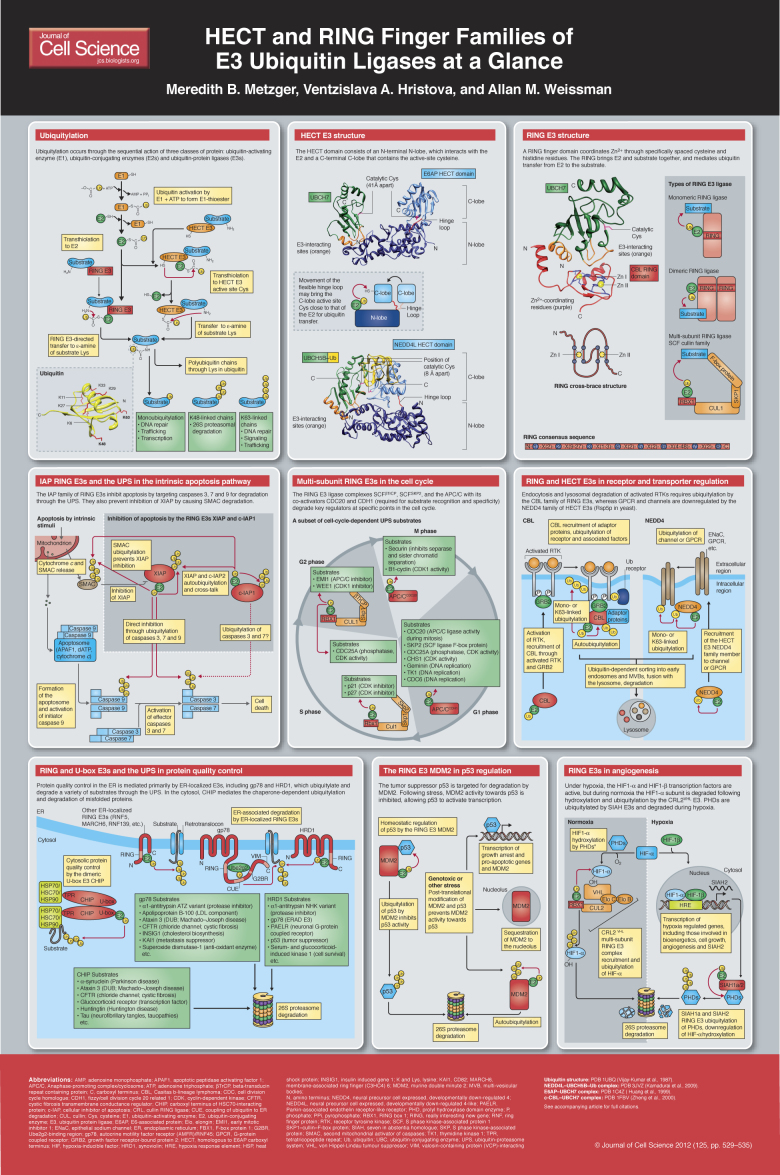
The post-translational attachment of ubiquitin, a highly conserved 76-amino-acid polypeptide, directs myriad eukaryotic proteins to a variety of fates and functions. Ubiquitylation is best-known for targeting proteins for degradation by the 26S proteasome. Other functions include internalization and lysosomal targeting, modulation of protein interactions, alteration of subcellular distribution, regulation of transcription, DNA repair and propagation of transmembrane signaling, most notably in the NF-κB pathway. Not surprisingly, ubiquitylation has been linked to virtually every cellular process.
Ubiquitylation occurs through the sequential action of three classes of protein: ubiquitin-activating enzymes (E1s), ubiquitin-conjugating enzymes (E2s, also referred to as ubiquitin carrier proteins or UBCs), and ubiquitin-protein ligases (E3s) (Komander, 2009). The C-terminal carboxyl group of ubiquitin is activated in an ATP-dependent process, which results in a high-energy thioester linkage with the active-site cysteine of E1. Ubiquitin is trans-thiolated from the E1 to the active-site cysteine of one of ~40 E2s (in mammals). Finally, ubiquitin is generally transferred from the E2 to the ε-amino group of a substrate lysine in an E3-dependent manner. For the E3 ligases of the homologous to the E6AP carboxyl terminus (HECT) domain family (Rotin and Kumar, 2009), this involves an obligate thioester intermediate with the active-site cysteine of the E3. The vast majority of E3 ligases belong to the group of really interesting new gene (RING) and RING-related E3s – such as plant homeodomain (PHD) and leukemia associated protein (LAP) finger proteins, and members of the U-box family (Deshaies and Joazeiro, 2009). These mediate the direct transfer of ubiquitin from E2 to substrate. Ubiquitin can also be added to the N-terminus of proteins (Ciechanover and Ben-Saadon, 2004), and the modification of other nucleophilic amino acids (threonine, serine, cysteine) by ubiquitylation has been described (Cadwell and Coscoy, 2005; Carvalho et al., 2007; Ishikura et al., 2010; Shimizu et al., 2010; Tait et al., 2007; Wang et al., 2007; Williams et al., 2007).
Substrates can be modified by a single ubiquitin on one or more sites (monoubiquitylation and multi-monoubiquitylation, respectively) or can be tagged with chains of ubiquitin (poly- or multiubiquitylation). The
majority of experimental evidence suggests that chains are built sequentially, beginning with the substrate-attached ubiquitin, although there is evidence for chains being built on E2 or E3 and transferred to substrates en bloc (Bazirgan and Hampton, 2008; Li et al., 2007; Ravid and Hochstrasser, 2007; Wang and Pickart, 2005). Chain formation can occur through the seven internal lysine residues on ubiquitin or its N-terminus (Vijay-Kumar et al., 1987; Behrends and Harper, 2011; Clague and Urbe, 2010). The type of ubiquitin modification specifies the function of the modification (i.e. monoubiquitylation or different chain linkages), together with other factors such as ubiquitin receptors and the opposing action of deubiquitylating enzymes (DUBs); see also the Commentary by Michael J. Clague, Judy M., Coulson and Sylvie Urbé in the previous issue (J. Cell Sci. 125, xxxx).
Ubiquitylation occurs with exquisite spatial, temporal and substrate specificity, much of which is dictated by the more than 600 E3s that are estimated to be encoded by the mammalian genome (Li et al., 2008). In addition to this cellular specificity and complexity, E3s are implicated in a number of pathophysiological conditions, which makes them attractive therapeutic targets (Kirkin and Dikic, 2011; Lipkowitz and Weissman, 2011). This Cell Science at a Glance article and the accompanying poster describe structural aspects of E3s, and highlight several mammalian pathways and cellular processes in which E3s have key roles. The roles of other specific E3s and cellular processes that are regulated by ubiquitylation are discussed in detail in accompanying review articles in this and the previous issue.
HECT domain E3s
In mammals, there are ~30 HECT domain E3s. Among their many functions, HECT E3s have prominent roles in protein trafficking, the immune response, and in several signaling pathways that regulate cellular growth and proliferation (Rotin and Kumar, 2009). The conserved HECT domain (which comprises ~350 amino acids) is located at the C-terminus of these enzymes, whereas their N-terminal domains are diverse and mediate substrate targeting. The HECT domain itself is bi-lobed, consisting of an N-terminal N-lobe that interacts with the E2 and a C-terminal C-lobe that contains the active-site cysteine that forms the thioester with ubiquitin (Huang et al., 1999). The crystal structure of the NEDD4L HECT domain in complex with ubiquitin-conjugated E2 [UBCH5B~Ub, officially known as UBE2D2] shows the C-lobe contacting the esterified ubiquitin and folding down onto UbcH5B, thereby making the distance between the E2 and E3 catalytic cysteines ~8Å (Kamadurai et al., 2009). By contrast, the first identified member of the HECT family, E6-associated protein (E6AP, officially known as UBE3A) (Huibregtse et al., 1995), in complex with its E2 UBCH7 (officially known as UBE2L3), shows the C-lobe in a more open architecture where the catalytic cysteine residues are 41Å apart (Huang et al., 1999). Taken together, these and other structural studies (Ogunjimi et al., 2005; Verdecia et al., 2003) suggest that the two lobes of the HECT domain are connected through a flexible hinge that allows them to come together during ubiquitin transfer. Interestingly, ubiquitin chain linkage specificity appears to be inherently dependent on the last 60 amino acids of the HECT domain C-lobe (Kim and Huibregtse, 2009).
