Understanding the second half of the genetic code, i.e., protein folding, is not only of immense importance for using the results of genome sequencing efforts, but also is required to understand how a subset of human proteins undergoes the conformational changes that render them pathogenic (1–11). The amyloid and prion diseases appear to result from the conversion of one of about 20 normally soluble and functional proteins into a β-sheet-rich quaternary structure that is often fibrilar (2, 5, 6, 9, 12–15). This conversion likely occurs in the partially denaturing environment of a cellular compartment such as a lysosome, where the lower pH (or otherwise denaturing) environment effects the conformational changes that facilitate amyloid and prion self-assembly (3, 5). This process does not challenge Christian Anfinsen’s hypothesis and demonstration that a proteins amino acid sequence is a strong determinant in specifying its fold, but it does enforce the point made by Anfinsen and by many others that the exact aqueous environment (pH, temperature, ionic strength, presence of chaotropic agents) also strongly influences the conformation adopted by the polypeptide (5). The mechanistic role that amyloid fibrils play in human amyloid disease is receiving attention from numerous groups. What is clear is that there is an overwhelming amount of evidence to support the cause and effect relationship in amyloid disease, but the amyloid hypothesis has not yet been proven (5, 10, 16–22). At the moment, it isn’t clear whether the neuropathology observed results from soluble or insoluble fibril-like assemblies operating from within the cell or from the extracellular deposits seen at autopsy. Prion diseases are even more remarkable in that the prion protein (Prp) scrapie isoform (PrPsc) deposits in an animal or human that facilitate neurodegeneration can be transmitted with species barriers that are reasonably stringent (1, 2, 7, 23–26). These strains or species barriers appear to result from variability in prion quaternary structures as discerned from their differing protease sensitivity (1, 2, 25, 26). The idea that a protein-only infectious particle exists and is responsible for prion diseases such as mad cow disease last year earned Stan Prusiner the Nobel Prize for his experiments, which strongly support this hypothesis (1, 3). Prion diseases occur sporadically with low frequency, appear with higher frequency in individuals possessing a susceptibility mutation, and can be transmitted by eating tissue harboring prion deposits. It now is believed that bovine prions can cross the species barrier with very low frequency and infect humans. Probably the most convincing experiment in favor of the prion hypothesis is that rodents with the cellular PrP isoform (PrPc) knocked out are not susceptible to prion infection (1, 3). Transmissibility presumably is caused by the infectious PrP quaternary structure recruiting more cellular protein into the prion quaternary form (2, 7). Treatments that selectively inactivate RNA and DNA do not change the infectivity of the prion particle, yet protein selective degradative procedures do reduce infectivity. Donne et al. (36) provide critical structural information on the likely cellular form of PrPc, which in the context of the other structural data suggests that the exact PrPc C-terminal structure is environmentally dependent and that the N-terminal portion of PrPc is largely unfolded.
Three different NMR groups recently have reported solution structural information on different constructs of the putative cellular form of the PrP. Although these structures are similar, they differ in detail, which is interesting and most likely demonstrates the subtle dependency that the prion sequence has on the exact aqueous environment in which the structures were recorded. The PrP, encoded by a single chromosomal gene, is composed of approximately 250 residues, which is proteolytically processed to remove the 22-residue N-terminal signal peptide and 23 C-terminal amino acids after addition of the glycosylphosphatidylinositol anchor to Ser-231 (1, 3). The processed protein contains a 179–214 disulfide crosslink and the glycosylation sites at Asn-181 and Asn-197. The protease resistant core of the PrP is composed of residues 90–231, which is sufficient to transmit infectivity. The Glockshuber and Wuthrich laboratories collaborated to report the structure of mouse PrPc (121–231), which exhibits three α-helices and two short antiparallel β-strands (Table 1) (27). Tom James, in collaboration with the Prusiner and Cohen laboratories, solved the structure of PrPc(90–231) of Syrian hamster, which contains three α-helices and a short irregular two-stranded sheet, the helices being longer than those observed in PrPc(121–231) (Table 1) (28). The N-terminal octarepeat region (PQ/HGGG(G/-)WGQ)5 missing from both of these structures is known to be associated with inherited prion disease. Hence, the Dyson, Wright, Prusiner, and Cohen laboratories have solved the structure of Syrian hamster PrPc(29–231), representing the entire PrP, including the octarepeats (Table 1). The most interesting feature of their structure is that almost half of the protein, residues 29–124, appear to be in a random coil conformation based on chemical shift indices and negative heteronuclear 1H-15N nuclear Overhauser effects (NOEs).
Table 1.
