Biol Evol. 2010;27:1436–1448. doi: 10.1093/molbev/msq029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alverson AJ, et al. Origins and recombination of the bacterial-sized multichromosomal mitochondrial genome of cucumber. Plant Cell. 2011a;23:2499–2513. doi: 10.1105/tpc.111.087189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alverson AJ, et al. The mitochondrial genome of the legume Vigna radiata and the analysis of recombination across short mitochondrial repeats. PLoS One. 2011b;6:e16404. doi: 10.1371/journal.pone.0016404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrieta-Montiel MP, Mackenzie SA. Plant mitochondrial genomes and recombination. In: Kempken Frank., editor. Plant mitochondria. New York: Springer; 2011. pp. 65–82. [Google Scholar]
- Arrieta-Montiel M, et al. Tracing evolutionary and developmental implications of mitochondrial stoichiometric shifting in the common bean. Genetics. 2001;158:851–864. doi: 10.1093/genetics/158.2.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrieta-Montiel MP, et al. Diversity of the Arabidopsis mitochondrial genome occurs via nuclear-controlled recombination activity. Genetics. 2009;183:1261–1268. doi: 10.1534/genetics.109.108514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backert S, Börner T. Phage T4-like intermediates of DNA replication and recombination in the mitochondria of the higher plant Chenopodium album (L.) Curr Genet. 2000;37:304–314. doi: 10.1007/s002940050532. [DOI] [PubMed] [Google Scholar]
- Backert S, Lynn Nielsen B, Börner T. The mystery of the rings: structure and replication of mitochondrial genomes from higher plants. Trends Plant Sci. 1997;2:477–483. [Google Scholar]
- Barr CM, Fishman L. The nuclear component of a cytonuclear hybrid incompatibility in Mimulus maps to a cluster of pentatricopeptide repeat genes. Genetics. 2010;184:455–465. doi: 10.1534/genetics.109.108175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonen L. RNA splicing in plant mitochondria. In: Kempken Frank., editor. Plant Mitochondria. New York: Springer; 2011. pp. 131–155. [Google Scholar]
- Case AL, Willis JH. Hybrid male sterility in Mimulus (Phrymaceae) is associated with a geographically restricted mitochondrial rearrangement. Evolution. 2008;62:1026–1039. doi: 10.1111/j.1558-5646.2008.00360.x. [DOI] [PubMed] [Google Scholar]
- Darracq A, et al. Structural and content diversity of mitochondrial genome in beet: a comparative genomic analysis. Genome Biol Evol. 2011;3:723–736. doi: 10.1093/gbe/evr042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davila JI, et al. Double-strand break repair processes drive evolution of the mitochondrial genome in Arabidopsis. BMC Biol. 2011;9:64. doi: 10.1186/1741-7007-9-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishman L, Willis JH. A cytonuclear incompatibility causes anther sterility in Mimulus hybrids. Evolution. 2006;60:1372–1381. doi: 10.1554/05-708.1. [DOI] [PubMed] [Google Scholar]
- Floro ER. Mitochondrial heteroplasmy in Mimulus guttatus [master’s thesis] [Kent (OH)]: Kent State University; 2011. OhioLink document number kent1302199999. [Google Scholar]
- Folkerts O, Hanson MR. Three copies of a single recombination repeat occur on the 443 Kb mastercircle of the Petunia hybrida 3704 mitochondrial genome. Nucleic Acids Res. 1989;17:7345–7357. doi: 10.1093/nar/17.18.7345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii S, Kazama T, Yamada M, Toriyama K. Discovery of global genomic re-organization based on comparison of two newly sequenced rice mitochondrial genomes with cytoplasmic male sterility-related genes. BMC Genomics. 2010;11:209. doi: 10.1186/1471-2164-11-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon D, Abajian C, Green P. Consed: a graphical tool for sequence finishing. Genome Res. 1998;8:195–202. doi: 10.1101/gr.8.3.195. [DOI] [PubMed] [Google Scholar]