RING finger E3s
The mammalian genome encodes more than 600 potential RING finger E3s (Li et al., 2008). A canonical RING finger is a Zn2+-coordinating domain that consists of a series of specifically spaced cysteine and histidine residues, and facilitates E2-dependent ubiquitylation (Lorick et al., 1999). The structure of the RING domain of Cbl in complex with an E2 illustrates several notable features of the RING domain (Zheng et al., 2000). The two zinc ions and the coordinating residues form a ‘cross-brace’ structure. Unlike the HECT domain, the RING finger domain does not form a catalytic intermediate with ubiquitin. Instead, the RING finger serves, at a minimum, as a scaffold that brings E2 and substrate together, and at least one study suggests that RING finger domains can also allosterically activate E2s (Ozkan et al., 2005).
Members of the RING finger ubiquitin ligase family can function as monomers, dimers or multi-subunit complexes. Dimerization generally occurs through the RING finger domain or surrounding regions and can result in homodimers [e.g. cIAP (cellular inhibitor of apoptosis; officially known as BIRC2), RNF4 (ring finger protein 4), SIAH (seven in absentia homologue 1), and TRAF2 (TNF receptor-associated factor 2)] (Liew et al., 2010; Mace et al., 2008; Park et al., 1999; Polekhina et al., 2002) or heterodimers [e.g. MDM2 (murine double minute 2, also known as HDM2 in human) and MDMX (officially known as MDM4, also known as HDMX or HDM4 in human), BRCA1 (breast cancer 1) and BARD1 (BRCA1-associated RING domain 1), RING1b (officially known RNF2) and BMI1 (BMI1 polycomb ring finger oncogene)] (Brzovic et al., 2001; Buchwald et al., 2006; Linke et al., 2008; Satijn and Otte, 1999; Sharp et al., 1999; Simons et al., 2006; Wu et al., 1996). For heterodimers, one RING domain (MDMX, BARD1, BMI1) often lacks ligase activity and might conform and/or stabilize the active E2-binding RING domain. Multi-subunit RING domains are exemplified by the cullin RING ligase (CRL) superfamily (Hua and Vierstra, 2011; Petroski and Deshaies, 2005), which includes the SCF complex, consisting of S-phase kinase-associated protein 1 (SKP1), cullin and F-box protein, and the more elaborate anaphase-promoting complex/cyclosome (APC/C).
Multi-subunit RING E3s and the cell cycle
Correct progression of the cell cycle is highly dependent on two families of multi-subunit RING E3s, the APC/C and the SCF; for a detailed discussion please also refer to the Commentary by Annamaria Mocchiaro and Michael Rape in the previous issue (J. Cell Sci. 125, xxxx). The APC/C consists of at least thirteen different subunits, including the E2-binding RING finger protein APC11. One of two co-activators, cell division cycle homologue 20 (CDC20) and CDH1 (officially known as FZR1) confers substrate specificity by associating with the APC/C at specific stages of the cell cycle (Nakayama and Nakayama, 2006; Peters, 2006). APC/CCDC20 is active from prometaphase to telophase. Among its ubiquitin-proteasome system (UPS) substrates are securin, which enables correct separation of sister chromatids, and cyclin B, whose degradation is required to prevent ongoing activity of cyclin-dependent kinase 1 (CDK1) after the completion of mitosis (Peters, 2006). In G1 phase, CDH1 replaces CDC20 in APC/C and a second set of substrates is targeted for degradation. Interestingly, these include CDC20 and a substrate recognition element of the SCF family, the S-phase kinase-associated protein 2 (SKP2), which is also involved in cell cycle regulation (see below). The phosphatase CDC25 is also degraded by the UPS through the activity of APC/CCDH1, which results in activation of the CDK inhibitors p21 and p27. Transition into S phase is facilitated by inactivation of CDH1 through phosphorylation by CDK2 (leading to its dissociation from the core APC/C) and targeting for degradation by both APCCDH1 and an unknown SCF E3 (Peters, 2006). APC/C activity is further regulated through association with its inhibitory pseudo-substrate early mitotic inhibitor (EMI1). EMI1 facilitates inactivation of APC/C from late G1 through S phase, and in some cells into G2 phase (Frescas and Pagano, 2008; Peters, 2006).
A second RING E3 complex that is required for cell cycle progression belongs to the SCF family of CRL E3s. SCF E3s utilize inter-changeable F-box substrate-recognition elements and, therefore, there are potentially 69 different mammalian SCF E3s. Two F-box proteins implicated in cell cycle regulation are SKP2 and beta-transducin repeat-containing protein (β-TrCP, officially known as BTRC) (Frescas and Pagano, 2008). SCFSKP2 is active during S and G2 phases, and is responsible for the UPS-mediated degradation of p21 and p27, thereby leading to activation of CDK1 and CDK2. SCFβ-TrCP targets CDC25A and WEE1 (another phosphatase) for degradation during G2, thereby exerting both a positive and a negative effect on CDK1 activity, respectively. SCFβ-TrCP is also responsible for the degradation of EMI1 in late G2 phase (Frescas and Pagano, 2008).
The role of the RING E3 ligase MDM2 in the regulation of p53
The tumor suppressor protein p53 is a transcription factor that induces cell cycle arrest and apoptosis in response to stress such as DNA damage. Maintaining low basal levels of p53 without disrupting cell cycle progression requires a high degree of regulation, which is mediated by the UPS. Over ten E3s have been associated with the regulation of p53, but the RING E3 MDM2 is of unquestionable importance (Lee and Gu, 2010; Wade et al., 2010). MDM2 ubiquitylates p53 in a RING-finger-dependent manner, which leads to the proteasomal degradation of p53 (Fang et al., 2000; Haupt et al., 1997; Honda et al., 1997; Honda and Yasuda, 2000; Kubbutat et al., 1997). MDM2 also autoubiquitylates, thereby regulating its own level in the cell (Fang et al., 2000). The functional MDM2 E3 ligase can be a MDM2 homodimer or a heterodimer with MDMX (Lee and Gu, 2010; Wade et al., 2010). MDMX, itself, is essential for the regulation of p53 and binds to p53 in a manner that is similar to its binding to MDM2.
In response to genotoxic and other stresses, MDM2 activity towards p53 is inhibited through several mechanisms, which include phosphorylation of MDM2 and p53; these lead to increased p53 signaling. Furthermore, levels of the tumor suppressor ARF increase in response to stress, which results in MDM2 being sequestered into the nucleolus and its interaction with p53 being inhibited (Linke et al., 2008).
HECT and RING finger E3s, and their function in receptor and transporter regulation
Another process that is essential for the correct regulation of cell growth is the downregulation of receptor tyrosine kinase (RTK) signaling, which is accomplished in part by internalization of the ligand-activated RTK and its trafficking to the lysosome for degradation. Although the precise endocytic mechanism varies for different mammalian receptors, ubiquitylation by the CBL family of RING finger E3s has crucial roles in this process (Acconcia et al., 2009). In addition to the RING finger domain E3, CBL proteins contain a tyrosine-kinase-binding (TKB) domain that mediates direct binding to activated RTKs. CBL proteins can be recruited to RTKs indirectly through interactions with the adaptor protein growth-factor-receptor-bound protein 2 (GRB2) (Peschard et al., 2001). CBL family members modify the activated RTK with mono- or K63-linked ubiquitin chains that – together with the direct recruitment of proteins to CBL itself – assemble a multi-protein complex that includes ubiquitin-binding domain (UBD)-containing proteins (so-called ubiquitin receptors) (Swaminathan and Tsygankov, 2006). The exact role for RTK ubiquitylation by CBL in the initial receptor internalization step is unclear, but ongoing ubiquitylation of RTKs by this E3 ligase is necessary for correct downstream sorting and degradation of the signaling complex by the lysosome (Marmor and Yarden, 2004; Williams and Urbe, 2007). As part of the process of RTK downregulation, CBL proteins are negatively regulated through autoubiquitylation. CBLs themselves are also subject to ubiquitylation by the HECT E3s, NEDD4 and ITCH (Magnifico et al., 2003), although the relationship between this ubiquitylation and RTK activation is unknown. Ubiquitylation, as well as deubiquitylation of components of the RTK complex and recruitment of ubiquitin-binding proteins, thus, have essential roles in the modulation of growth factor signaling.