The residues of PrP found in secondary structural elements are construct and environment dependent
| PrPc construct | Helix A | Helix B | Helix C | B1 | B2 | Buffer pH |
|---|---|---|---|---|---|---|
| (29–231) | 144–156 | 172–193 | 200–227 | ?137–140? | ?160–163? | 5.2 |
| (23–231) | 144–155 | 175–193 | 200–219 | 128–131 | 161–164 | 4.5 |
| (90–231) | 144–156 | 172–193 | 200–227 | 128–131 | 161–164 | 5.2 |
| (121–231) | 144–154 | 170–193 | 200–217 | 128–131 | 161–164 | 4.5 |
The C terminus contains three α-helices and possibly two short β-strands based on chemical shift indexes. The helical secondary structure is confirmed by medium-range NOEs and supported by continuous NN NOEs, however, the strand or sheet structures are not clearly indicated by the expected NOE patterns. Interestingly, chemical shift differences suggest that the flexible N terminus is interacting with residues 187–193 in the B helix of PrPc(29–231). Wuthrich and Glockshuber et al. (29) also recently reported the structure of PrPc(23–231), finding that the N-terminal region (23–121) is largely disordered whereas the previously determined structure of PrPc(121–231) is preserved in the C terminus of PrPc (Table 1). One thus can conclude from the four relevant NMR structures that Prpc adopts a disulfide linked three-helix bundle with arguably some environmentally dependent β-sheet structure present. It is important to remember that the PrPc proteins studied thus far are not glycosylated. One can’t help but wonder whether glycosylating PrPc would further strengthen the apparently weak interactions between the B-helix portion and the C terminus reported by Donne et al. (36) and impart some order in the N terminus. However, they point out that the disordered N terminus could prove to be important in lowering the activation barrier for the conversion of PrPc into Prpsc. A partially unfolded intermediate has been suggested previously for the conversion of PrPc to PrPsc (2, 5, 7, 15). Unfortunately, it has not yet proven possible to convert PrPc into infectious Prpsc in vitro despite significant effort. Hence at the present time it is not possible to test this hypothesis, but it is observed that residues 90–121, which are largely disordered in the structure of PrPc, become protease resistant in PrPsc. The inability to convert PrPc into an infectious quaternary structure suggests that the attached carbohydrate may be required for the PrPc to PrPsc conversion. Of course it is also possible that the appropriate conditions for this conversion in the absence of carbohydrate have not yet been found. Nonetheless, the importance of carbohydrate in the PrPc to PrPsc tertiary and quaternary structure conversion needs to be evaluated. Both PrPc and PrPsc have a glycosylphosphatidylinositol anchor at the C terminus and are glycosylated at both Asn-181 and Asn-197.
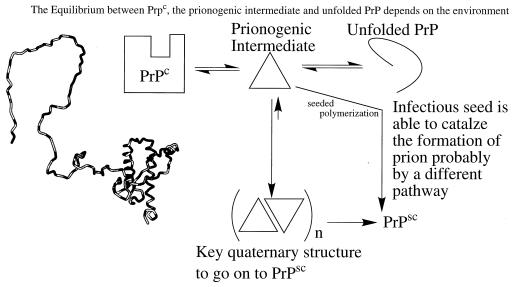
Interestingly, James and coworkers (28) report that a weak dimerization of PrPc(90–231) can occur according to analytical equilibrium ultracentrifugation studies. Such an assembly process could be very important in converting PrPc into the prionogenic state by linked tertiary and quaternary structural changes (Fig. 1). Alternatively it may be necessary to partially unfold the C-terminal half of PrPc to form the prionogenic intermediate as in the conversion of transthyretin to amyloid fibrils or nearly completely unfold PrPc as in the conversion of the lysozyme variants into amyloid (Fig. 1; refs. 5, 9, 12, and 20). Little is known about the structural details of the prion particle other than it is richer in β-sheet structure than PrPc. Interestingly, PrPsc can adopt a fibrilar structure similar to amyloid, implying the two structures could be similar (1, 3, 6, 30). Unfortunately, we are just beginning to understand some of the features of the amyloid structure so this isn’t much help (6, 31–34). It seems likely that the different quaternary structures that appear to responsible for strain specificity in prions will have similar structures, perhaps with differing numbers of protofilaments making up the particle as in amyloid deposits. The carbohydrate accessibility and conformation also may prove important in determining species barriers for infectivity (strains).
Figure 1.
Possible pathways for the formation of PrPsc in the sporadic case and in the seeded infectious case, based on the presence of an intermediate that is critical for prion formation.
