Abstract
Patients with systemic lupus erythematosus (SLE) show an over-expression of Type I Interferon (IFN) responsive genes called “Interferon Signature”. We found that the B6.NZMSle1/Sle2/Sle3 (Sle1,2,3) lupus-prone mice also express an Interferon Signature compared to non autoimmune C57BL/6 mice. In vitro, myeloid dendritic cells (mDCs)(GM-CSF bone marrow-derived BMDCs) from Sle1,2,3 mice constitutively over-expressed IFN responsive genes such as IFNb, Oas-3, Mx-1, ISG-15 and CXCL10, and the members of IFN signaling pathway STAT1, STAT2, and IRF7. The Interferon Signature was similar in Sle1,2,3 BMDCs from young, pre-autoimmune mice and from mice with high titers of autoantibodies, suggesting that the Interferon Signature in mDCs precedes disease onset and it is independent from the autoantibodies. Sle1,2,3 BMDCs hyper-responded to stimulation with IFNa and the TLR7 and TLR9 agonists R848 and CpGs. We propose that this hyper-response is induced by the Interferon Signature and only partially contributes to the Signature, since oligonucleotides inhibitory for TLR7 and TLR9 only partially suppressed the constitutive Interferon Signature and pre-exposure to IFNa induced the same hyper-response in wild type BMDCs than in Sle1,2,3 BMDCs. In vivo, mDCs and with lesser extent T and B cells from young pre-diseased Sle1,2,3 mice also expressed the Interferon Signature, although they lacked the strength that BMDCs showed in vitro. Sle1,2,3 plasmacytoid DCs expressed the Interferon Signature in vitro but not in vivo, suggesting that mDCs may be more relevant before disease onset. We propose that Sle1,2,3 mice are useful tools to study the role of the Interferon Signature in lupus pathogenesis.
Keywords: Myeloid Dendritic cells, Type I Interferon, systemic lupus erythematosus, TLR, gene expression
Introduction
Several reports support an important role for Type I Interferons (IFNs) (1) in the pathogenesis of the autoimmune disease systemic lupus erythematosus (SLE) (2). Type I IFNs are a family of cytokines, pivotal in the activation of the innate (3) and adaptive immune systems (4-6) and in the antiviral response (7).
It is has been known for a long time that many SLE patients have high serum levels of IFN alpha (IFNa)(8, 9) (10). Several groups have found a pattern of elevated expression of IFN-responsive genes in peripheral blood cells of SLE patients, as measured by microarray analysis, now known as “Interferon Signature” (11-13). Since this Interferon Signature is frequent in pediatric patients (12) and in adults with central nervous system involvement and nephritis (13), it has been associated with the early and most acute phases of the disease (14, 15).
Animal models also support a prominent role for Type I IFNs in the initiation of autoimmunity in lupus. The administration of IFNa or inducers of Type I IFNs like polyI:C, accelerated disease onset in polygenic lupus prone mice (16-19), mirroring the sporadic development of lupus in patients treated with IFNa (20). Lupus prone mice, made resistant to Type I IFNs by the genetic deletion of the receptor common to all the Type I IFNs (IFNAR), had milder disease in four out of five reports. This was true for the lupus prone mice NZB (21), C57BL/lpr (22), (B6.Nba2 × NZW)F1 (23) and NZM2328 (17). In contrast, the MRL/lpr mice deficient in IFNAR showed an aggravation of the disease (24), although the group of T.W. Behrens reported that MRL/lpr have an increased expression of Type I IFN responsive genes (25). In a model of chemically induced lupus, following administration of pristane, IFNAR deficient mice failed to develop autoimmunity (26) and wild type mice exhibited an Interferon Signature just before disease onset (27), indicating the importance of Type I IFNs in the initiation of chemically-induced lupus. It remains to be demonstrated whether an Interferon Signature is expressed by all the genetically determined models of murine lupus.