The nine members of the NEDD4 family of HECT E3 ligases include the NEDD4 isoforms, ITCH and members of the SMAD-specific E3 ubiquitin protein ligases (SMURFs), which – collectively – are the mammalian orthologues of yeast Rsp5p. Early discoveries regarding endocytosis and vacuolar targeting in yeast advanced our understanding of the mechanisms that are involved in the downregulation of proteins from the plasma membrane in mammalian cells. The NEDD4 isoforms in particular mediate ubiquitylation that leads to the cell-surface downregulation of transporters and receptors, such as the epithelial sodium channel (ENaC) (Rotin and Kumar, 2009; Staub et al., 2000) and the β2-adrenergic receptor (ADRB2) (Shenoy et al., 2008). A common feature of the NEDD4 family is the presence of two to four double tryptophan residue (WW) domains in the N-terminal half of the molecule that recognize proline-containing (so-called PY) motifs on substrates (Rotin and Kumar, 2009). Most NEDD4 family members also contain an N-terminal C2 domain that facilitates interactions with the plasma membrane (Rotin and Kumar, 2009). Mutations in ENaC subunits that decrease the interaction with the WW domains of NEDDL are associated with Liddle syndrome, an autosomal dominant disorder that is characterized by severe hypertension (Rotin and Staub, 2011).
RING finger and U-box E3s, and the UPS in protein quality control
RING E3s mediate protein quality control by ubiquitylating an array of misfolded and unassembled proteins, as well as those whose levels must be regulated. In the endoplasmic reticulum (ER), the polytopic mammalian ER-associated degradation (ERAD) RING E3s gp78 (officially known as AMFR) and HRD1 (officially known as SYVN1) tag proteins with polyubiquitin chains and, thereby, direct them for cytosolic degradation through proteasomes (Bernasconi and Molinari, 2011).
gp78 and HRD1 are orthologues of the well-characterized yeast ER-resident ERAD E3 Hrd1p (also known as Der3p) (Fang et al., 2001; Kikkert et al., 2004). gp78 contains a complex domain structure in its C-terminal cytosolic half. In addition to its RING finger domain, it includes a coupling of ubiquitin to ER degradation (CUE) domain that can bind ubiquitin (Chen et al., 2006), and the UBE2G2-binding region (G2BR) that binds to and imparts allosteric effects on its cognate E2 (Chen et al., 2006; Li et al., 2009; Das et al., 2009). All of those domains are essential for gp78 function. Moreover, gp78 also contains a p97/valosin-containing protein (VCP)-interacting motif (VIM) that recruits the AAA-ATPase p97/VCP to ubiquitylated substrates (Ballar and Fang, 2008; Zhong et al., 2004).
gp78 is a pro-metastatic E3 ligase that mediates the degradation of the metastasis suppressor KAI1 (officially known as CD82) (Tsai et al., 2007). Also, gp78 regulates cholesterol metabolism by targeting insulin-induced gene 1 (INSIG1) to the UPS (Lee et al., 2006) (for additional gp78 substrates, please refer to the poster). HRD1 has been implicated in arthropathies including rheumatoid arthritis (Amano et al., 2003) and has substrates that include the Parkinson-disease-associated ‘orphan’ G-protein-coupled receptor PaelR (officially known as GPR37) (Omura et al., 2006), p53 (Yamasaki et al., 2007), and other substrates (see poster). Adding further complexity to ERAD pathways, gp78 can be targeted for degradation by autoubiquitylation and by ubiquitylation through HRD1 that – in turn – stabilizes gp78 substrates (Ballar et al., 2010; Shmueli et al., 2009). Other ER-localized E3s, including RNF5, MARCH6 and RNF139, have also been implicated in ERAD (Morito et al., 2008; Stagg et al., 2009; Tcherpakov et al., 2009; Zavacki et al., 2009).
In mammals, the best-characterized mediator of cytosolic protein quality control is the U-box E3 CHIP (officially known as STUB1) (Cyr et al., 2002; Hohfeld et al., 2001). The U-box creates an E2 binding surface that resembles a RING finger, but lacks Zn2+-coordinating sites and, instead, contains stabilizing hydrogen bonds and salt bridges (Aravind and Koonin, 2000; Hatakeyama et al., 2001; Pringa et al., 2001). CHIP contains N-terminal tetratricopeptide repeat (TPR) domains that mediate binding to HSP70, HSC70 and HSP90 chaperones (Ballinger et al., 1999; Connell et al., 2001; Nikolay et al., 2004). The CHIP U-box exists as an asymmetric homodimer that only binds one E2 molecule (Kundrat and Regan, 2010; Zhang et al., 2005).
CHIP is a chaperone-dependent E3 that modulates the switch from chaperone-assisted protein folding to UPS degradation (Connell et al., 2001; Hohfeld et al., 2001) and, thus, potentially targets any chaperone-bound protein for degradation. Of note, several CHIP substrates (e.g. α-synuclein, huntingtin, Tau) form protein aggregates that have been implicated in neurodegenerative diseases (Hatakeyama et al., 2004; Jana et al., 2005; Petrucelli et al., 2004; Shimura et al., 2004; Tetzlaff et al., 2008). Furthermore, some substrates (for example, cytochrome P450 3A4, the cystic fibrosis transmembrane conductance regulator, and others) are targeted by both ERAD E3s and CHIP (Pabarcus et al., 2009; Younger et al., 2006), which suggests cooperativity between different quality control pathways. In addition, studies in yeast have implicated other E3s in protein quality control within the cytosol as well as throughout the cell (Bengtson and Joazeiro, 2010; Eisele and Wolf, 2008; Gardner et al., 2005; Heck et al., 2010; Metzger et al., 2008; Nillegoda et al., 2010; Swanson et al., 2001); the same is likely to be true in mammalian cells.
IAP RING finger E3s and UPS in the intrinsic apoptosis pathway
Chronic protein misfolding, as well as oxidative or genotoxic stress and developmental cues, can lead to apoptosis – another process during which E3s have crucial roles (Vucic et al., 2011). In the intrinsic apoptotic pathway, stimuli trigger release of cytochrome c from mitochondria, leading to formation of apoptosomes. These activate caspase 9, which – in turn – activates the effector caspases 3 and 7 that mediate cell death.
Inhibitor of apoptosis proteins (IAPs) are RING E3s that prevent apoptosis at the level of caspase activation (Eckelman et al., 2006). In addition to their RING finger domains, IAPs contain baculoviral IAP repeats (BIRs) that mediate their caspase interaction. X-chromosome-linked IAP (XIAP) binds to caspases 3, 7 and 9, directly blocks their catalytic activity and, additionally, regulates their protein levels through ubiquitylation and subsequent degradation (Morizane et al., 2005; Schile et al., 2008; Suzuki et al., 2001). Cellular IAP1 (c-IAP1, officially known as BIRC2) might also promote the ubiquitylation of caspases 3 and 7, although the physiological significance of this is less clear (Choi et al., 2009).
Second mitochondrial activator of caspases (SMAC, officially known as DIABLO) is also released from mitochondria following apoptotic stimuli. SMAC prevents XIAP from interacting with and inhibiting caspases (Du et al., 2000; Verhagen et al., 2000), but SMAC itself is targeted for degradation by XIAP and cIAPs (Hu and Yang, 2003; MacFarlane et al., 2002). Further, cIAP1 can ubiquitylate XIAP, and both XIAP and c-IAP1 have autoubiquitylation activity (Cheung et al., 2008; Silke et al., 2005; Yang et al., 2000). Other RING E3s, such as SIAH1, also can target XIAP for degradation (Garrison et al., 2011), adding further complexity to the finely tuned regulation of apoptosis.
RING finger E3s in angiogenesis
Angiogenesis is a hallmark of tumor growth and progression (Hanahan and Weinberg, 2011). A key mediator of this is the hypoxia-inducible factor (HIF) family of transcription factors that induce transcription of genes involved in bioenergetics, cell growth and angiogenesis (Kaelin and Ratcliffe, 2008; Majmundar et al., 2010). Under hypoxic conditions, HIF proteins are found as transcriptionally active HIF1-α and HIF1-β (also known as HIF1A and ARNT, respectively) heterodimers that bind hypoxia response elements (HREs) upstream of target genes (Benita et al., 2009). Under normoxic conditions, HIF1-β is stable, whereas the HIF1-α proteins are degraded by the UPS (Huang et al., 1998), thereby preventing transcriptional activation. The key to oxygen-dependent regulation of HIF1-α is the von Hippel-Lindau (VHL) CRL (CRL2VHL), which comprises the VHL tumor suppressor protein, the elongins B and C, cullin 2 and the RING protein RBX1 (Iwai et al., 1999; Kamura et al., 1999; Kibel et al., 1995; Lisztwan et al., 1999; Lonergan et al., 1998; Pause et al., 1997). VHL mediates recognition of HIF1-α proteins by binding to hydroxylated prolines, a modification that is catalyzed by the oxygen-sensing prolyl hydroxylases (PHDs) (Jaakkola et al., 2001; Ohh, 2006; Yu et al., 2001). The SIAH family of RING E3s also regulates the HIF-dependent response to hypoxia. Under hypoxic conditions, SIAH2 transcription is induced by the HIF transcription factor. In turn, SIAH1a and SIAH2 ubiquitylate PHDs, which targets them to the UPS (Nakayama et al., 2004) and, thus, stabilizes HIF-α proteins.
