Abstract
The diversity and abundance of non–long terminal repeat (LTR) retrotransposons (nLTR-RT) differ drastically among vertebrate genomes. At one extreme, the genome of placental mammals is littered with hundreds of thousands of copies resulting from the activity of a single clade of nLTR-RT, the L1 clade. In contrast, fish genomes contain a much more diverse repertoire of nLTR-RT, represented by numerous active clades and families. Yet, the number of nLTR-RT copies in teleostean fish is two orders of magnitude smaller than in mammals. The vast majority of insertions appear to be very recent, suggesting that nLTR-RT do not accumulate in fish genomes. This pattern had previously been explained by a high rate of turnover, in which the insertion of new elements is offset by the selective loss of deleterious inserts. The turnover model was proposed because of the similarity between fish and Drosophila genomes with regard to their nLTR-RT profile. However, it is unclear if this model applies to fish. In fact, a previous study performed on the puffer fish suggested that transposable element insertions behave as neutral alleles. Here we examined the dynamics of amplification of nLTR-RT in the three-spine stickleback (Gasterosteus aculeatus). In this species, the vast majority of nLTR-RT insertions are relatively young, as suggested by their low level of divergence. Contrary to expectations, a majority of these insertions are fixed in lake and oceanic populations; thus, nLTR-RT do indeed accumulate in the genome of their fish host. This is not to say that nLTR-RTs are fully neutral, as the lack of fixed long elements in this genome suggests a deleterious effect related to their length. This analysis does not support the turnover model and strongly suggests that a much higher rate of DNA loss in fish than in mammals is responsible for the relatively small number of nLTR-RT copies and for the scarcity of ancient elements in fish genomes. We further demonstrate that nLTR-RT decay in fish occurs mostly through large deletions and not by the accumulation of small deletions.
Keywords: non-LTR retrotransposon, retroposon, Gasterosteus aculeatus, three-spine stickleback, genome size evolution
Introduction
Non–long terminal repeat (LTR) retrotransposons (nLTR-RT) are mobile elements in the genome that replicate using an RNA intermediate and lack LTRs. They have considerably affected the size, structure, and function of vertebrate genomes. In fact, the abundance of nLTR-RT is one of the major determinants of genome size differences among vertebrates. The impact nLTR-RTs have on their host is directly related to their diversity and abundance, which differ considerably among vertebrate groups. In mammals, nLTR-RTs are extremely abundant and account for as much as 30% of genome size (Lander et al. 2001; Waterston et al. 2002). Mammalian genomes are dominated by a single clade of nLTR-RT called L1 (Furano 2000). L1 has been amplifying since the origin of the eutharian radiation and has accumulated to considerable numbers, accounting for the large genome size of mammals (2.0–3.6 GB). In stark contrast, the genomes of teleostean fish and squamate reptiles tend to be small and to contain an extraordinary diversity of active nLTR-RT, generally representing multiple clades (Volff et al. 2003; Duvernell et al. 2004; Furano et al. 2004; Novick et al. 2009). These clades are generally represented by multiple and distinct groups of sequences, called families, that are concurrently active. Families of elements are usually represented by small numbers (10 to a few hundreds) of very similar copies, suggesting that the majority of insertions are recent and do not accumulate in the genome of the host (Duvernell et al. 2004; Furano et al. 2004). The young age and small copy number of nLTR-RT in fish is suggestive of a rapid turnover of elements, in which the insertion of new elements is offset by the selective loss of element-containing loci. However, the turnover model has not been rigorously tested in fish and was proposed because of the similarity between fish and Drosophila with regard to their nLTR-RT profile (Duvernell et al. 2004; Furano et al. 2004). In fact, the only population study done on a fish, the puffer fish, found a high number of fixed and high frequency insertions, suggesting that nLTR-RT are neutral, at least in this fish species (Neafsey et al. 2004).
Teleostean fish constitute the most diverse vertebrate group, and this diversity is also reflected in the diversity of their genome size and structure (Volff 2005). A bioinformatic exploration of teleostean genomes has revealed considerable differences in the diversity and abundance of nLTR-RT among species (Basta et al. 2007). The factors responsible for these differences are not well understood. The copy number and family diversity in a given genome result from the interactions between the rate of transposition, the control of transposition by the host, competition between families of elements for host-encoded resources, the intensity of selection against new inserts, and the demographic history of populations. How these different factors interact remains unclear because empirical studies in natural populations are limited to a very small number of taxa and comparative studies are lacking. Here we present a detailed analysis of nLTR-RT in the three-spine stickleback (Gasterosteus aculeatus).
Gasterosteus aculeatus is a small teleostean fish that has become one of the premier animal models in evolutionary biology. It is found in the coastal waters of the northern Atlantic and Pacific Oceans. It is originally an oceanic species, but it has colonized innumerable freshwater habitats where it has undergone an extremely rapid adaptive radiation resulting in morphologically diverse populations (Bell and Foster 1994). A draft of the stickleback genome has been available since February 2006 on the University of California—Santa Cruz (UCSC) genome browser (http://genome.ucsc.edu). The individual that was sequenced comes from the Bear Paw Lake population in Alaska. It was chosen because of the low heterozygosity of this population due to isolation since the lake was colonized less than 14,000 years ago. We performed a bioinformatic analysis of the stickleback genome to assess the diversity of nLTR-RT in this species. We also determined the frequency of nLTR-RT in oceanic and lake populations, in particular from the population of origin of the sequenced genome. We found that short nLTR-RTs accumulate readily in the stickleback genome, whereas full-length copies appear to be under purifying selection. However, the near absence of ancient nLTR-RT copies suggests that a post-insertional mechanism is controlling nLTR-RT copy number in this species. We found that a much higher rate of DNA loss in fish than in mammals is responsible for the relatively small number of nLTR-RT copies and for the paucity of ancient elements in fish genomes.
Materials and Methods
Coordinates for all nLTR-RT elements were extracted from the February 2006 version of the stickleback genome (v1.0) using the RepeatMasker table available from the UCSC genome browser (www.genome.ucsc.edu). Elements were then collected using the coordinates of the elements to which 500 bp of downstream and upstream sequences were added. In the case of the Maui elements, RepeatMasker did not identify accurately the 5′ end of the elements; thus, 2 kb of upstream sequences were collected in this case. The length of each insertion as well as its start and end points were determined.
Within each clade, elements were aligned to each other using ClustalW in BioEdit (Hall 1999) to identify subsets of sequences that would represent distinct families. To this end, only elements at least 300 bp in length were included. Once the elements were aligned, a phylogenetic analysis using the neighbor joining and maximum likelihood methods implemented in MEGA5.0 was performed. Groups of sequences that were well supported by a bootstrap procedure (1,000 iterations; at least 80% bootstrap support) were considered valid families. A consensus sequence was determined for each family. Each family was characterized by its copy number (using a 100-bp cutoff) and its divergence used as a proxy of its age. Within-family divergences were estimated using the mean pairwise divergence between members of the families or the mean divergence between each member and the family consensus. Divergences and their standard deviation were calculated using MEGA5.0.