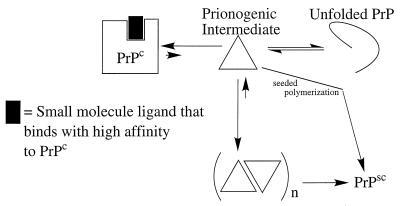
The structural information available from the Dyson, James, and Wuthrich laboratories should prove to be very important for developing a therapeutic strategy for prion diseases (Fig. 2). An approach developed by the Kelly laboratory for preventing transthyretin amyloid fibril formation should apply to the prion diseases as well (35). This strategy uses the structure of the normally folded form of transthyretin to design high-affinity ligands to this fold, thus making it very difficult for the protein-small molecule complex to undergo the quaternary and tertiary structural changes required for amyloid fibril formation. With four PrPc structures now in hand, it should be possible to design ligands that stabilize the C-terminal helical structure and make it very difficult to convert the PrPc structure into PrPsc (Fig. 2). Alternatively, it may be possible to design ligands that bind with high affinity to the octarepeats and thus make the conversion of the N terminus of PrPc into the prion structure difficult or impossible. Another approach is to design peptidomimetics that prevent the assembly of the prionogenic intermediate into a prion particle. An analogy can be made here with Alzheimer’s disease peptidomimetics, i.e., Phe-rich peptidomimetics, which have been shown to be effective in preventing the assembly of the β-peptide into amyloid fibrils. Apart from the obvious therapeutic benefits, compounds that prevent prion particle formation will be very important in further demonstrating the validity of the prion hypothesis that earned Stan Prusiner the Nobel Prize in medicine.
Figure 2.
Prion formation can be inhibited either by ligand binding to PrPc, which shifts the equilibrium toward Prpc under cellular conditions that normally would form the prionogenic intermediate (Upper; also see ref. 35), or by using peptidomimetics that bind to the prionogenic intermediate or other intermediates that precede prion formation (not shown).
Footnotes
The companion to this commentary is published on page 13452 in issue 25 of volume 94.
References
- 1.Prusiner S B. Science. 1997;278:245–251. doi: 10.1126/science.278.5336.245. [DOI] [PubMed] [Google Scholar]
- 2.Huang Z, Prusiner S B, Cohen F E. Curr Top Microbiol Immunol. 1996;207:49–67. doi: 10.1007/978-3-642-60983-1_5. [DOI] [PubMed] [Google Scholar]
- 3.Prusiner S B. Trends Biochem Sci. 1996;21:482–487. doi: 10.1016/s0968-0004(96)10063-3. [DOI] [PubMed] [Google Scholar]
- 4.Kelly J W, Lansbury P T J. Amyloid Int J Exp Clin Invest. 1994;1:186–205. [Google Scholar]
- 5.Kelly J W. Curr Opin Struct Biol. 1996;6:11–17. doi: 10.1016/s0959-440x(96)80089-3. [DOI] [PubMed] [Google Scholar]
- 6.Kelly J W. Structure. 1997;5:595–600. doi: 10.1016/s0969-2126(97)00215-3. [DOI] [PubMed] [Google Scholar]
- 7.Caughey B, Kocisko D A, Raymond G J, Lansbury P T. Chem Biol. 1995;2:807–817. doi: 10.1016/1074-5521(95)90087-x. [DOI] [PubMed] [Google Scholar]
- 8.Lansbury P T. Chem Biol. 1995;2:1–5. doi: 10.1016/1074-5521(95)90074-8. [DOI] [PubMed] [Google Scholar]
- 9.Colon W, Kelly J W. Biochemistry. 1992;31:8654–8660. doi: 10.1021/bi00151a036. [DOI] [PubMed] [Google Scholar]
- 10.Lai Z, Colon W, Kelly J W. Biochemistry. 1995;35:6470–6482. doi: 10.1021/bi952501g. [DOI] [PubMed] [Google Scholar]
- 11.Mihara H, Takahashi Y. Curr Opin Struct Biol. 1997;7:501–508. doi: 10.1016/s0959-440x(97)80113-3. [DOI] [PubMed] [Google Scholar]
- 12.Colon W, Kelly J W. In: Applications of Enzyme Biotechnology. Kelly J W, Balwin T O, editors. New York: Plenum; 1991. pp. 99–108. [Google Scholar]
- 13.Wetzel R. Trends Biotech. 1994;12:193–198. doi: 10.1016/0167-7799(94)90082-5. [DOI] [PubMed] [Google Scholar]