Perspectives
E3s have crucial roles in cellular homeostasis and development, and frequently contribute to pathophysiological states. As our appreciation of the complexity of the ubiquitylation system increases, so do the number of questions regarding E3s, including their cellular functions, mechanisms of action and possible redundancy. Whereas humans have 600–700 putative E3s, the Arabidopsis thaliana genome encodes over 700 F-box proteins alone (Gagne et al., 2002), some of which have novel roles as components of SCF hormone receptors (Vierstra, 2009). Whereas, intuitively, it makes sense that sessile organisms have more varied ways of adapting to their environments, these numbers are still extraordinary.
In addition to the ligase domains discussed above, we now know that mammalian A20 zinc fingers also possess ligase activity (Mattera et al., 2006; Wertz et al., 2004). Several bacterial virulence proteins also function as ubiquitin ligases in eukaryotic host cells, despite having no sequence or structural similarity to mammalian E3s (Diao et al., 2008; Levin et al., 2010; Quezada et al., 2009). The potential for additional domains in mammals, as well as in other organisms, to have E3 activity cannot be discounted. Further complexity is added by the fact that some E3s that are believed to contain two RING finger domains (RING-in-between-RING) might actually function through a covalent thioester in a manner similar to HECTs (Wenzel et al., 2011). It is also becoming apparent that E3 domains that bind E2s in areas that are distinct from the overlapping RING or HECT and E1-binding sites – such as the G2BR of gp78, the Rad6 binding domain (R6BD) of RAD18, and the basic canyon of CUL1 (Das et al., 2009; Hibbert et al., 2011; Kleiger et al., 2009; Li et al., 2009) – might aid in the activation or regulation of ubiquitylation. Additionally, there is recent evidence that other domains within E3s, including various ubiquitin-binding domains and as-yet-uncharacterized regions, might have distinct effects on ubiquitylation. As exemplified by the discussion of E3s in this Cell Science at a Glance article, as well as other recent works (Weissman et al., 2011), autoubiquitylation and crossubiquitylation of E3s serves to regulate E3 fate and function, and is likely to be involved in many more processes than currently appreciated. The discovery and characterization of ubiquitin ligases and their cognate substrates promises to be a fruitful and clinically important area of investigation for the foreseeable future.
Supplementary Material
Acknowledgements
We apologize to the many scientists whose contributions to this vast area of research could not be directly cited because of space limitations. We thank Stan Lipkowitz, Yien Che Tsai and Daniel Stringer for insightful comments on this manuscript.
Footnotes
This article is part of a Minifocus on Ubiquitin. For further reading, please see related articles: ‘Ubiquitin and SUMO in DNA repair at a glance’ by Helle D. Ulrich (J. Cell Sci. 125, 249-254), ‘Emerging regulatory mechanisms in ubiquitin-dependent cell cycle control’ by Annamaria Mocciaro and Michael Rape (J. Cell Sci. 125, 255-263), ‘The role of ubiquitylation in receptor endocytosis and endosomal sorting’ by Kaisa Haglund and Ivan Dikic (J. Cell Sci. 125, 265-275), ‘Cellular functions of the DUBs’ by Michael J. Clague et al. (J. Cell Sci. 125, 277-286), ‘Non-canonical ubiquitin-based signals for proteasomal degradation’ by Yelena Kravtsova-Ivantsiv and Aaron Ciechanover (J. Cell Sci. 125, 539-548) and ‘No one can whistle a symphony alone – how different ubiquitin linkages cooperate to orchestrate NF-κB activity’ by Anna C. Schmukle and Henning Walczak (J. Cell Sci. 125, 549-559).
Funding
Research in the authors' laboratory is supported by the National Institutes of Health, National Cancer Institute and the Center for Cancer Research.
A high-resolution version of the poster is available for downloading in the online version of this article at jcs.biologists.com. Individual poster panels are available as JPEG files at http://jcs.biologists.org/lookup/suppl/doi:10.1242/jcs.091777/-/DC1
References
- Acconcia F., Sigismund S., Polo S. (2009). Ubiquitin in trafficking: the network at work. Exp. Cell Res. 315, 1610-1618 [DOI] [PubMed] [Google Scholar]
- Amano T., Yamasaki S., Yagishita N., Tsuchimochi K., Shin H., Kawahara K., Aratani S., Fujita H., Zhang L., Ikeda R., et al. (2003). Synoviolin/Hrd1, an E3 ubiquitin ligase, as a novel pathogenic factor for arthropathy. Genes Dev. 17, 2436-2449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind L., Koonin E. V. (2000). The U box is a modified RING finger-a common domain in ubiquitination. Curr. Biol. 10, R132-R134 [DOI] [PubMed] [Google Scholar]
- Ballar P., Fang S. (2008). Regulation of ER-associated degradation via p97/VCP-interacting motif. Biochem. Soc. Trans. 36, 818-822 [DOI] [PubMed] [Google Scholar]
- Ballar P., Ors A. U., Yang H., Fang S. (2010). Differential regulation of CFTRDeltaF508 degradation by ubiquitin ligases gp78 and Hrd1. Int. J. Biochem. Cell Biol. 42, 167-173 [DOI] [PubMed] [Google Scholar]
- Ballinger C. A., Connell P., Wu Y., Hu Z., Thompson L. J., Yin L. Y., Patterson C. (1999). Identification of CHIP, a novel tetratricopeptide repeat-containing protein that interacts with heat shock proteins and negatively regulates chaperone functions. Mol. Cell. Biol. 19, 4535-4545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazirgan O. A., Hampton R. Y. (2008). Cue1p is an activator of Ubc7p E2 activity in vitro and in vivo. J. Biol. Chem. 283, 12797-12810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrends C., Harper J. W. (2011). Constructing and decoding unconventional ubiquitin chains. Nat. Struct. Mol. Biol. 18, 520-528 [DOI] [PubMed] [Google Scholar]
- Bengtson M. H., Joazeiro C. A. (2010). Role of a ribosome-associated E3 ubiquitin ligase in protein quality control. Nature 467, 470-473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benita Y., Kikuchi H., Smith A. D., Zhang M. Q., Chung D. C., Xavier R. J. (2009). An integrative genomics approach identifies Hypoxia Inducible Factor-1 (HIF-1)-target genes that form the core response to hypoxia. Nucleic Acids Res. 37, 4587-4602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernasconi R., Molinari M. (2011). ERAD and ERAD tuning: disposal of cargo and of ERAD regulators from the mammalian ER. Curr. Opin. Cell Biol. 23, 176-183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzovic P. S., Rajagopal P., Hoyt D. W., King M. C., Klevit R. E. (2001). Structure of a BRCA1-BARD1 heterodimeric RING-RING complex. Nat. Struct. Biol. 8, 833-837 [DOI] [PubMed] [Google Scholar]
- Buchwald G., van der Stoop P., Weichenrieder O., Perrakis A., van Lohuizen M., Sixma T. K. (2006). Structure and E3-ligase activity of the Ring-Ring complex of polycomb proteins Bmi1 and Ring1b. EMBO J. 25, 2465-2474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadwell K., Coscoy L. (2005). Ubiquitination on nonlysine residues by a viral E3 ubiquitin ligase. Science 309, 127-130 [DOI] [PubMed] [Google Scholar]
- Carvalho A. F., Pinto M. P., Grou C. P., Alencastre I. S., Fransen M., Sa-Miranda C., Azevedo J. E. (2007). Ubiquitination of mammalian Pex5p, the peroxisomal import receptor. J. Biol. Chem. 282, 31267-31272 [DOI] [PubMed] [Google Scholar]