Consensus sequences were aligned to each other. The National Center for Biotechnology Information ORF-Finder and Conserved Domains tools were used to identify the reverse transcriptase (RTase) domain, which was translated into amino acid by ORF-Finder. The RTase domains were then aligned with the RTase domains of other nLTR-RT representative of the major clades of nLTR-RT. Phylogenies of the RTase domains were then constructed using the maximum likelihood method implemented in MEGA5.0.
The frequency of RTE insertions was determined experimentally on ten stickleback populations. The Geographic Information System coordinates of the populations are provided as Supplementary Material online. The fraction of fixed and polymorphic (for presence/absence) insertions was determined experimentally. DNA was extracted from the muscle or fin of either frozen or ethanol-preserved fish. Tissues were digested with proteinase K followed by a phenol/chloroform extraction and ethanol precipitation. The quality of the DNA extraction was verified by electrophoresis on a 1% agarose gel followed by ethidium bromide staining. The presence or absence of specific nLTR-RT insertions was determined using polymerase chain reaction (PCR). Primers in the flanking sequence of the insertions were designed manually or using the Primer3 program (Rozen and Skaletsky 2000). The specificity of the primers was verified using the in silico PCR tool from the UCSC web page (www.genome.ucsc.edu). For inserts longer than 1.5 kb, a second PCR was performed using a primer cognate to the flank and an internal primer. PCR products were run on 1% agarose gels. The sequence of the primers is provided as Supplementary Material online.
Results
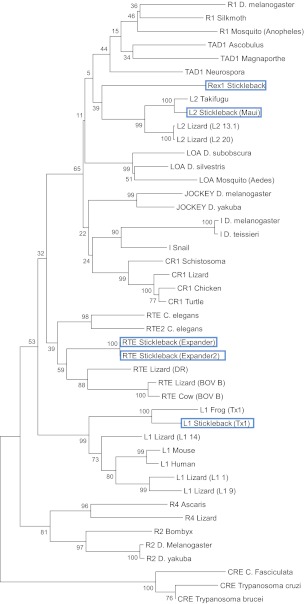
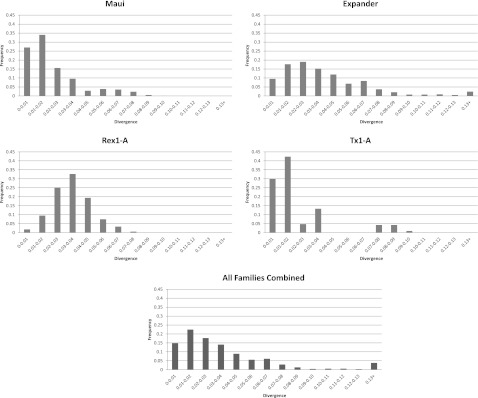
The stickleback genome contains 11 families of nLTR-RT belonging to 4 of the 28 clades identified previously (Kapitonov et al. 2009): the L1/Tx1, L2, Rex/Babar, and RTE clades (fig. 1). This level of clade diversity is consistent with the analysis of Basta et al. (2007) who used a completely different approach to identify retrotransposons (McClure et al. 2005). With ∼2,396 elements, but only 12 full-length copies, the most abundant clade, L2, is represented by a single family with high similarity to the Maui family previously described in Takifugu rubripes (Poulter et al. 1999) (table 1). Notably, about a third of the elements are shorter than 100 bp, indicating a high level of fragmentation of these elements. Figure 2A depicts a phylogenetic tree of Maui elements. This tree has the typical cascade structure expected when a single family of closely related elements is active in a genome. Elements closer to the root represent older copies, whereas clusters of very similar sequences indicate recent activity of the family. In fact, the presence of groups of elements that are identical to each other (reflected by the branches of null length) strongly suggests that Maui is active in the stickleback. The recent activity of Maui is reflected in the relatively low average divergence of the family (2.2% pairwise divergence; table 1) as well as the distribution of pairwise divergence (fig. 3), where most values fall under 4% and no values are above 10%.
FIG. 1.—
Phylogenetic position of the three-spine stickleback elements among the diversity of nLTR-RTs. The stickleback consensus sequences are framed in blue. This maximum likelihood tree was constructed from a portion of the translated RTase domain using the rtREV + G + I + F model of substitution. The robustness of the nodes was assessed using a bootstrap procedure (500 iterations).
Table 1.
Copy Number and Divergence of Stickleback nLTR-RT
| Clade | Family | Copy Number (>100 bp) | Copy Number (>300 bp) | Full-Length Copy Number | Average Pairwise Divergence (±Standard Deviation) |
| L2 | Maui | 2,396 | 1,691 | 12 | 2.2 ± 0.4 |
| RTE | Total | 2,253 | 1,070 | ||
| Expander “recent” | — | 930 | 28 | 4.7 ± 0.5 | |
| Expander “old” | — | 140 | 0 | 35.6 ± 2.3 | |
| Rex1 | Total | 1,266 | 740 | ||
| Rex1-A | — | 570 | 4 | 3.5 ± 0.4 | |
| Rex1-B | — | 40 | 0 | 19.6 ± 1.8 | |
| Rex1-C | — | 130 | 0 | 18.5 ± 1.6 | |
| L1/Tx1 | Total | 406 | 268 | ||
| A | — | — | 5 | 1.0 ± 0.2 | |
| B | — | — | 4 | 3.0 ± 0.4 | |
| C | — | — | 0 | 4.0 ± 0.5 | |
| D | — | — | 0 | 20.0 ± 2.0 | |
| E | — | — | 0 | 12.2 ± 1.5 | |
| F | — | — | 0 | 27.4 ± 2.4 |
FIG. 2.—
Phylogenetic relationships among Maui (A) and Expander (B) elements from the three-spine stickleback genome. The trees were constructed with the maximum likelihood method using the K2 + G model. Only bootstrap (1,000 iterations) values >80% are shown.
FIG. 3.—
Pairwise divergence of families belonging to the four clades recovered from the three-spine stickleback genome, Maui, Expander, Rex1-A, and Tx1-A, and combined for all families.