The Interferon Signature in SLE patients has a complex etiology, in which genetic and environmental factors play a role (28). The current view considers the high levels of Type I IFNs a complex inheritable disease risk factor that can be triggered/amplified by the lupus-associated autoantibodies (29). Indeed, genetic studies in SLE patients have identified gene variants in pathways connected to Type I IFNs that result in the excessive release of IFNa (30). For example, polymorphisms in IRF5, IRF7 and STAT4, which are part of the signaling pathway of Type I IFNs, are associated with a higher risk of developing SLE (31)(32)(33)(34-36). In animal models, knocking out the genes STAT4 and IRAK-1, which were highlighted by GWAS in SLE patients, decreased disease severity in lupus prone mice (37, 38) and some IFN-related genes, such as Ifi202 (39), are located in susceptibility loci of polygenic lupus prone mice.
The levels of Type I IFNs determined by this genetic make-up can be up-regulated by the activation of TLR7 and TLR9 (40, 41) triggered by nucleic acids coming from recurrent viral infections (42), endogenous retroviruses (43) or from badly scavenged apoptotic cells (44). A duplication of TLR7 accelerated lupus in BXSB-Yaa male mice (45); treatment with TLR7 and TLR9 inhibitors ameliorated severity (46) and TLR7 deficiency inhibited disease development in lupus prone mice (47). These reports suggest that TLR7/9 are involved in lupus development, possibly as an inducer of Type I IFNs.
TLR7/9 can also be triggered by immune complexes (IC) containing nucleic acids and anti-DNA and anti-ribonucleoprotein antibodies, the immunological hallmarks of lupus. Indeed, ICs can bind FcgRIIa on the surface of immune cells and shuttle the nucleic acids to the endosomal compartment containing TLR7/9, leading to induction of Type I IFNs (41, 48)(51). A novel mechanism of Type I IFN production involves the release of neutrophil extracellular traps (NETs): normally a way to kill and trap pathogen (49), in lupus patients NETs stimulate IFNa production by plasmacytoid dendritic cells (pDCs) via TLR9 triggering (50, 51).
Although current advances in lupus pathogenesis have provided many insights in the genetic aspects and progression of the disease, there is yet an incomplete understanding of the cellular sources and the triggers of the Interferon Signature in SLE. The identification of a mouse model of polygenic lupus that expresses the Interferon Signature would be an important tool to perform mechanistic studies and test novel therapeutic approaches. With this goal, we investigated the lupus prone mice B6.NZM Sle1/Sle2/Sle3 (Sle1,2,3) (52). Sle1,2,3 mice are congenic mice in which three susceptibility loci from NZM2410 lupus prone mice were introgressed into the non-autoimmune mice C57BL/6. These mice spontaneously develop a form of lupus characterized by high titers of autoantibodies against double stranded DNA and chromatin, and by the development of glomerulonephritis (53, 54). We found that Sle1,2,3 mice express an Interferon Signature in vivo before the onset of the disease, compared to gender- and age-matched non autoimmune C57BL/6 (B6) mice. To determine the original cellular source of this IFN response, we investigated the dendritic cells (DCs) because of their pivotal role in lupus (55, 56), in which they are abnormally activated (57) and are able to induce autoimmunity (58), and also their ability to activate upon Type I IFNs (3) and produce large amounts of IFNa/b (59). We investigated the expression of Type I IFNs and IFN responsive genes in Sle1,2,3 bone marrow-derived myeloid DCs (mDCs) and plasmacytoid DCs (pDCs) in young pre-diseased mice, in absence of autoantibodies, and we measured Sle1,2,3 BMDCs’ response to stimulation with IFNa and TLR7 and TLR9 agonists and inhibitors. In summary, our results demonstrate that Sle1,2,3 lupus prone mice express an Interferon Signature, similar to SLE patients and mDCs are one important source of the Interferon Signature. We propose that Sle1,2,3 mice are useful tools to study the role of the Interferon Signature in lupus pathogenesis.