- Chen B., Mariano J., Tsai Y. C., Chan A. H., Cohen M., Weissman A. M. (2006). The activity of a human endoplasmic reticulum-associated degradation E3, gp78, requires its Cue domain, RING finger, and an E2-binding site. Proc. Natl. Acad. Sci. USA 103, 341-346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung H. H., Plenchette S., Kern C. J., Mahoney D. J., Korneluk R. G. (2008). The RING domain of cIAP1 mediates the degradation of RING-bearing inhibitor of apoptosis proteins by distinct pathways. Mol. Biol. Cell 19, 2729-2740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y. E., Butterworth M., Malladi S., Duckett C. S., Cohen G. M., Bratton S. B. (2009). The E3 ubiquitin ligase cIAP1 binds and ubiquitinates caspase-3 and -7 via unique mechanisms at distinct steps in their processing. J. Biol. Chem. 284, 12772-12782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciechanover A., Ben-Saadon R. (2004). N-terminal ubiquitination: more protein substrates join in. Trends Cell. Biol. 14, 103-106 [DOI] [PubMed] [Google Scholar]
- Clague M. J., Urbe S. (2010). Ubiquitin: same molecule, different degradation pathways. Cell 143, 682-685 [DOI] [PubMed] [Google Scholar]
- Connell P., Ballinger C. A., Jiang J., Wu Y., Thompson L. J., Hohfeld J., Patterson C. (2001). The co-chaperone CHIP regulates protein triage decisions mediated by heat-shock proteins. Nat. Cell Biol. 3, 93-96 [DOI] [PubMed] [Google Scholar]
- Cyr D. M., Hohfeld J., Patterson C. (2002). Protein quality control: U-box-containing E3 ubiquitin ligases join the fold. Trends Biochem. Sci. 27, 368-375 [DOI] [PubMed] [Google Scholar]
- Das R., Mariano J., Tsai Y. C., Kalathur R. C., Kostova Z., Li J., Tarasov S. G., McFeeters R. L., Altieri A. S., Ji X., et al. (2009). Allosteric activation of E2-RING finger-mediated ubiquitylation by a structurally defined specific E2-binding region of gp78. Mol. Cell 34, 674-685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshaies R. J., Joazeiro C. A. (2009). RING domain E3 ubiquitin ligases. Annu. Rev. Biochem. 78, 399-434 [DOI] [PubMed] [Google Scholar]
- Diao J., Zhang Y., Huibregtse J. M., Zhou D., Chen J. (2008). Crystal structure of SopA, a Salmonella effector protein mimicking a eukaryotic ubiquitin ligase. Nat. Struct. Mol. Biol. 15, 65-70 [DOI] [PubMed] [Google Scholar]
- Du C., Fang M., Li Y., Li L., Wang X. (2000). Smac, a mitochondrial protein that promotes cytochrome c-dependent caspase activation by eliminating IAP inhibition. Cell 102, 33-42 [DOI] [PubMed] [Google Scholar]
- Eckelman B. P., Salvesen G. S., Scott F. L. (2006). Human inhibitor of apoptosis proteins: why XIAP is the black sheep of the family. EMBO Rep. 7, 988-994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisele F., Wolf D. H. (2008). Degradation of misfolded protein in the cytoplasm is mediated by the ubiquitin ligase Ubr1. FEBS Lett. 582, 4143-4146 [DOI] [PubMed] [Google Scholar]
- Fang S., Jensen J. P., Ludwig R. L., Vousden K. H., Weissman A. M. (2000). Mdm2 is a RING finger-dependent ubiquitin protein ligase for itself and p53. J. Biol. Chem. 275, 8945-8951 [DOI] [PubMed] [Google Scholar]
- Fang S., Ferrone M., Yang C., Jensen J. P., Tiwari S., Weissman A. M. (2001). The tumor autocrine motility factor receptor, gp78, is a ubiquitin protein ligase implicated in degradation from the endoplasmic reticulum. Proc. Natl. Acad. Sci. USA 98, 14422-14427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frescas D., Pagano M. (2008). Deregulated proteolysis by the F-box proteins SKP2 and beta-TrCP: tipping the scales of cancer. Nat. Rev. Cancer 8, 438-449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagne J. M., Downes B. P., Shiu S. H., Durski A. M., Vierstra R. D. (2002). The F-box subunit of the SCF E3 complex is encoded by a diverse superfamily of genes in Arabidopsis. Proc. Natl. Acad. Sci. USA 99, 11519-11524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner R. G., Nelson Z. W., Gottschling D. E. (2005). Degradation-mediated protein quality control in the nucleus. Cell 120, 803-815 [DOI] [PubMed] [Google Scholar]
- Garrison J. B., Correa R. G., Gerlic M., Yip K. W., Krieg A., Tamble C. M., Shi R., Welsh K., Duggineni S., Huang Z., et al. (2011). ARTS and Siah collaborate in a pathway for XIAP degradation. Mol. Cell 41, 107-116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D., Weinberg R. A. (2011). Hallmarks of cancer: the next generation. Cell 144, 646-674 [DOI] [PubMed] [Google Scholar]
- Hatakeyama S., Yada M., Matsumoto M., Ishida N., Nakayama K. I. (2001). U box proteins as a new family of ubiquitin-protein ligases. J. Biol. Chem. 276, 33111-33120 [DOI] [PubMed] [Google Scholar]
- Hatakeyama S., Matsumoto M., Kamura T., Murayama M., Chui D. H., Planel E., Takahashi R., Nakayama K. I., Takashima A. (2004). U-box protein carboxyl terminus of Hsc70-interacting protein (CHIP) mediates poly-ubiquitylation preferentially on four-repeat Tau and is involved in neurodegeneration of tauopathy. J. Neurochem. 91, 299-307 [DOI] [PubMed] [Google Scholar]
- Haupt Y., Maya R., Kazaz A., Oren M. (1997). Mdm2 promotes the rapid degradation of p53. Nature 387, 296-299 [DOI] [PubMed] [Google Scholar]
- Heck J. W., Cheung S. K., Hampton R. Y. (2010). Cytoplasmic protein quality control degradation mediated by parallel actions of the E3 ubiquitin ligases Ubr1 and San1. Proc. Natl. Acad. Sci. USA 107, 1106-1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibbert R. G., Huang A., Boelens R., Sixma T. K. (2011). E3 ligase Rad18 promotes monoubiquitination rather than ubiquitin chain formation by E2 enzyme Rad6. Proc. Natl. Acad. Sci. USA 108, 5590-5595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohfeld J., Cyr D. M., Patterson C. (2001). From the cradle to the grave: molecular chaperones that may choose between folding and degradation. EMBO Rep. 2, 885-890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda R., Yasuda H. (2000). Activity of MDM2, a ubiquitin ligase, toward p53 or itself is dependent on the RING finger domain of the ligase. Oncogene 19, 1473-1476 [DOI] [PubMed] [Google Scholar]
- Honda R., Tanaka H., Yasuda H. (1997). Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett. 420, 25-27 [DOI] [PubMed] [Google Scholar]
- Hu S., Yang X. (2003). Cellular inhibitor of apoptosis 1 and 2 are ubiquitin ligases for the apoptosis inducer Smac/DIABLO. J. Biol. Chem. 278, 10055-10060 [DOI] [PubMed] [Google Scholar]
- Hua Z., Vierstra R. D. (2011). The cullin-RING ubiquitin-protein ligases. Annu. Rev. Plant Biol. 62, 299-334 [DOI] [PubMed] [Google Scholar]
- Huang L., Kinnucan E., Wang G., Beaudenon S., Howley P. M., Huibregtse J. M., Pavletich N. P. (1999). Structure of an E6AP-UbcH7 complex: insights into ubiquitination by the E2-E3 enzyme cascade. Science 286, 1321-1326 [DOI] [PubMed] [Google Scholar]
- Huang L. E., Gu J., Schau M., Bunn H. F. (1998). Regulation of hypoxia-inducible factor 1alpha is mediated by an O2-dependent degradation domain via the ubiquitin-proteasome pathway. Proc. Natl. Acad. Sci. USA 95, 7987-7992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huibregtse J. M., Scheffner M., Beaudenon S., Howley P. M. (1995). A family of proteins structurally and functionally related to the E6-AP ubiquitin-protein ligase. Proc. Natl. Acad. Sci. USA 92, 2563-2567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikura S., Weissman A. M., Bonifacino J. S. (2010). Serine residues in the cytosolic tail of the T-cell antigen receptor alpha-chain mediate ubiquitination and endoplasmic reticulum-associated degradation of the unassembled protein. J. Biol. Chem. 285, 23916-23924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwai K., Yamanaka K., Kamura T., Minato N., Conaway R. C., Conaway J. W., Klausner R. D., Pause A. (1999). Identification of the von Hippel-lindau tumor-suppressor protein as part of an active E3 ubiquitin ligase complex. Proc. Natl. Acad. Sci. USA 96, 12436-12441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaakkola P., Mole D. R., Tian Y. M., Wilson M. I., Gielbert J., Gaskell S. J., Kriegsheim A., Hebestreit H. F., Mukherji M., Schofield C. J., et al. (2001). Targeting of HIF-alpha to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science 292, 468-472 [DOI] [PubMed] [Google Scholar]