The RTE clade is the second most abundant clade of nLTR-RT with ∼2,253 copies including 28 full-length insertions. It is represented by the Expander family, which was originally described from T. rubripes (Kapitonov and Jurka 1999). The RepeatMasker output indicates the presence of two subsets of Expander: Expander and Expander2. However, alignments and phylogenetic analysis of Expander and Expander2 reveal that these two putatively different groups of RTE are in fact indistinguishable in stickleback and correspond to the same family of elements. Thus, they were combined in our analysis. The pattern of evolution of Expander is similar to Maui as shown on figure 2B. The tree strongly indicates that a single family of Expander elements has been active in stickleback and probably still is, as suggested by the high level of similarity between the most recent elements. This recent activity is also reflected in the analysis of pairwise divergence between Expander elements (fig. 3), which shows a distribution shifted toward low values (<5%), suggesting that the vast majority of Expander elements have inserted recently in this genome. However, we also uncovered a smaller group of elements (14% of the total) with much higher divergence (∼35% average pairwise divergence), indicating that a wave of amplification occurred in the stickleback genome a long time ago (Expander old in table 1).
The Rex/Babar clade is represented in the stickleback by Rex1, which was originally discovered in Xiphophorus maculatus (Volff et al. 2000). More than 1,200 Rex1 copies are found in the stickleback genome. We identified three well-supported families we call Rex1-A, Rex1-B, and Rex1-C (fig. 4). As only elements at least 300 bp long can be accurately classified, we estimated the copy number for each subset using a 300-bp cutoff. Rex1-A is the dominant family with ∼570 copies, including four full-length elements, whereas Rex1-B and Rex1-C are represented by ∼40 and ∼130 copies, respectively, and no full-length copies. Rex1-B and C appear to have been unable to transpose for a long time and are likely to be extinct as suggested by their high level of divergence, 19.6% and 18.5%, respectively. The divergence distribution of Rex1-A is characterized by a peak at ∼4%, suggestive of a recent activity. Yet, the small number of values under 1% suggests that this family has a very low activity in extant stickleback populations, which is consistent with the very small number of full-length elements detected (fig. 3).
FIG. 4.—
Phylogenetic relationships among Rex1 elements from the three-spine stickleback genome. The tree was constructed with the maximum likelihood method using the K2 + G model. The three Rex1 families are indicated in brackets. Only bootstrap (1,000 iterations) values >80% are shown.
The most diverse, yet least abundant, clade is L1/Tx1, represented by six well-supported families (fig. 5A). Families D, E, and F are clearly monophyletic. They are represented by highly fragmented elements and are characterized by high level of divergence (12.2%–27.4% divergence), suggesting they have long been extinct. Because elements belonging to families D, E, and F are extremely fragmented, it is impossible to determine their copy number accurately. We can only determine that the stickleback genome does not contain any full-length element from any of these families. Families A, B, and C have a more complex history. Families B and C are reciprocally monophyletic, but depending on the section of the element used for the phylogenetic reconstruction, the position of family A varies. The tree based on the 3′ end of the element (fig. 5A) suggests that A is closer to B, but family A is closer to C on the tree built with the 5′ region (fig. 5B). This suggests that family A resulted from a recombination event between families B and C. Elements belonging to families A, B, and C are very similar to each other resulting in mean divergences of ∼1.0%, 3.0%, and 4.0%, respectively (fig. 3). These low values indicate that these three closely related families are still active or recently have been active in the stickleback. In fact, we identified 5 and 4 full-length elements in family A and B, respectively, that show very high level of similarity, suggesting they could represent active progenitors.
FIG. 5.—
Phylogenetic relationships among L1/Tx1 elements using sequences from the 3′ terminus (A) and the 5′ end (B) of the elements. The trees were constructed with the maximum likelihood method using the K2 + G model. Only bootstrap (1,000 iterations) values >80% are shown.
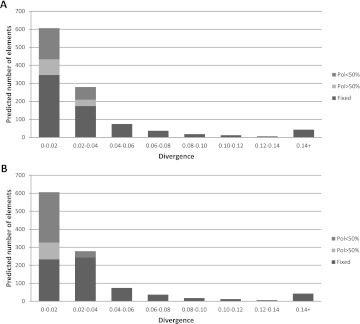
Although there are some differences of diversity among nLTR-RT clades, the vast majority of nLTR-RT insertions tend to be recent, with a striking lack of ancient (i.e., divergent) elements (fig. 3, bottom panel) and an extreme paucity of full-length copies (table 1). There are two nonexclusive explanations for this observation. First, nLTR-RT insertions could fail to accumulate in the stickleback genome due to a high rate of turnover in which the insertion of new elements is offset by the selective loss of deleterious elements. This model is identical to the one proposed for the evolution of transposable element copy number in Drosophila (Charlesworth B and Charlesworth D 1983; Montgomery and Langley 1983; Montgomery et al. 1987). Second, nLTR-RT could decay rapidly, before or after fixation, because of a high rate of DNA loss. To determine if nLTR-RT insertions do reach fixation, we experimentally assessed the polymorphism of 50 Expander insertions representing a wide range of divergence in 16 individuals from Bear Paw Lake, the population from which fish used for the genome project came (table 2). The presence/absence of inserts was determined by PCR using primers located in the flank of the elements and/or a primer cognate to the flank and a primer internal to the element (for long inserts). We found that in this population, all insertions diverging from their consensus by more than 3% are fixed. Although the fraction of elements that are fixed is proportionally lower in elements that have a low divergence from the family consensus, a significant proportion of those low divergence elements are also fixed. For instance, out of eight elements with divergence between 1% and 2%, six are fixed. To estimate the number of fixed Expander elements in the stickleback genome, we drew the curve of divergence from consensus for all ∼1,070 Expander elements (fig. 6, top panel). We then extrapolated the fraction of fixed elements in each divergence category to the entire Expander family. Using this approach, we estimated that 710 Expander elements (i.e., 66% of the insertions) are fixed in stickleback. Assuming that all nLTR-RT evolve at the same rate, we determined that 72.3% of all nLTR-RT insertions are fixed in the Bear Paw Lake population, which corresponds to 2,725 copies out of 3,769. Although this is a rough estimate, a large majority of nLTR-RT is undoubtedly fixed in this population.
Table 2.