Materials and Methods
Mice
C57BL/6 mice (Jackson Laboratory) and B6.NZM Sle1/Sle2/Sle3 (Sle1,2,3) (kind gift from Laurence Morel and later purchased from Jackson Laboratory) were bred and maintained in our colony in accordance with the guidelines of the Institutional Animal Care and Use Committees of the Children’s Hospital of Philadelphia and Temple University, both of which are American Association for the Accreditation of Laboratory Animal Care-accredited facilities. Female mice were used between 7 and 14 weeks of age with the exception of the experiments in which one week old pups were used.
Measurement of anti-DNA and anti-chromatin antibodies
Sera of young mice were tested for anti-DNA and anti-chromatin antibodies by ELISA as described previously (60). Briefly, the plates were coated with chicken erythrocyte-derived chromatin at 3 ug/ml, or with calf thymus-derived dsDNA at 2.5 ug/ml in borate-buffered saline (BBS). Following addition of blocking buffer (3% BSA and 1% Tween 80 in 1× BBS), serum samples diluted 1/250 in BBT (BBS, 0.4% Tween 80, 0.5% BSA) were added in duplicate and incubated overnight at 4°C. For the anti-dsDNA ELISA, plates were coated with poly(l-lysine) (1 ug/ml; Sigma-Aldrich) before coating with Ag. Alkaline phosphatase-conjugated goat anti-mouse IgG (Fcg specific; Jackson ImmunoResearch Laboratories) was used as secondary Ab. The plates were developed using 1 mg/ml paranitrophenyl phosphate substrate (Sigma- Aldrich) in 0.01 M diethanolamine (pH 9.8). To generate a standard curve for these assays, serum from an older MRL/lpr mouse with high titer of autoantibodies was also assayed at serial 2-fold dilutions from 1/250 to 1/128,000 (60). Pooled sera from 5 B6 mice that were individually negative for autoantibodies were used as negative controls.
In vitro mDC cultures
As a model of mDCs, bone marrow-derived dendritic cells (BMDCs) were generated as previously described (61). Briefly, BM precursors from young (between 7-14 wks) age-matched female Sle1,2,3 and B6 were negatively selected using anti-CD19, Thy 1.2 and MHC Class II magnetic beads (Miltenyi Biotech) to deplete B, T, macrophages and DCs that differentiated in vivo, and seeded at 1 × 106/ml in complete IMDM (10% FBS, penicillin/streptomycin, gentamicin, and beta-mercaptoethanol) enriched with 3.3 ng/ml GM-CSF (BD Biosciences) in 24-well plates. One milliliter of medium was added on day 2 and 1 ml of the medium was replaced on day 5 and subsequently each day until the culture was used (day 6 or 7). We also cultured BMDCs from 7 days old female mouse pups. The gender of these very young mice was determined by PCR using primers for the male specific gene Sry as described elsewhere (62). Bone marrow was flushed and cells were plated as 1×10^6 cells per ml per well in 24 well plate. Resting DC cultures were stimulated at day 6 or 7 of culture with each of the following: 100 ng/ml LPS (Sigma-Aldrich), 2500U/ml IFNa (HyCult Biotechnology), 10ug/ml CpG-B 1826 (synthesized from IDT biotechnologies) 1ug/ml R848 (Invivogen). When TLR 7/9 inhibitors (ODN 2088, ODN 954, ODN 661 (20ug/ml) (46), INH1, INH18 (10ug/ml)(63)) were used (synthesized from IDT biotechnologies), cells were first treated with the inhibitors for 30 minutes and then stimulated with CpG, R848 or medium alone. BMDCs were harvested after 5 h or 24h of stimulation for RNA analysis, and 24 h for FACS analysis of surface activation markers. Culture supernatants were collected at the same time points for cytokine analysis by ELISA.
In vitro pDC cultures
Bone marrow precursors were differentiated into pDC in medium containing Flt3L (64). Briefly, bone marrow cells were flushed and cells were plated as 5 × 10^6 cells/ well in a 6-well plate in complete RPMI medium containing 10%FBS, L-glutamine, Pencillin/Streptomycin, 2-mercaptoethanol and 7.5% supernatant from Flt3L cell line (kind gift from Dr. Terri Laufer, U Penn). Cells were harvested on day 8 or 9 and stained and sorted for B220+CD11c+ pDCs. RNA was extracted from the sorted cells and analyzed for the Interferon Signature.