- Jana N. R., Dikshit P., Goswami A., Kotliarova S., Murata S., Tanaka K., Nukina N. (2005). Co-chaperone CHIP associates with expanded polyglutamine protein and promotes their degradation by proteasomes. J. Biol. Chem. 280, 11635-11640 [DOI] [PubMed] [Google Scholar]
- Kaelin W. G., Jr, Ratcliffe P. J. (2008). Oxygen sensing by metazoans: the central role of the HIF hydroxylase pathway. Mol. Cell 30, 393-402 [DOI] [PubMed] [Google Scholar]
- Kamadurai H. B., Souphron J., Scott D. C., Duda D. M., Miller D. J., Stringer D., Piper R. C., Schulman B. A. (2009). Insights into ubiquitin transfer cascades from a structure of a UbcH5B approximately ubiquitin-HECT(NEDD4L) complex. Mol. Cell 36, 1095-1102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamura T., Koepp D. M., Conrad M. N., Skowyra D., Moreland R. J., Iliopoulos O., Lane W. S., Kaelin W. G., Jr, Elledge S. J., Conaway R. C., et al. (1999). Rbx1, a component of the VHL tumor suppressor complex and SCF ubiquitin ligase. Science 284, 657-661 [DOI] [PubMed] [Google Scholar]
- Kibel A., Iliopoulos O., DeCaprio J. A., Kaelin W. G., Jr (1995). Binding of the von Hippel-Lindau tumor suppressor protein to Elongin B and C. Science 269, 1444-1446 [DOI] [PubMed] [Google Scholar]
- Kikkert M., Doolman R., Dai M., Avner R., Hassink G., van Voorden S., Thanedar S., Roitelman J., Chau V., Wiertz E. (2004). Human HRD1 is an E3 ubiquitin ligase involved in degradation of proteins from the endoplasmic reticulum. J. Biol. Chem. 279, 3525-3534 [DOI] [PubMed] [Google Scholar]
- Kim H. C., Huibregtse J. M. (2009). Polyubiquitination by HECT E3s and the determinants of chain type specificity. Mol. Cell. Biol. 29, 3307-3318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkin V., Dikic I. (2011). Ubiquitin networks in cancer. Curr. Opin. Genet. Dev. 21, 21-28 [DOI] [PubMed] [Google Scholar]
- Kleiger G., Saha A., Lewis S., Kuhlman B., Deshaies R. J. (2009). Rapid E2-E3 assembly and disassembly enable processive ubiquitylation of cullin-RING ubiquitin ligase substrates. Cell 139, 957-968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komander D. (2009). The emerging complexity of protein ubiquitination. Biochem. Soc. Trans. 37, 937-953 [DOI] [PubMed] [Google Scholar]
- Kubbutat M. H., Jones S. N., Vousden K. H. (1997). Regulation of p53 stability by Mdm2. Nature 387, 299-303 [DOI] [PubMed] [Google Scholar]
- Kundrat L., Regan L. (2010). Balance between folding and degradation for Hsp90-dependent client proteins: a key role for CHIP. Biochemistry 49, 7428-7438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. N., Song B., DeBose-Boyd R. A., Ye J. (2006). Sterol-regulated degradation of Insig-1 mediated by the membrane-bound ubiquitin ligase gp78. J. Biol. Chem. 281, 39308-39315 [DOI] [PubMed] [Google Scholar]
- Lee J. T., Gu W. (2010). The multiple levels of regulation by p53 ubiquitination. Cell Death Differ. 17, 86-92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin I., Eakin C., Blanc M. P., Klevit R. E., Miller S. I., Brzovic P. S. (2010). Identification of an unconventional E3 binding surface on the UbcH5 ~ Ub conjugate recognized by a pathogenic bacterial E3 ligase. Proc. Natl. Acad. Sci. USA 107, 2848-2853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Tu D., Brunger A. T., Ye Y. (2007). A ubiquitin ligase transfers preformed polyubiquitin chains from a conjugating enzyme to a substrate. Nature 446, 333-337 [DOI] [PubMed] [Google Scholar]
- Li W., Bengtson M. H., Ulbrich A., Matsuda A., Reddy V. A., Orth A., Chanda S. K., Batalov S., Joazeiro C. A. (2008). Genome-wide and functional annotation of human E3 ubiquitin ligases identifies MULAN, a mitochondrial E3 that regulates the organelle's dynamics and signaling. PLoS ONE 3, e1487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Tu D., Li L., Wollert T., Ghirlando R., Brunger A. T., Ye Y. (2009). Mechanistic insights into active site-associated polyubiquitination by the ubiquitin-conjugating enzyme Ube2g2. Proc. Natl. Acad. Sci. USA 106, 3722-3727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liew C. W., Sun H., Hunter T., Day C. L. (2010). RING domain dimerization is essential for RNF4 function. Biochem. J. 431, 23-29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linke K., Mace P. D., Smith C. A., Vaux D. L., Silke J., Day C. L. (2008). Structure of the MDM2/MDMX RING domain heterodimer reveals dimerization is required for their ubiquitylation in trans. Cell Death Differ. 15, 841-848 [DOI] [PubMed] [Google Scholar]
- Lipkowitz S., Weissman A. M. (2011). RINGs of good and evil: RING finger ubiquitin ligases at the crossroads of tumour suppression and oncogenesis. Nat. Rev. Cancer 11, 629-643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisztwan J., Imbert G., Wirbelauer C., Gstaiger M., Krek W. (1999). The von Hippel-Lindau tumor suppressor protein is a component of an E3 ubiquitin-protein ligase activity. Genes Dev. 13, 1822-1833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonergan K. M., Iliopoulos O., Ohh M., Kamura T., Conaway R. C., Conaway J. W., Kaelin W. G., Jr (1998). Regulation of hypoxia-inducible mRNAs by the von Hippel-Lindau tumor suppressor protein requires binding to complexes containing elongins B/C and Cul2. Mol. Cell. Biol. 18, 732-741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorick K. L., Jensen J. P., Fang S., Ong A. M., Hatakeyama S., Weissman A. M. (1999). RING fingers mediate ubiquitin-conjugating enzyme (E2)-dependent ubiquitination. Proc. Natl. Acad. Sci. USA 96, 11364-11369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mace P. D., Linke K., Feltham R., Schumacher F. R., Smith C. A., Vaux D. L., Silke J., Day C. L. (2008). Structures of the cIAP2 RING domain reveal conformational changes associated with ubiquitin-conjugating enzyme (E2) recruitment. J. Biol. Chem. 283, 31633-31640 [DOI] [PubMed] [Google Scholar]
- MacFarlane M., Merrison W., Bratton S. B., Cohen G. M. (2002). Proteasome-mediated degradation of Smac during apoptosis: XIAP promotes Smac ubiquitination in vitro. J. Biol. Chem. 277, 36611-36616 [DOI] [PubMed] [Google Scholar]
- Magnifico A., Ettenberg S., Yang C., Mariano J., Tiwari S., Fang S., Lipkowitz S., Weissman A. M. (2003). WW domain HECT E3s target Cbl RING finger E3s for proteasomal degradation. J. Biol. Chem. 278, 43169-43177 [DOI] [PubMed] [Google Scholar]
- Majmundar A. J., Wong W. J., Simon M. C. (2010). Hypoxia-inducible factors and the response to hypoxic stress. Mol. Cell 40, 294-309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marmor M. D., Yarden Y. (2004). Role of protein ubiquitylation in regulating endocytosis of receptor tyrosine kinases. Oncogene 23, 2057-2070 [DOI] [PubMed] [Google Scholar]
- Mattera R., Tsai Y. C., Weissman A. M., Bonifacino J. S. (2006). The Rab5 guanine nucleotide exchange factor Rabex-5 binds ubiquitin (Ub) and functions as a Ub ligase through an atypical Ub-interacting motif and a zinc finger domain. J. Biol. Chem. 281, 6874-6883 [DOI] [PubMed] [Google Scholar]