Frequency of Insertions Tested by PCR in the Bear Paw Lake and Rabbit Slough Populations
| Locus Number | Coordinates of Locus | Length of Insertion | Divergence from Consensus (%) | Bear Paw Lake | Rabbit Slough |
| Loc1 | chrIX:20177336–20177676 | 340 | 0.00 | 0.80 | 0.95 |
| Loc2 | chrVII:228754–231552 | 2,798 | 0.00 | 0.10 | 0.00 |
| Loc3 | chrIV:20985857–20989180 | 3,323 | 0.00 | 1.00 | 0.00 |
| Loc4 | chrIII:15771865–15775195 | 3,330 | 0.00 | 0.00 | 0.37 |
| Loc5 | chrVII:12188102–12191449 | 3,347 | 0.00 | 1.00 | 0.18 |
| Loc6 | chrIV:24308317–24311680 | 3,363 | 0.00 | 0.00 | 0.00 |
| Loc7 | chrVII:15556265–15559605 | 3,340 | 0.34 | 0.33 | 0.00 |
| Loc8 | chrXV:2980897–2981317 | 420 | 0.34 | 0.70 | 0.82 |
| Loc9 | chrIII:6449280–6449749 | 469 | 0.34 | 1.00 | 1.00 |
| Loc10 | chrIV:23261707–23264743 | 3,036 | 0.34 | 0.20 | 0.00 |
| Loc11 | chrII:725502–725889 | 387 | 0.34 | 1.00 | 1.00 |
| Loc12 | chrXX:131232–131647 | 415 | 0.35 | 0.00 | 0.04 |
| Loc13 | chrVII:13330653–13331112 | 459 | 0.68 | 1.00 | 0.00 |
| Loc14 | chrV:7725501–7725980 | 479 | 0.68 | 0.67 | 1.00 |
| Loc15 | chrI:11778807–11779368 | 561 | 0.68 | 1.00 | 1.00 |
| Loc16 | chrI:16849656–16850343 | 687 | 0.68 | 1.00 | 1.00 |
| Loc17 | chrI:12839403–12842746 | 3,343 | 0.69 | 0.00 | 0.51 |
| Loc18 | chrI:21573012–21574283 | 1,271 | 0.69 | 1.00 | 0.00 |
| Loc19 | chrIV:27086216–27089151 | 2,935 | 0.70 | 1.00 | 0.50 |
| Loc20 | chrIV:26298715–26299022 | 307 | 0.78 | 1.00 | NA |
| Loc21 | chrVII:11214917–11217225 | 2,308 | 1.02 | 1.00 | 1.00 |
| Loc22 | chrVI:17025662–17026098 | 436 | 1.03 | 0.70 | 1.00 |
| Loc23 | chrVIII:7969513–7969829 | 316 | 1.03 | 1.00 | 0.00 |
| Loc24 | chrIV:25767136–25770441 | 3,305 | 1.03 | 0.43 | 0.45 |
| Loc25 | chrXIII:15925609–15926017 | 408 | 1.05 | 1.00 | 1.00 |
| Loc26 | chrIV:23957871–23958211 | 340 | 1.37 | 1.00 | 1.00 |
| Loc27 | chrIX:2238963–2239452 | 489 | 1.40 | 1.00 | NA |
| Loc28 | chrII:1134048–1134688 | 640 | 1.81 | 1.00 | 1.00 |
| Loc29 | chrI:19360848–19361301 | 453 | 2.44 | 0.96 | 1.00 |
| Loc30 | chrII:21806245–21807448 | 1,203 | 2.46 | 0.00 | 1.00 |
| Loc31 | chrVII:9967491–9968887 | 1,396 | 2.51 | 0.00 | 1.00 |
| Loc32 | chrXIV:10308068–10308391 | 323 | 3.16 | 1.00 | 1.00 |
| Loc33 | chrI:20009150–20009606 | 456 | 3.37 | 1.00 | 0.00 |
| Loc34 | chrI:17570285–17570923 | 638 | 3.51 | 1.00 | 1.00 |
| Loc35 | chrI:8606080–8606552 | 472 | 3.53 | 1.00 | 1.00 |
| Loc36 | chrIV:99671–100126 | 455 | 3.54 | 1.00 | 1.00 |
| Loc37 | chrXVI:5421357–5421768 | 411 | 4.22 | 1.00 | 1.00 |
| Loc38 | chrV:5918311–5918748 | 437 | 4.68 | 1.00 | 1.00 |
| Loc39 | chrVII:11640817–11642283 | 1,466 | 5.52 | 1.00 | 1.00 |
| Loc40 | chrXIV:14881766–14882111 | 345 | 7.10 | 1.00 | 1.00 |
| Loc41 | chrIII:6205638–6208177 | 2,539 | 7.20 | 1.00 | 1.00 |
| Loc42 | chrIV:26239365–26239741 | 376 | 9.17 | 1.00 | 1.00 |
| Loc43 | chrII:9308974–9309348 | 374 | 14.12 | 1.00 | 1.00 |
| Loc44 | chrII:20575935–20576507 | 572 | 20.69 | 1.00 | 1.00 |
| Loc45 | chrIII:4938664–4939000 | 336 | 21.25 | 1.00 | 1.00 |
| Loc46 | chrI:9011472–9011777 | 305 | 24.21 | 1.00 | 1.00 |
| Loc47 | chrII:9734544–9734844 | 300 | 25.17 | 1.00 | 1.00 |
| Loc48 | chrVII:12290981–12291438 | 457 | 26.10 | 1.00 | 1.00 |
| Loc49 | chrVI:5415002–5415472 | 470 | 35.41 | 1.00 | 1.00 |
| Loc50 | chrIII:6418419–6419108 | 689 | 35.70 | 1.00 | 1.00 |
Note.—NA, no amplification.
FIG. 6.—
Fraction of fixed and polymorphic Expander elements extrapolated from population data. The analysis was performed separately for the Bear Paw Lake (A) and the anadromous Rabbit Slough (B) populations. Polymorphic elements were split into elements found at frequencies higher and lower than 50%.
It is plausible, however, that the large number of fixed insertions in the Bear Paw Lake population results from the demographic history of this population. The Bear Paw Lake population is characterized by a lower level of genetic variation than marine and stream populations, suggesting it has a lower effective population size (Aguirre 2007). Smaller population size decreases the efficiency of purifying selection, allowing the fixation of insertions that otherwise would have been eliminated in a population with a large effective size. To test this hypothesis, we estimated the frequency of the same Expander insertions in nine other populations including lake, stream, and oceanic populations (see Supplementary Material online). Of particular interest is a comparison with the anadromous (sea-run) Rabbit Slough population (N = 43), which has apparently not suffered any reduction in population size (table 2). This population exhibits a level of genetic variation (based on microsatellite variation) similar to the one reported in other marine species, which is consistent with a large effective population size (Aguirre 2007). We also found that a majority of insertions are fixed in this population, and using the same calculation as above, we estimated that ∼670 Expander insertions are fixed (fig. 6, bottom panel), which is very close to the estimate obtained for Bear Paw Lake (710 fixed insertions). We extrapolated these calculations to all nLTR-RT families, and we estimated that 73.3% (i.e., 2,765 copies out of 3,769) of the elements are fixed, a result remarkably close to the estimate for the Bear Paw Lake population. Similar calculations performed on the other populations provided consistent estimates, suggesting that most insertions reached fixation before these different populations separated.