Flow cytometry
BMDCs were harvested from the wells, washed in cold PBS, incubated with rat anti-mouse CD16/CD32 (clone 2.4G2) mAb for 10 min to block FcRs, and then stained for 30 min on ice with APC-conjugated hamster anti-mouse CD11c, PE-conjugated rat anti-mouse CD86, FITC conjugated mouse anti-mouse H2Kb, Alexa-488 conjugated rat anti-mouse B220 and PE-conjugated rat anti-mouse CD8a antibodies (BD Biosciences). Cells were fixed in 1% formaldehyde and analyzed on a FACS Canto cytometer (BD Biosciences). FlowJo software was used for data analysis.
Ex-vivo immune populations sorted from lymphoid organs
Bone marrow and spleen of young pre-diseased (negative for autoantibodies) Sle 1,2,3 lupus-prone female mice and age-gender matched B6 were processed for sorting by FACS to analyze the IFN gene signature in the different immune populations ex vivo. Briefly, spleens were incubated in medium containing 8mg/ml collagenase and 1000U/ml DNase for 45 min and single-cell suspensions were prepared by passing through a 100 micron mesh filter. Bone marrow cells were flushed from femur and tibia. Cells were stained for sorting with the following antibodies: Rat anti-mouse CD19 PercpCy5.5, CD3 FITC, CD11b PeCy7, B220 PE and hamster anti-mouse CD11c APC (BD Pharmingen and eBioscience). The immune populations were sorted as B cells (CD19 positive), T cells (CD3 positive), Macrophages (CD19−CD3−CD11b+CD11c−), mDCs (CD19−CD3−CD11c+CD11b+) and pDC (CD19−CD3−B220+CD11c+). RNA was extracted from these samples and analyzed by real-time RT-PCR for the Interferon Signature.
Quantitative RT-PCR
Gene expression in BMDCs was analyzed by real-time RT-PCR using Taqman probes. Briefly, RNA was extracted by Trizol method and repurified using Qiagen columns (Qiagen Inc. USA). cDNA was synthesized using the cDNA archive kit followed by a preamplification reaction (Applied Biosystems). Premade TaqMan primers and probes from Applied Biosystems were used to study the expression of seventeen IFN responsive genes (IFNb, IRF7, ISG-15, CXCL10, Oas-3, Mx-1, STAT1, STAT2, IFNa, IL-6, STAT4, IL-15, IFNAR1, IFNAR2, IRF3, IRF5, PKR). Cyclophilin was used as the reference gene for normalization. In each independent experiment, one BMDC culture generated from one Sle1,2,3 mouse was compared to one BMDC culture generated from one B6 mouse. The Ct method of relative quantification of gene expression was used for these TaqMan PCRs, and the normalized Ct values (against cyclophilin) were calibrated against the control sample (untreated C57BL/6 BMDCs) in each experiment (61). TLR7, TLR9 and TNF-alpha were analyzed by the same method.
For the analysis of the BMDCs generated from the 7 days old pups, 6 BMDC cultures were analyzed that had been generated from three Sle1,2,3 mice and three B6 mice: the average of the normalized Ct values against cyclophilin for three BMDCs from B6 pups was used to calculate the results of three BMDCs from three Sle1,2,3 pups. For the analysis of the Interferon Signature in the populations sorted ex vivo from bone marrows and spleens, normalized Cts (against cyclophilin housekeeping gene) from control B6 mice were averaged and used for normalization against each Sle tested.
ELISA
We used ELISA kits to measure the levels of TNF-a and CXCL10 (R&D Biosystems) in the supernatants of BMDC cultures stimulated for 24 hours with IFNa, TLR ligands or medium alone.