- Metzger M. B., Maurer M. J., Dancy B. M., Michaelis S. (2008). Degradation of a cytosolic protein requires endoplasmic reticulum-associated degradation machinery. J. Biol. Chem. 283, 32302-32316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morito D., Hirao K., Oda Y., Hosokawa N., Tokunaga F., Cyr D. M., Tanaka K., Iwai K., Nagata K. (2008). Gp78 cooperates with RMA1 in endoplasmic reticulum-associated degradation of CFTRDeltaF508. Mol. Biol. Cell 19, 1328-1336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morizane Y., Honda R., Fukami K., Yasuda H. (2005). X-linked inhibitor of apoptosis functions as ubiquitin ligase toward mature caspase-9 and cytosolic Smac/DIABLO. J. Biochem. 137, 125-132 [DOI] [PubMed] [Google Scholar]
- Nakayama K., Frew I. J., Hagensen M., Skals M., Habelhah H., Bhoumik A., Kadoya T., Erdjument-Bromage H., Tempst P., Frappell P. B., et al. (2004). Siah2 regulates stability of prolyl-hydroxylases, controls HIF1alpha abundance, and modulates physiological responses to hypoxia. Cell 117, 941-952 [DOI] [PubMed] [Google Scholar]
- Nakayama K. I., Nakayama K. (2006). Ubiquitin ligases: cell-cycle control and cancer. Nat. Rev. Cancer 6, 369-381 [DOI] [PubMed] [Google Scholar]
- Nikolay R., Wiederkehr T., Rist W., Kramer G., Mayer M. P., Bukau B. (2004). Dimerization of the human E3 ligase CHIP via a coiled-coil domain is essential for its activity. J. Biol. Chem. 279, 2673-2678 [DOI] [PubMed] [Google Scholar]
- Nillegoda N. B., Theodoraki M. A., Mandal A. K., Mayo K. J., Ren H. Y., Sultana R., Wu K., Johnson J., Cyr D. M., Caplan A. J. (2010). Ubr1 and Ubr2 function in a quality control pathway for degradation of unfolded cytosolic proteins. Mol. Biol. Cell 21, 2102-2116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogunjimi A. A., Briant D. J., Pece-Barbara N., Le Roy C., Di Guglielmo G. M., Kavsak P., Rasmussen R. K., Seet B. T., Sicheri F., Wrana J. L. (2005). Regulation of Smurf2 ubiquitin ligase activity by anchoring the E2 to the HECT domain. Mol. Cell 19, 297-308 [DOI] [PubMed] [Google Scholar]
- Ohh M. (2006). Ubiquitin pathway in VHL cancer syndrome. Neoplasia 8, 623-629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omura T., Kaneko M., Okuma Y., Orba Y., Nagashima K., Takahashi R., Fujitani N., Matsumura S., Hata A., Kubota K., et al. (2006). A ubiquitin ligase HRD1 promotes the degradation of Pael receptor, a substrate of Parkin. J. Neurochem. 99, 1456-1469 [DOI] [PubMed] [Google Scholar]
- Ozkan E., Yu H., Deisenhofer J. (2005). Mechanistic insight into the allosteric activation of a ubiquitin-conjugating enzyme by RING-type ubiquitin ligases. Proc. Natl. Acad. Sci. USA 102, 18890-18895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pabarcus M. K., Hoe N., Sadeghi S., Patterson C., Wiertz E., Correia M. A. (2009). CYP3A4 ubiquitination by gp78 (the tumor autocrine motility factor receptor, AMFR) and CHIP E3 ligases. Arch. Biochem. Biophys. 483, 66-74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park Y. C., Burkitt V., Villa A. R., Tong L., Wu H. (1999). Structural basis for self-association and receptor recognition of human TRAF2. Nature 398, 533-538 [DOI] [PubMed] [Google Scholar]
- Pause A., Lee S., Worrell R. A., Chen D. Y., Burgess W. H., Linehan W. M., Klausner R. D. (1997). The von Hippel-Lindau tumor-suppressor gene product forms a stable complex with human CUL-2, a member of the Cdc53 family of proteins. Proc. Natl. Acad. Sci. USA 94, 2156-2161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peschard P., Fournier T. M., Lamorte L., Naujokas M. A., Band H., Langdon W. Y., Park M. (2001). Mutation of the c-Cbl TKB domain binding site on the Met receptor tyrosine kinase converts it into a transforming protein. Mol. Cell 8, 995-1004 [DOI] [PubMed] [Google Scholar]
- Peters J. M. (2006). The anaphase promoting complex/cyclosome: a machine designed to destroy. Nat. Rev. Mol. Cell Biol. 7, 644-656 [DOI] [PubMed] [Google Scholar]
- Petroski M. D., Deshaies R. J. (2005). Function and regulation of cullin-RING ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 6, 9-20 [DOI] [PubMed] [Google Scholar]
- Petrucelli L., Dickson D., Kehoe K., Taylor J., Snyder H., Grover A., De Lucia M., McGowan E., Lewis J., Prihar G., et al. (2004). CHIP and Hsp70 regulate tau ubiquitination, degradation and aggregation. Hum. Mol. Genet. 13, 703-714 [DOI] [PubMed] [Google Scholar]
- Polekhina G., House C. M., Traficante N., Mackay J. P., Relaix F., Sassoon D. A., Parker M. W., Bowtell D. D. (2002). Siah ubiquitin ligase is structurally related to TRAF and modulates TNF-alpha signaling. Nat. Struct. Biol. 9, 68-75 [DOI] [PubMed] [Google Scholar]
- Pringa E., Martinez-Noel G., Muller U., Harbers K. (2001). Interaction of the ring finger-related U-box motif of a nuclear dot protein with ubiquitin-conjugating enzymes. J. Biol. Chem. 276, 19617-19623 [DOI] [PubMed] [Google Scholar]
- Quezada C. M., Hicks S. W., Galan J. E., Stebbins C. E. (2009). A family of Salmonella virulence factors functions as a distinct class of autoregulated E3 ubiquitin ligases. Proc. Natl. Acad. Sci. USA 106, 4864-4869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravid T., Hochstrasser M. (2007). Autoregulation of an E2 enzyme by ubiquitin-chain assembly on its catalytic residue. Nat. Cell Biol. 9, 422-427 [DOI] [PubMed] [Google Scholar]
- Rotin D., Kumar S. (2009). Physiological functions of the HECT family of ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 10, 398-409 [DOI] [PubMed] [Google Scholar]
- Rotin D., Staub O. (2011). Role of the ubiquitin system in regulating ion transport. Pflugers Arch. 461, 1-21 [DOI] [PubMed] [Google Scholar]
- Satijn D. P., Otte A. P. (1999). RING1 interacts with multiple Polycomb-group proteins and displays tumorigenic activity. Mol. Cell. Biol. 19, 57-68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schile A. J., Garcia-Fernandez M., Steller H. (2008). Regulation of apoptosis by XIAP ubiquitin-ligase activity. Genes Dev. 22, 2256-2266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp D. A., Kratowicz S. A., Sank M. J., George D. L. (1999). Stabilization of the MDM2 oncoprotein by interaction with the structurally related MDMX protein. J. Biol. Chem. 274, 38189-38196 [DOI] [PubMed] [Google Scholar]
- Shenoy S. K., Xiao K., Venkataramanan V., Snyder P. M., Freedman N. J., Weissman A. M. (2008). Nedd4 mediates agonist-dependent ubiquitination, lysosomal targeting, and degradation of the beta2-adrenergic receptor. J. Biol. Chem. 283, 22166-22176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu Y., Okuda-Shimizu Y., Hendershot L. M. (2010). Ubiquitylation of an ERAD substrate occurs on multiple types of amino acids. Mol. Cell 40, 917-926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimura H., Schwartz D., Gygi S. P., Kosik K. S. (2004). CHIP-Hsc70 complex ubiquitinates phosphorylated tau and enhances cell survival. J. Biol. Chem. 279, 4869-4876 [DOI] [PubMed] [Google Scholar]
- Shmueli A., Tsai Y. C., Yang M., Braun M. A., Weissman A. M. (2009). Targeting of gp78 for ubiquitin-mediated proteasomal degradation by Hrd1: cross-talk between E3s in the endoplasmic reticulum. Biochem. Biophys. Res. Commun. 390, 758-762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silke J., Kratina T., Chu D., Ekert P. G., Day C. L., Pakusch M., Huang D. C., Vaux D. L. (2005). Determination of cell survival by RING-mediated regulation of inhibitor of apoptosis (IAP) protein abundance. Proc. Natl. Acad. Sci. USA 102, 16182-16187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons A. M., Horwitz A. A., Starita L. M., Griffin K., Williams R. S., Glover J. N., Parvin J. D. (2006). BRCA1 DNA-binding activity is stimulated by BARD1. Cancer Res. 66, 2012-2018 [DOI] [PubMed] [Google Scholar]