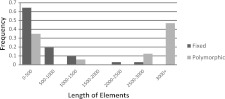
These estimates strongly indicate that nLTR-RTs accumulate readily in the stickleback genome; yet, they do not imply that insertions are fully neutral in this species. Although the number of insertions we screened here is too small to estimate accurately selection coefficients, our data suggest that some insertions are indeed likely to be deleterious. Figure 7 shows the proportion of fixed and truncated insertions relative to the length of the elements. To avoid the confounding effect of demography, this figure was estimated using only the Rabbit Slough data. The vast majority (∼85%) of fixed insertions is severely truncated (<1 kb); fixed long (>1 kb) insertions are rare, and we failed to find a single fixed full-length insertion. Full-length and truncated insertions are produced by target-primed reverse transcription and truncations of the 5′ end occur at the time of insertion. Thus, the deficiency in fixed full-length elements is likely due to a post-insertional process. Although the full-length elements could be rapidly lost because of a high rate of DNA deletion (see below), it is also possible that the lack of fixed full-length elements reflect the differential fixation of elements of different lengths. This would imply that purifying selection is acting on long elements, thus preventing their fixation, and suggests that Expander elements could be imposing a fitness cost related to the insertion length on their host. It remains true, however, that purifying selection is insufficient to prevent the fixation of truncated elements, which constitute the majority of the inserts.
FIG. 7.—
Fraction of polymorphic and fixed Expander elements relative to their length. The distribution is based on 48 insertions screened in the Rabbit Slough population.
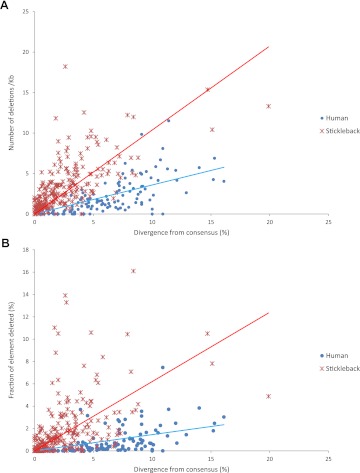
We then examined the second explanation for the low copy number and the low divergence of nLTR-RT, namely, the DNA loss hypothesis. DNA can be lost in two ways, either by the accumulation of small (<50 bp) internal deletions or by deletions of large segments of sequence. We first examined the occurrence of short deletions in elements belonging to the Maui and Expander families. For comparison, we collected ∼120 L1 elements from the human genome representing a similar range of divergence to the stickleback elements. Figure 8A shows the number of small deletions per kilo base pairs relative to the age of elements. Small deletions occur readily in stickleback, at a rate of about 1 deletion/kb per unit of divergence. This rate of deletion is about three times higher than the rate in humans (∼0.3 deletion/kb per unit of divergence), suggesting that nLTR-RT sequences are much less stable in fish than in humans. However, the accumulation of small deletions is insufficient to account for the extreme scarcity of elements with divergence higher than 10%. The fraction of elements deleted through the accumulation of small deletions is ∼0.6% per unit of divergence, meaning that an element with a 10% divergence from consensus will have, on average, lost only 6% of its length (fig. 8B). Although this value is four times higher than the rate of deletion in humans, it is clearly insufficient to explain the lack of ancient elements in the stickleback genome.
FIG. 8.—
(A) Relationship between the number of small deletions and the divergence from consensus for stickleback Expander (y = 1.037×; R2 = 0.3581) and human L1 elements (y = 0.3584×; R2 = 0.4446). (B) Relationship between the fraction of element lost through small deletions and the divergence from consensus for stickleback Expander (y = 0.6202×; R2 = 0.2253) and human L1 elements (y = 0.145×; R2 = 0.282).
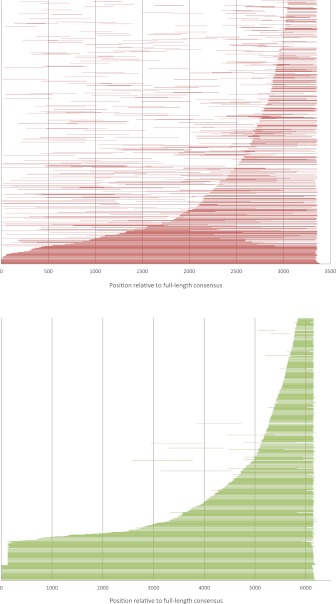
We then examined the impact of large deletions on the decay of nLTR-RT sequences. Large deletions will produce highly fragmented elements, particularly elements that will lack one or both of their termini. The difficulty in assessing the occurrence of large deletions in nLTR-RT results from the diversity of structure that can be generated at the time of insertion. In particular, a majority of nLTR-RT insertions are truncated in 5′ at the time of insertion, possibly because of premature base pairing with the target site (Martin et al. 2005). Thus, when an element is missing its 5′ end, it is nearly impossible to determine if this is the result of a truncation at the time of insertion or of a large deletion. Conversely, the loss of the 3′ extremity can only be caused by a DNA deletion. We collected 683 intact Expander elements, and for each of them, we scored the beginning and end of the sequence relative to the full-length consensus of the family. Elements interrupted by gaps in the draft sequence were eliminated. These elements are presented on the top panel of figure 9. We first verified that elements missing their 3′ ends are on average more divergent than those with intact 3′ ends, which is expected if 3′ termini are lost post-insertionally and not at the time of insertion. As predicted, we found that elements missing their 3′ ends are more divergent (4.73%) than elements with an intact 3′ end (2.60%). Figure 9 shows that a large number of elements (51.5% of the total) are missing their 3′ end and that most of them (46.9%) are missing both their 5′ and 3′ ends. The remaining 48.5% can be considered to be intact and have presumably not suffered a deletion. Of those, 4% are full length and 44.5% are truncated in 5′. Assuming conservatively that all missing 5′ termini were due to truncation and that missing 3′ ends were caused by post-insertional deletions, we estimated that at least 37% of the DNA generated by the Expander family has been lost by large deletions. This is certainly an underestimate as a number of missing 5′ ends probably resulted from deletion and not truncation. This rate of DNA loss was unexpected, considering the age distribution of Expander inserts (fig. 3), but it is consistent with the large fraction of elements shorter than 300 bp (table 1). For comparison, we performed the same analysis in human sequences using 584 L1 elements with a range of divergence similar to the one of Expander. We found that a tiny fraction of L1 elements (<1%) are missing their 3′ end and that the vast majority of elements are structurally intact. This difference in fragmentation between fish and human nLTR-RT is even more striking when one considers that a full-length L1 is almost twice as long as a full-length Expander and thus should be more likely to experience deletions. This analysis demonstrated that large deletions occur much more often in stickleback than in humans and are sufficiently common to account for the extreme scarcity of ancient elements in the stickleback genome.
FIG. 9.—
Length distribution of stickleback Expander elements (top) and human L1 elements (bottom). Elements are ordered by length from the shortest one at the top of the graphs to the longest one above the x axis. Note that the scale is different as a full-length Expander element is ∼3.3 kb, whereas a human full-length L1 is ∼6 kb.