- Goremykin VV, Salamini F, Velasco R, Viola R. Mitochondrial DNA of Vitis vinifera and the issue of rampant horizontal gene transfer. Mol Biol Evol. 2009;26:99–110. doi: 10.1093/molbev/msn226. [DOI] [PubMed] [Google Scholar]
- Handa H. The complete nucleotide sequence and RNA editing content of the mitochondrial genome of rapeseed (Brassica napus L.): comparative analysis of the mitochondrial genomes of rapeseed and Arabidopsis thaliana. Nucleic Acids Res. 2003;31:5907–5916. doi: 10.1093/nar/gkg795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson MR, Bentolila S. Interactions of mitochondrial and nuclear genes that affect male gametophyte development. Plant Cell. 2004;16(Suppl):S154–S169. doi: 10.1105/tpc.015966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht J, Grewe F, Knoop V. Extreme RNA editing in coding islands and abundant microsatellites in repeat sequences of Selaginella moellendorffii mitochondria: the root of frequent plant mtDNA recombination in early tracheophytes. Genome Biol Evol. 2011;3:344–358. doi: 10.1093/gbe/evr027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, et al. PCAP: a whole-genome assembly program. Genome Res. 2003;13:2164–2170. doi: 10.1101/gr.1390403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janska H, Woloszynska M. The dynamic nature of plant mitochondrial genome organization. Acta Biochim Pol. 1997;44:239–250. [PubMed] [Google Scholar]
- Kaul MLH. Male sterility in higher plants. New York: Springer-Verlag; 1988. [Google Scholar]
- Kempken F, Pring DR. Male sterility in higher plants—fundamentals and applications. Prog Bot. 1999;60:139–166. [Google Scholar]
- Kitazaki K, et al. A mitochondrial gene involved in cytochrome c maturation (ccmC) is expressed as a precursor with a long NH2-terminal extension in sugar beet. J Plant Physiol. 2009;166:775–780. doi: 10.1016/j.jplph.2008.09.002. [DOI] [PubMed] [Google Scholar]
- Klein M, et al. Physical mapping of the mitochondrial genome of Arabidopsis thaliana by cosmid and YAC clones. Plant J. 1994;6:447–455. doi: 10.1046/j.1365-313x.1994.06030447.x. [DOI] [PubMed] [Google Scholar]
- Kmiec B, Woloszynska M, Janska H. Heteroplasmy as a common state of mitochondrial genetic information in plants and animals. Curr Genet. 2006;50:149–159. doi: 10.1007/s00294-006-0082-1. [DOI] [PubMed] [Google Scholar]
- Knoop V, et al. copia-, gypsy- and LINE-like retrotransposon fragments in the mitochondrial genome of Arabidopsis thaliana. Genetics. 1996;142:579–585. doi: 10.1093/genetics/142.2.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo N, Arimura S. Discovery of the rpl10 gene in diverse plant mitochondrial genomes and its probable replacement by the nuclear gene for chloroplast RPL10 in two lineages of angiosperms. DNA Res. 2010;17:1–9. doi: 10.1093/dnares/dsp024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo T, et al. The complete nucleotide sequence of the mitochondrial genome of sugar beet (Beta vulgaris L.) reveals a novel gene for tRNACys(GCA) Nucleic Acids Res. 2000;28:2571–2576. doi: 10.1093/nar/28.13.2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laser K, Lersten N. Anatomy and cytology of microsporogenesis in cytoplasmic male sterile angiosperms. Bot Rev. 1972;38:425–454. [Google Scholar]
- Lonsdale DM, Hodge TP, Fauron CM. The physical map and organisation of the mitochondrial genome from the fertile cytoplasm of maize. Nucleic Acids Res. 1984;12:9249–9261. doi: 10.1093/nar/12.24.9249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonsdale DM, et al. The plant mitochondrial genome: homologous recombination as a mechanism for generating heterogeneity. Phil Trans R Soc B. 1988;319:149–163. [Google Scholar]
- Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie SA. The unique biology of mitochondrial genome instability in plants. In: Logan DC, editor. Plant mitochondria. Oxford: Blackwell Publishing, Ltd; 2007. pp. 36–49. [Google Scholar]
- Manchekar M, et al. DNA recombination activity in soybean mitochondria. J Mol Biol. 2006;356:288–299. doi: 10.1016/j.jmb.2005.11.070. [DOI] [PubMed] [Google Scholar]
- Maréchal A, Brisson N. Recombination and the maintenance of plant organelle genome stability. New Phytol. 2010;186:299–317. doi: 10.1111/j.1469-8137.2010.03195.x. [DOI] [PubMed] [Google Scholar]
- McCauley DE, Olson MS. Do recent findings in plant mitochondrial molecular and population genetics have implications for the study of gynodioecy and cytonuclear conflict? Evolution. 2008;62:1013–1025. doi: 10.1111/j.1558-5646.2008.00363.x. [DOI] [PubMed] [Google Scholar]
- Mollier P, Hoffmann B, Debast C, Small I. The gene encoding Arabidopsis thaliana mitochondrial ribosomal protein S13 is a recent duplication of the gene encoding plastid S13. Curr Genet. 2002;40:405–409. doi: 10.1007/s00294-002-0271-5. [DOI] [PubMed] [Google Scholar]
- Mower JP. The PREP suite: predictive RNA editors for plant mitochondrial genes, chloroplast genes and user-defined alignments. Nucleic Acids Res. 2009;37:W253–W259. doi: 10.1093/nar/gkp337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mower JP, Bonen L. Ribosomal protein L10 is encoded in the mitochondrial genome of many land plants and green algae. BMC Evol Biol. 2009;9:265. doi: 10.1186/1471-2148-9-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mower JP, Palmer JD. Patterns of partial RNA editing in mitochondrial genes of Beta vulgaris. Mol Genet Genomics. 2006;276:285–293. doi: 10.1007/s00438-006-0139-3. [DOI] [PubMed] [Google Scholar]
- Mower JP, Sloan DB, Alverson AJ. Plant mitochondrial genome diversity: the genomics revolution. In: Wendel JH, editor. Plant genome diversity volume 1: plant genomes, their residents, and their evolutionary dynamics. New York: Springer; 2012. pp. 123–144. [Google Scholar]
- Notsu Y, et al. The complete sequence of the rice (Oryza sativa L.) mitochondrial genome: frequent DNA sequence acquisition and loss during the evolution of flowering plants. Mol Genet Genomics. 2002;268:434–445. doi: 10.1007/s00438-002-0767-1. [DOI] [PubMed] [Google Scholar]
- Oda K, et al. Gene organization deduced from the complete sequence of liverwort Marchantia polymorpha mitochondrial DNA. A primitive form of plant mitochondrial genome. J Mol Biol. 1992;223:1–7. doi: 10.1016/0022-2836(92)90708-r. [DOI] [PubMed] [Google Scholar]
- Ogihara Y, et al. Structural dynamics of cereal mitochondrial genomes as revealed by complete nucleotide sequencing of the wheat mitochondrial genome. Nucleic Acids Res. 2005;33:6235–6250. doi: 10.1093/nar/gki925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldenburg DJ, Bendich AJ. Size and structure of replicating mitochondrial DNA in cultured tobacco cells. Plant Cell. 1996;8:447–461. doi: 10.1105/tpc.8.3.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldenburg DJ, Bendich AJ. The structure of mitochondrial DNA from the liverwort, Marchantia polymorpha. J Mol Biol. 1998;276:745–758. doi: 10.1006/jmbi.1997.1581. [DOI] [PubMed] [Google Scholar]