- 14.Wetzel R. Cell. 1996;86:699–702. doi: 10.1016/s0092-8674(00)80143-9. [DOI] [PubMed] [Google Scholar]
- 15.Safar J. Microbiol Immunol. 1996;207:69–76. doi: 10.1007/978-3-642-60983-1_6. [DOI] [PubMed] [Google Scholar]
- 16.Selkoe D J. Science. 1997;275:630–631. doi: 10.1126/science.275.5300.630. [DOI] [PubMed] [Google Scholar]
- 17.Selkoe D J. J Biol Chem. 1996;271:18295–18298. doi: 10.1074/jbc.271.31.18295. [DOI] [PubMed] [Google Scholar]
- 18.Selkoe D J. Nature (London) 1995;375:734–735. doi: 10.1038/375734a0. [DOI] [PubMed] [Google Scholar]
- 19.Selkoe D J. Annu Rev Cell Biol. 1994;10:373–403. doi: 10.1146/annurev.cb.10.110194.002105. [DOI] [PubMed] [Google Scholar]
- 20.Booth D R, Sunde M, Bellotti V, Robinson C V, Hutchinson W L, Fraser P E, Hawkins P N, Dobson C M, Radford S E, Blake C C F, Pepys M B. Nature (London) 1997;385:787–793. doi: 10.1038/385787a0. [DOI] [PubMed] [Google Scholar]
- 21.Hurle M R, Helms L R, Li L, Chan W, Wetzel R. Proc Natl Acad Sci USA. 1994;91:5446–5450. doi: 10.1073/pnas.91.12.5446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jarrett J, Lansbury P T. Cell. 1993;73:1055–1058. doi: 10.1016/0092-8674(93)90635-4. [DOI] [PubMed] [Google Scholar]
- 23.Prusiner S B. Science. 1991;252:1515–1522. doi: 10.1126/science.1675487. [DOI] [PubMed] [Google Scholar]
- 24.Kocisko D A, Come J H, Priola S A, Chesebro B, Raymond G J, Lansbury P T, Caughey B. Nature (London) 1994;370:471–474. doi: 10.1038/370471a0. [DOI] [PubMed] [Google Scholar]
- 25.Kocisko D A, Priola S A, Raymond G J, Chesebro B, Lansbury P T, Jr, Caughey B. Proc Natl Acad Sci USA. 1995;92:3923–3927. doi: 10.1073/pnas.92.9.3923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bessen R A, Kocisko D A, Raymond G J, Nandan S, Lansbury P T, Caughey B. Nature (London) 1995;375:698–700. doi: 10.1038/375698a0. [DOI] [PubMed] [Google Scholar]
- 27.Riek R, Hornemann S, Wider G, Billeter M, Glockshuber R, Wuthrich K. Nature (London) 1996;382:180–182. doi: 10.1038/382180a0. [DOI] [PubMed] [Google Scholar]
- 28.James T L, Liu H, Ulyanov N B, Farr-Jones S, Zhang H, Donne D G, Kaneko K, Groth D, Mehlhorn I, Prusiner S B, Cohen F E. Proc Natl Acad Sci USA. 1997;94:10086–10091. doi: 10.1073/pnas.94.19.10086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Riek R, Hornemann S, Wider G, Glockshuber R, Wuthrich K. FEBS Lett. 1997;413:282–288. doi: 10.1016/s0014-5793(97)00920-4. [DOI] [PubMed] [Google Scholar]
- 30.Lansbury P T. Biochemistry. 1992;31:6865–6870. doi: 10.1021/bi00145a001. [DOI] [PubMed] [Google Scholar]
- 31.Blake C, Serpell L. Structure. 1996;4:989–998. doi: 10.1016/s0969-2126(96)00104-9. [DOI] [PubMed] [Google Scholar]
- 32.Blake C C F, Serpell L C, Sunde M, Sandgren O, Lundgren E. In: The Nature and Origin Of Amyloid Fibrils. Bock G R, editor. Vol. 199. New York: Wiley; 1996. , Ciba Symposium, pp. 6–21. [Google Scholar]
- 33.Serpell L C, Sunde M, Fraser P E, Luther P K, Morris E P, Sandgren O, Lundgren E, Blake C C F. J Mol Biol. 1995;254:113–118. doi: 10.1006/jmbi.1995.0604. [DOI] [PubMed] [Google Scholar]
- 34.Inouye H, Fraser P E, Kirschner D A. Biophys J. 1993;64:502–519. doi: 10.1016/S0006-3495(93)81393-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miroy G J, Lai Z, Lashuel H, Peterson S A, Strang C, Kelly J W. Proc Natl Acad Sci USA. 1996;93:15051–15056. doi: 10.1073/pnas.93.26.15051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Donne D G, Viles J H, Groth D, Mehlhorn I, James T L, Cohen F E, Prusiner S B, Wright P E, Dyson H J. Proc Natl Acad Sci USA. 1997;94:13452–13457. doi: 10.1073/pnas.94.25.13452. [DOI] [PMC free article] [PubMed] [Google Scholar]