Discussion
The stickleback genome contains four active clades of nLTR-RTs, some of which are represented by multiple families of elements. There are, however, some interesting differences among nLTR-RT clades: the RTE and L2 clades are represented by a single family but there are three Rex1 and six L1/Tx1 families. How does this level of diversity compare with that of other nonmammalian vertebrates? A previous study showed that the stickleback has reduced clade diversity compared with other teleosteans (Basta et al. 2007). Here we showed that this low level of diversity is also found at the family level. With six families including only three active ones, the L1/Tx1 clade in stickleback is considerably less diverse than the L1/Tx1 clade in killifish (Duvernell et al. 2004), zebra fish, which harbor at least 32 distinct families (Furano et al. 2004), or in the lizard Anolis carolinensis (Novick et al. 2009). Similarly, the L2 clade is represented by the sole Maui family, whereas the zebra fish genome contains more than 40 L2-related families (based on the annotations of the zebra fish genome at http://genome.ucsc.edu) and the lizard has 17 families (Novick et al. 2009). The low level of diversity of Rex1 and RTE on the other hand is similar to that reported in other taxa as these two clades do not seem to diversify to the same extent as the L1 or L2 clade (Kordis and Gubensek 1998; Volff et al. 2000; Zupunski et al. 2001).
The relatively low copy number and the very recent age of nLTR-RT elements in stickleback are reminiscent of the situation in the other teleostean genomes examined so far (Volff et al. 2003; Duvernell et al. 2004; Furano et al. 2004; Neafsey et al. 2004). Because of the similarities with Drosophila, it was originally proposed that nLTR-RT elements in teleosteans are subjected to a high rate of turnover in which the insertion of new elements is offset by the selective loss of insertions (Duvernell et al. 2004; Furano et al. 2004). This model predicts that most elements are deleterious and segregate at low frequency in populations. However, the high number of fixed insertions in stickleback is not consistent with the turnover model as it applies to Drosophila. There are two nonexclusive explanations for the accumulation of nLTR-RT insertions in stickleback. First, it is possible that most nLTR-RT insertions have no impact on host fitness. This hypothesis is consistent with the population genetic analysis of the spotted puffer fish (Tetraodon nigroviridis) performed by Neafsey et al. (2004), who found that most elements segregated at high frequency or were fixed in this species and behaved as neutral alleles. It is notable that in stickleback, the vast majority of fixed insertions are truncated, suggesting that truncated insertions could be neutral. Similarly, in Drosophila and humans, purifying selection acts preferentially against long elements, and severely truncated elements behave as neutral or nearly neutral alleles (Petrov et al. 2003; Boissinot et al. 2006).
In contrast, the number of full-length elements is extremely small in stickleback for all nLTR-RT families, and we failed to find a single fixed full-length insertion. The number of full-length insertions found in other teleostean genomes is also extremely small, suggesting that a common mechanism might limit fixation of full-length insertions in all teleosteans (Basta et al. 2007). It is possible that the rate of DNA loss in stickleback (see below) is sufficiently high to eliminate full-length elements soon after or even before they reach fixation. However, the general scarcity of full-length elements and the apparent absence of fixed full-length insertions could also be interpreted as evidence for a strongly deleterious effect of these elements, which would prevent their fixation. Thus, the turnover model might apply in teleosts but only to full-length elements. A deleterious impact of such long elements was not detected in the T. nigroviridis study, possibly because only severely truncated elements were examined in this study (Neafsey et al. 2004). A deleterious effect of nLTR-RT related to the length of the elements has previously been described in Drosophila and in humans (Boissinot et al. 2001, 2006; Petrov et al. 2003, 2011) and results from the greater ability of long elements to mediate ectopic recombination events, which are extremely deleterious (Langley et al. 1988; Song and Boissinot 2007). Although our results in stickleback are consistent with the ectopic recombination model, it is possible that selection acts specifically against full-length elements because of a deleterious effect related to the transcription or translation of these elements (Nuzhdin et al. 1996; Brookfield and Badge 1997). Whatever the exact mechanism, it is clear that the number of full-length elements in fish genomes is strictly limited. As full-length elements are the only elements capable of transposition, selection limiting the spread of full-length copies could reduce the transposition rate and the number of new nLTR-RT copies, contributing to the low copy number of most families. This could, in part, explain the much greater copy number in mammals than in teleosts. Eutherian genomes harbor much larger number of active copies than fish genomes. For instance, the number of full-length L1 active or potentially active copies in human and mouse is 80–100 and 2,000–3,000, respectively (Brouha et al. 2003; Akagi et al. 2008). Thus, the strength of selection against full-length copies in mammals, although significant, does not prevent the fixation of a large number of full-length copies, which in turn could yield to greater transposition rate and larger families in mammals than in fish.
The high fraction of fixed insertions in stickleback could also result from the demographic history of the species. As nLTR-RTs are obligatory parasites, their dynamics in the genome is affected by the evolution and natural history of their host. Thus, any factor that affects the effective population size (Ne) of the host will modify the equilibrium between drift and selection. When Ne is large, like in Drosophila, selection dominates over drift, but any factor that decreases Ne (e.g., bottleneck, mating system) will strengthen drift. In populations with a small Ne, purifying selection against deleterious insertions is not acting as efficiently as in large population. Thus, we expect a higher rate of fixation in population that went through a bottleneck or a founder effect, as was observed in populations of the plant Arabidopsis lyrata and in Drosophila subobscura (Garcia Guerreiro et al. 2008; Lockton et al. 2008). A number of recent studies have examined the amount of genetic variation in three-spine stickleback (Hohenlohe et al. 2010; Deagle et al. 2011; Jones et al. 2012). Three-spine stickleback populations are genetically very diverse, and there is no evidence for a reduced effective population at the level of the species that could have favored the fixation of a large number of nLTR-RT. Thus, it is very unlikely that the large proportion of fixed insertions in this genome could be due to a reduction in population size.
Whatever the cause, it remains that a very large number of elements reached fixation in three-spine stickleback, and it is likely that it has been the case for a long time. Thus, the relatively young age of nLTR-RT families and the extreme rarity of ancient elements imply that a second mechanism, DNA loss, has played a significant role in limiting nLTR-RT copy number. Accumulation of small deletions cannot account for the rapid decay of insertions, but large deletions were frequent enough to rapidly eliminate a large fraction of the DNA sequence generated by nLTR-RT activity. The loss of long fragments by large-scale deletion had previously been reported in a lizard (Novick et al. 2009) and is apparently the major cause of genome shrinkage in plants (Devos et al. 2002; Ma et al. 2004; Hawkins et al. 2009). The high rate of DNA loss by large deletions reported in these taxa is certainly sufficient to counteract the amplification of transposable elements and to limit genome size expansion. In contrast, large deletions seem to occur very rarely in mammals, and this could contribute to the extremely large size of mammalian genomes.