- Palmer JD. Intraspecific variation and multicircularity in Brassica mitochondrial DNAs. Genetics. 1988;118:341–351. doi: 10.1093/genetics/118.2.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer JD, Herbon LA. Tricircular mitochondrial genomes of Brassica and Raphanus: reversal of repeat configurations by inversion. Nucleic Acids Res. 1986;14:9755–9764. doi: 10.1093/nar/14.24.9755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer JD, Herbon LA. Plant mitochondrial DNA evolves rapidly in structure, but slowly in sequence. J Mol Evol. 1988;28:87–97. doi: 10.1007/BF02143500. [DOI] [PubMed] [Google Scholar]
- Palmer JD, Shields CR. Tripartite structure of the Brassica campestris mitochondrial genome. Nature. 1984;307:437–440. [Google Scholar]
- Rodriguez-Moreno L, et al. Determination of the melon chloroplast and mitochondrial genome sequences reveals that the largest reported mitochondrial genome in plants contains a significant amount of DNA having a nuclear origin. BMC Genomics. 2011;12:424. doi: 10.1186/1471-2164-12-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai A, Takano H, Kuroiwa T. Organelle nuclei in higher plants: structure, composition, function, and evolution. Int Rev Cytol. 2004;238:59–118. doi: 10.1016/S0074-7696(04)38002-2. [DOI] [PubMed] [Google Scholar]
- Schnable PS, Wise RP. The molecular basis of cytoplasmic male sterility and fertility restoration. Trends Plant Sci. 1998;3:175–180. [Google Scholar]
- Siculella L, et al. Gene content and organization of the oat mitochondrial genome. Theor Appl Genet. 2001;103:359–365. [Google Scholar]
- Sloan DB, et al. Extensive loss of translational genes in the structurally dynamic mitochondrial genome of the angiosperm Silene latifolia. BMC Evol Biol. 2010;10:274. doi: 10.1186/1471-2148-10-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DR, Hua J, Lee RW. Evolution of linear mitochondrial DNA in three known lineages of Polytomella. Curr Genet. 2010;56:427–438. doi: 10.1007/s00294-010-0311-5. [DOI] [PubMed] [Google Scholar]
- Stern DB, Palmer JD. Tripartite mitochondrial genome of spinach: physical structure, mitochondrial gene mapping, and locations of transposed chloroplast DNA sequences. Nucleic Acids Res. 1986;14:5651–5666. doi: 10.1093/nar/14.14.5651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama Y, et al. The complete nucleotide sequence and multipartite organization of the tobacco mitochondrial genome: comparative analysis of mitochondrial genomes in higher plants. Mol Genet Genomics. 2005;272:603–615. doi: 10.1007/s00438-004-1075-8. [DOI] [PubMed] [Google Scholar]
- Sunkel S, Brennicke A, Knoop V. RNA editing of a conserved reading frame in plant mitochondria increases its similarity to two overlapping reading frames in Escherichia coli. Mol Gen Genet. 1994;242:65–72. doi: 10.1007/BF00277349. [DOI] [PubMed] [Google Scholar]
- Terasawa K, et al. The mitochondrial genome of the moss Physcomitrella patens sheds new light on mitochondrial evolution in land plants. Mol Biol Evol. 2007;24:699–709. doi: 10.1093/molbev/msl198. [DOI] [PubMed] [Google Scholar]
- Tiffin P, Olson MS, Moyle LC. Asymmetrical crossing barriers in angiosperms. Proc Biol Sci. 2001;268:861–867. doi: 10.1098/rspb.2000.1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmis JN, Ayliffe MA, Huang CY, Martin W. Endosymbiotic gene transfer: organelle genomes forge eukaryotic chromosomes. Nat Rev Genet. 2004;5:123–135. doi: 10.1038/nrg1271. [DOI] [PubMed] [Google Scholar]
- Woloszynska M. Heteroplasmy and stoichiometric complexity of plant mitochondrial genomes-though this be madness, yet there's method in't. J Exp Bot. 2010;61:657–671. doi: 10.1093/jxb/erp361. [DOI] [PubMed] [Google Scholar]
- Wu CA, et al. Mimulus is an emerging model system for the integration of ecological and genomic studies. Heredity. 2008;100:220–230. doi: 10.1038/sj.hdy.6801018. [DOI] [PubMed] [Google Scholar]