- Stagg H. R., Thomas M., van den Boomen D., Wiertz E. J., Drabkin H. A., Gemmill R. M., Lehner P. J. (2009). The TRC8 E3 ligase ubiquitinates MHC class I molecules before dislocation from the ER. J. Cell Biol. 186, 685-692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staub O., Abriel H., Plant P., Ishikawa T., Kanelis V., Saleki R., Horisberger J. D., Schild L., Rotin D. (2000). Regulation of the epithelial Na+ channel by Nedd4 and ubiquitination. Kidney Int. 57, 809-815 [DOI] [PubMed] [Google Scholar]
- Suzuki Y., Nakabayashi Y., Takahashi R. (2001). Ubiquitin-protein ligase activity of X-linked inhibitor of apoptosis protein promotes proteasomal degradation of caspase-3 and enhances its anti-apoptotic effect in Fas-induced cell death. Proc. Natl. Acad. Sci. USA 98, 8662-8667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaminathan G., Tsygankov A. Y. (2006). The Cbl family proteins: ring leaders in regulation of cell signaling. J. Cell Physiol. 209, 21-43 [DOI] [PubMed] [Google Scholar]
- Swanson R., Locher M., Hochstrasser M. (2001). A conserved ubiquitin ligase of the nuclear envelope/endoplasmic reticulum that functions in both ER-associated and Matalpha2 repressor degradation. Genes Dev. 15, 2660-2674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tait S. W., de Vries E., Maas C., Keller A. M., D'Santos C. S., Borst J. (2007). Apoptosis induction by Bid requires unconventional ubiquitination and degradation of its N-terminal fragment. J. Cell Biol. 179, 1453-1466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tcherpakov M., Delaunay A., Toth J., Kadoya T., Petroski M. D., Ronai Z. A. (2009). Regulation of endoplasmic reticulum-associated degradation by RNF5-dependent ubiquitination of JNK-associated membrane protein (JAMP). J. Biol. Chem. 284, 12099-12109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tetzlaff J. E., Putcha P., Outeiro T. F., Ivanov A., Berezovska O., Hyman B. T., McLean P. J. (2008). CHIP targets toxic alpha-Synuclein oligomers for degradation. J. Biol. Chem. 283, 17962-17968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai Y. C., Mendoza A., Mariano J. M., Zhou M., Kostova Z., Chen B., Veenstra T., Hewitt S. M., Helman L. J., Khanna C., et al. (2007). The ubiquitin ligase gp78 promotes sarcoma metastasis by targeting KAI1 for degradation. Nat. Med. 13, 1504-1509 [DOI] [PubMed] [Google Scholar]
- Verdecia M. A., Joazeiro C. A., Wells N. J., Ferrer J. L., Bowman M. E., Hunter T., Noel J. P. (2003). Conformational flexibility underlies ubiquitin ligation mediated by the WWP1 HECT domain E3 ligase. Mol. Cell 11, 249-259 [DOI] [PubMed] [Google Scholar]
- Verhagen A. M., Ekert P. G., Pakusch M., Silke J., Connolly L. M., Reid G. E., Moritz R. L., Simpson R. J., Vaux D. L. (2000). Identification of DIABLO, a mammalian protein that promotes apoptosis by binding to and antagonizing IAP proteins. Cell 102, 43-53 [DOI] [PubMed] [Google Scholar]
- Vierstra R. D. (2009). The ubiquitin-26S proteasome system at the nexus of plant biology. Nat. Rev. Mol. Cell Biol. 10, 385-397 [DOI] [PubMed] [Google Scholar]
- Vijay-Kumar S., Bugg C. E., Cook W. J. (1987). Structure of ubiquitin refined at 1.8 A resolution. J. Mol. Biol. 194, 531-544 [DOI] [PubMed] [Google Scholar]
- Vucic D., Dixit V. M., Wertz I. E. (2011). Ubiquitylation in apoptosis: a post-translational modification at the edge of life and death. Nat. Rev. Mol. Cell Biol. 12, 439-452 [DOI] [PubMed] [Google Scholar]
- Wade M., Wang Y. V., Wahl G. M. (2010). The p53 orchestra: Mdm2 and Mdmx set the tone. Trends Cell Biol. 20, 299-309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Pickart C. M. (2005). Different HECT domain ubiquitin ligases employ distinct mechanisms of polyubiquitin chain synthesis. EMBO J. 24, 4324-4333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Herr R. A., Chua W. J., Lybarger L., Wiertz E. J., Hansen T. H. (2007). Ubiquitination of serine, threonine, or lysine residues on the cytoplasmic tail can induce ERAD of MHC-I by viral E3 ligase mK3. J. Cell Biol. 177, 613-624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissman A., Shabek N., Ciechanover A. (2011). The predator becomes the prey: regulating the ubiquitin system by ubiquitylation and degradation. Nat. Rev. Mol. Cell Biol. 12, 605-620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenzel D. M., Lissounov A., Brzovic P. S., Klevit R. E. (2011). UBCH7 reactivity profile reveals parkin and HHARI to be RING/HECT hybrids. Nature 474, 105-108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wertz I. E., O'Rourke K. M., Zhou H., Eby M., Aravind L., Seshagiri S., Wu P., Wiesmann C., Baker R., Boone D. L., et al. (2004). De-ubiquitination and ubiquitin ligase domains of A20 downregulate NF-kappaB signalling. Nature 430, 694-699 [DOI] [PubMed] [Google Scholar]
- Williams C., van den Berg M., Sprenger R. R., Distel B. (2007). A conserved cysteine is essential for Pex4p-dependent ubiquitination of the peroxisomal import receptor Pex5p. J. Biol. Chem. 282, 22534-22543 [DOI] [PubMed] [Google Scholar]
- Williams R. L., Urbe S. (2007). The emerging shape of the ESCRT machinery. Nat. Rev. Mol. Cell Biol. 8, 355-368 [DOI] [PubMed] [Google Scholar]
- Wu L. C., Wang Z. W., Tsan J. T., Spillman M. A., Phung A., Xu X. L., Yang M. C., Hwang L. Y., Bowcock A. M., Baer R. (1996). Identification of a RING protein that can interact in vivo with the BRCA1 gene product. Nat. Genet. 14, 430-440 [DOI] [PubMed] [Google Scholar]
- Yamasaki S., Yagishita N., Sasaki T., Nakazawa M., Kato Y., Yamadera T., Bae E., Toriyama S., Ikeda R., Zhang L., et al. (2007). Cytoplasmic destruction of p53 by the endoplasmic reticulum-resident ubiquitin ligase ‘Synoviolin’. EMBO J. 26, 113-122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Fang S., Jensen J. P., Weissman A. M., Ashwell J. D. (2000). Ubiquitin protein ligase activity of IAPs and their degradation in proteasomes in response to apoptotic stimuli. Science 288, 874-877 [DOI] [PubMed] [Google Scholar]
- Younger J. M., Chen L., Ren H. Y., Rosser M. F., Turnbull E. L., Fan C. Y., Patterson C., Cyr D. M. (2006). Sequential quality-control checkpoints triage misfolded cystic fibrosis transmembrane conductance regulator. Cell 126, 571-582 [DOI] [PubMed] [Google Scholar]
- Yu F., White S. B., Zhao Q., Lee F. S. (2001). HIF-1alpha binding to VHL is regulated by stimulus-sensitive proline hydroxylation. Proc. Natl. Acad. Sci. USA 98, 9630-9635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavacki A. M., Arrojo E. D. R., Freitas B. C., Chung M., Harney J. W., Egri P., Wittmann G., Fekete C., Gereben B., Bianco A. C. (2009). The E3 ubiquitin ligase TEB4 mediates degradation of type 2 iodothyronine deiodinase. Mol. Cell. Biol. 29, 5339-5347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M., Windheim M., Roe S. M., Peggie M., Cohen P., Prodromou C., Pearl L. H. (2005). Chaperoned ubiquitylation-crystal structures of the CHIP U box E3 ubiquitin ligase and a CHIP-Ubc13-Uev1a complex. Mol. Cell 20, 525-538 [DOI] [PubMed] [Google Scholar]
- Zheng N., Wang P., Jeffrey P. D., Pavletich N. P. (2000). Structure of a c-Cbl-UbcH7 complex: RING domain function in ubiquitin-protein ligases. Cell 102, 533-539 [DOI] [PubMed] [Google Scholar]
- Zhong X., Shen Y., Ballar P., Apostolou A., Agami R., Fang S. (2004). AAA ATPase p97/valosin-containing protein interacts with gp78, a ubiquitin ligase for endoplasmic reticulum-associated degradation. J. Biol. Chem. 279, 45676-45684 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.