This analysis of nLTR-RT decay in stickleback sheds new light on the controversial question of genome size evolution. In a landmark paper, Petrov (2002) proposed that the genome size reflects an equilibrium between large insertions that increase genome size and accumulation of small deletions that decrease it. This model was based on the observation that small deletions occur more frequently in insect species with small genomes than in species with large genomes (Petrov and Hartl 1997; Petrov et al. 2000; Bensasson et al. 2001). Petrov’s model has been controversial because even in species where small deletions occur frequently, this process appears to be too slow to account for the small size of these genomes (Gregory 2003, 2004). In the original description of the model, large deletions were discounted as a significant source of DNA loss because they should be very deleterious, particularly in compact genomes such as the Drosophila genome. However, it seems that in plants and in nonmammalian vertebrates, large deletions do occur readily and, based on their frequency, are unlikely to be very deleterious. It is indeed surprising that large deletions are tolerated in these organisms because they could affect regulatory or protein coding regions. It is, however, possible that these deletions preferentially target repetitive DNA and that coding regions are protected from them. Clearly more work on the mechanisms and distribution of large deletions in vertebrates is required. It should be noted that the occurrence of large deletions in other groups, such as insects, has yet to be examined in detail. Early studies relied on the amplification and cloning of transposable element insertions or pseudogenes to infer the indel spectrum and consequently could not capture large deletion (Petrov et al. 2000; Bensasson et al. 2001). In conclusion, our analysis does not contradict the general idea behind the mutational equilibrium model, but we suggest that large deletions certainly play a far greater role in the process of DNA loss than originally thought, at least in teleostean fish.
Supplementary Material
Supplementary materials are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
This research was supported by PSC-CUNY grant 63799-00-41 and NIH grant R15GM096267-01 to S.B. E.B. received support from the Summer Program in Undergraduate Research at Queens College. The work was conducted in part with equipment from the Core facilities for Imaging, Cellular and Molecular Biology at Queens College. Samples were collected with the support of DEB-0211391 and DEB-0919184 to M.A.B. and DEB-0509070 to W.E. Aguirre. This is contribution 1214 from Ecology and Evolution at Stony Brook University. We thank the Broad Institute Genome Sequencing Platform and Genome Sequencing and Analysis Program for making the data for G. aculeatus available.
References
- Aguirre WE. The pattern and process of evolutionary diversification: lessons from a threespine stickleback adaptive radiation. Stony Brook (NY): Stony Brook University; 2007. p. 205. [Google Scholar]
- Akagi K, Li J, Stephens RM, Volfovsky N, Symer DE. Extensive variation between inbred mouse strains due to endogenous L1 retrotransposition. Genome Res. 2008;18:869–880. doi: 10.1101/gr.075770.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basta HA, Buzak AJ, McClure MA. Identification of novel retroid agents in Danio rerio, Oryzias latipes, Gasterosteus aculeatus and Tetraodon nigroviridis. Evol Bioinform Online. 2007;3:179–195. [PMC free article] [PubMed] [Google Scholar]
- Bell MA, Foster SA, editors. The evolutionary biology of the threespine stickleback. Oxford: Oxford University Press; 1994. [Google Scholar]
- Bensasson D, Petrov DA, Zhang DX, Hartl DL, Hewitt GM. Genomic gigantism: DNA loss is slow in mountain grasshoppers. Mol Biol Evol. 2001;18:246–253. doi: 10.1093/oxfordjournals.molbev.a003798. [DOI] [PubMed] [Google Scholar]
- Boissinot S, Davis J, Entezam A, Petrov D, Furano AV. Fitness cost of LINE-1 (L1) activity in humans. Proc Natl Acad Sci U S A. 2006;103:9590–9594. doi: 10.1073/pnas.0603334103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boissinot S, Entezam A, Furano AV. Selection against deleterious LINE-1-containing loci in the human lineage. Mol Biol Evol. 2001;18:926–935. doi: 10.1093/oxfordjournals.molbev.a003893. [DOI] [PubMed] [Google Scholar]
- Brookfield JF, Badge RM. Population genetics models of transposable elements. Genetica. 1997;100:281–294. [PubMed] [Google Scholar]
- Brouha B, et al. Hot L1s account for the bulk of retrotransposition in the human population. Proc Natl Acad Sci U S A. 2003;100:5280–5285. doi: 10.1073/pnas.0831042100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B, Charlesworth D. The population dynamics of transposable elements. Genet Res. 1983;42:1–27. [Google Scholar]
- Deagle BE, et al. Population genomics of parallel phenotypic evolution in stickleback across stream-lake ecological transitions. Proc Biol Sci. 2011;279:1277–1286. doi: 10.1098/rspb.2011.1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devos KM, Brown JK, Bennetzen JL. Genome size reduction through illegitimate recombination counteracts genome expansion in Arabidopsis. Genome Res. 2002;12:1075–1079. doi: 10.1101/gr.132102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duvernell DD, Pryor SR, Adams SM. Teleost fish genomes contain a diverse array of L1 retrotransposon lineages that exhibit a low copy number and high rate of turnover. J Mol Evol. 2004;59:298–308. doi: 10.1007/s00239-004-2625-8. [DOI] [PubMed] [Google Scholar]
- Furano AV. The biological properties and evolutionary dynamics of mammalian LINE-1 retrotransposons. Prog Nucleic Acid Res Mol Biol. 2000;64:255–294. doi: 10.1016/s0079-6603(00)64007-2. [DOI] [PubMed] [Google Scholar]
- Furano AV, Duvernell D, Boissinot S. L1 (LINE-1) retrotransposon diversity differs dramatically between mammals and fish. Trends Genet. 2004;20:9–14. doi: 10.1016/j.tig.2003.11.006. [DOI] [PubMed] [Google Scholar]
- Garcia Guerreiro MP, Chavez-Sandoval BE, Balanya J, Serra L, Fontdevila A. Distribution of the transposable elements bilbo and gypsy in original and colonizing populations of Drosophila subobscura. BMC Evol Biol. 2008;8:234. doi: 10.1186/1471-2148-8-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory TR. Is small indel bias a determinant of genome size? Trends Genet. 2003;19:485–488. doi: 10.1016/S0168-9525(03)00192-6. [DOI] [PubMed] [Google Scholar]
- Gregory TR. Insertion-deletion biases and the evolution of genome size. Gene. 2004;324:15–34. doi: 10.1016/j.gene.2003.09.030. [DOI] [PubMed] [Google Scholar]
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- Hawkins JS, Proulx SR, Rapp RA, Wendel JF. Rapid DNA loss as a counterbalance to genome expansion through retrotransposon proliferation in plants. Proc Natl Acad Sci U S A. 2009;106:17811–17816. doi: 10.1073/pnas.0904339106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohenlohe PA, et al. Population genomics of parallel adaptation in threespine stickleback using sequenced RAD tags. PLoS Genet. 2010;6:e1000862. doi: 10.1371/journal.pgen.1000862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones FC, Chan YF, Schmutz J, et al. A genome-wide SNP genotyping array reveals patterns of global and repeated species-pair divergence in sticklebacks. Curr Biol. 2012;22:83–90. doi: 10.1016/j.cub.2011.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapitonov V, Jurka J. 1999. Expander, Repbase Update, release 4.07. Available from: http://www.girinst.org/repbase/update. [Google Scholar]
- Kapitonov VV, Tempel S, Jurka J. Simple and fast classification of non-LTR retrotransposons based on phylogeny of their RT domain protein sequences. Gene. 2009;448:207–213. doi: 10.1016/j.gene.2009.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kordis D, Gubensek F. Unusual horizontal transfer of a long interspersed nuclear element between distant vertebrate classes. Proc Natl Acad Sci U S A. 1998;95:10704–10709. doi: 10.1073/pnas.95.18.10704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Langley CH, Montgomery E, Hudson R, Kaplan N, Charlesworth B. On the role of unequal exchange in the containment of transposable element copy number. Genet Res. 1988;52:223–235. doi: 10.1017/s0016672300027695. [DOI] [PubMed] [Google Scholar]
- Lockton S, Ross-Ibarra J, Gaut BS. Demography and weak selection drive patterns of transposable element diversity in natural populations of Arabidopsis lyrata. Proc Natl Acad Sci U S A. 2008;105:13965–13970. doi: 10.1073/pnas.0804671105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Devos KM, Bennetzen JL. Analyses of LTR-retrotransposon structures reveal recent and rapid genomic DNA loss in rice. Genome Res. 2004;14:860–869. doi: 10.1101/gr.1466204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin SL, Li W-HP, Furano AV, Boissinot S. The structures of mouse and human L1 elements reflect their insertion mechanism. Cytogenet Genome Res. 2005;110:223–228. doi: 10.1159/000084956. [DOI] [PubMed] [Google Scholar]
- McClure MA, et al. Automated characterization of potentially active retroid agents in the human genome. Genomics. 2005;85:512–523. doi: 10.1016/j.ygeno.2004.12.006. [DOI] [PubMed] [Google Scholar]
- Montgomery E, Charlesworth B, Langley CH. A test for the role of natural selection in the stabilization of transposable element copy number in a population of Drosophila melanogaster. Genet Res. 1987;49:31–41. doi: 10.1017/s0016672300026707. [DOI] [PubMed] [Google Scholar]
- Montgomery EA, Langley CH. Transposable elements in Mendelian populations. II. Distribution of three COPIA-like elements in a natural population of Drosophila melanogaster. Genetics. 1983;104:473–483. doi: 10.1093/genetics/104.3.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neafsey DE, Blumenstiel JP, Hartl DL. Different regulatory mechanisms underlie similar transposable element profiles in pufferfish and fruitflies. Mol Biol Evol. 2004;21:2310–2318. doi: 10.1093/molbev/msh243. [DOI] [PubMed] [Google Scholar]
- Novick PA, Basta H, Floumanhaft M, McClure MA, Boissinot S. The evolutionary dynamics of autonomous non-LTR retrotransposons in the lizard Anolis carolinensis shows more similarity to fish than mammals. Mol Biol Evol. 2009;26:1811–1822. doi: 10.1093/molbev/msp090. [DOI] [PubMed] [Google Scholar]
- Nuzhdin SV, Pasyukova EG, Mackay TF. Positive association between copia transposition rate and copy number in Drosophila melanogaster. Proc Biol Sci. 1996;263:823–831. doi: 10.1098/rspb.1996.0122. [DOI] [PubMed] [Google Scholar]
- Petrov D, Aminetzach YT, Davis JC, Bensasson D, Hirsh AE. Size matters: non-LTR retrotransposable elements and ectopic recombination in Drosophila. Mol Biol Evol. 2003;20:880–892. doi: 10.1093/molbev/msg102. [DOI] [PubMed] [Google Scholar]
- Petrov DA. Mutational equilibrium model of genome size evolution. Theor Popul Biol. 2002;61:531–544. doi: 10.1006/tpbi.2002.1605. [DOI] [PubMed] [Google Scholar]
- Petrov DA, Fiston-Lavier AS, Lipatov M, Lenkov K, Gonzalez J. Population genomics of transposable elements in Drosophila melanogaster. Mol Biol Evol. 2011;28:1633–1644. doi: 10.1093/molbev/msq337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrov DA, Hartl DL. Trash DNA is what gets thrown away: high rate of DNA loss in Drosophila. Gene. 1997;205:279–289. doi: 10.1016/s0378-1119(97)00516-7. [DOI] [PubMed] [Google Scholar]
- Petrov DA, Sangster TA, Johnston JS, Hartl DL, Shaw KL. Evidence for DNA loss as a determinant of genome size. Science. 2000;287:1060–1062. doi: 10.1126/science.287.5455.1060. [DOI] [PubMed] [Google Scholar]
- Poulter R, Butler M, Ormandy J. A LINE element from the pufferfish (fugu) Fugu rubripes which shows similarity to the CR1 family of non-LTR retrotransposons. Gene. 1999;227:169–179. doi: 10.1016/s0378-1119(98)00600-3. [DOI] [PubMed] [Google Scholar]
- Rozen S, Skaletsky HJ. Primer3 on the WWW for general users and for biologist programmers. In: Krawetz S, Misener S, editors. Bioinformatics methods and protocols: methods in molecular biology. Totowa (NJ): Humana Press; 2000. pp. 365–386. [DOI] [PubMed] [Google Scholar]
- Song M, Boissinot S. Selection against LINE-1 retrotransposons results principally from their ability to mediate ectopic recombination. Gene. 2007;390:206–213. doi: 10.1016/j.gene.2006.09.033. [DOI] [PubMed] [Google Scholar]
- Volff JN. Genome evolution and biodiversity in teleost fish. Heredity. 2005;94:280–294. doi: 10.1038/sj.hdy.6800635. [DOI] [PubMed] [Google Scholar]
- Volff JN, Bouneau L, Ozouf-Costaz C, Fischer C. Diversity of retrotransposable elements in compact pufferfish genomes. Trends Genet. 2003;19:674–678. doi: 10.1016/j.tig.2003.10.006. [DOI] [PubMed] [Google Scholar]
- Volff JN, Korting C, Schartl M. Multiple lineages of the non-LTR retrotransposon Rex1 with varying success in invading fish genomes. Mol Biol Evol. 2000;17:1673–1684. doi: 10.1093/oxfordjournals.molbev.a026266. [DOI] [PubMed] [Google Scholar]
- Waterston RHK, et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- Zupunski V, Gubensek F, Kordis D. Evolutionary dynamics and evolutionary history in the RTE clade of non-LTR retrotransposons. Mol Biol Evol. 2001;18:1849–1863. doi: 10.1093/oxfordjournals.molbev.a003727. [DOI] [PubMed] [Google Scholar]