Western blot analysis
We performed Western blotting using 30–50 μg of total DC cell protein. In brief, protein samples were denatured by boiling for 5 minutes and loaded onto 10% Bis-Tris gels. After electrophoresis, proteins were transferred to nitrocellulose membranes. Membranes were blocked for 1 hour with blocking buffer (2% non-fat milk in PBS), then incubated overnight at 4°C with the primary antibodies diluted in blocking buffer with 0.1% Tween 20 simultaneously. We used rabbit polyclonal anti-STAT1 and STAT2, (Upstate Biotechnology). Mouse anti-GAPDH (Santa-Cruz Biotech) was used as a loading control. After incubating with primary antibodies, the membranes were washed with PBS containing 0.1% Tween 20 (PBST) three times. Then the membranes were incubated for 1 hour with IR Dye 800 goat anti-rabbit and IR Dye 680 goat anti-mouse (LI-COR Biosciences) diluted in blocking buffer plus 0.1% Tween 20. The blots were then washed three times with PBST and rinsed with PBS. Proteins were visualized by scanning the membrane on an Odyssey Infrared Imaging System (LI-COR Biosciences) in both 700nm and 800nm channels.
Statistical analysis
We analyzed the data using Prism software (GraphPad, San Diego) and used as statistical tests one sample t-test, two-tailed Student’s t test or nonparametric Mann-Whitney U test, as appropriate, for the different sets of experiments and considered significant values of p < 0.05 (marked in the figures as * p < 0.05; **p < 0.01; ***p < 0.001).
Results
Constitutive expression of the Interferon Signature in Sle1,2,3 BMDCs
Previous studies have demonstrated hyper-activated DCs from bone marrow of lupus patients (65). To determine whether DCs from lupus prone mice have an Interferon Signature, we cultured bone marrow precursors, depleted of mature T and B cells, macrophages, and dendritic cells, in the presence of GM-CSF to obtain mDCs in 6-7 days of culture (61). We grew BMDCs from young (7-14 weeks old) female Sle1,2,3 mice and compared them with BMDCs from gender and age matched B6 mice. We found that Sle1,2,3 BMDCs were not different from B6 BMDCs in terms of absolute numbers of cells and percentages of CD11c+ cells (60-80% of the culture) (Supplemental Fig. 1A, B). The analysis by Flow Cytometry of the major differentiation markers revealed that more than 95% of the BMDCs were CD11c+CD11b+ mDCs, while no CD8a or pDCs (CD11c+CD8a+ and CD11cint B220+, respectively) could be detected in the cultures from both strains (Supplemental Fig 1C), as expected because of the culture conditions.
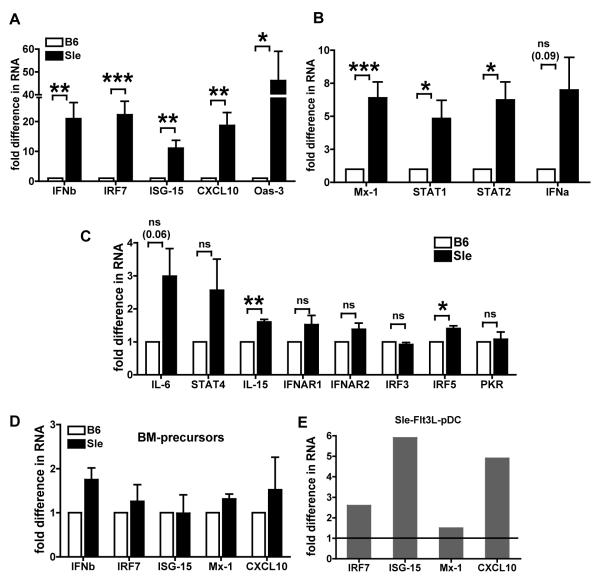
We analyzed by real time qRT-PCR a repertoire of 17 Type I IFN responsive genes selected because they represent the different kinds of responses triggered by Type I IFNs: the amplification of the IFN response itself, the anti-viral response and the immune activation. We found that the expression levels of some genes – IFNb, IRF7, ISG-15, CXCL10 and Oas-3 genes (Fig. 1A) were very high in Sle1,2,3 BMDCs (over 10 fold as compared to B6): these are genes that are induced by Type I IFNs as an autocrine activation (IFNb and IRF7) and are implicated in the innate anti-viral response and immune activation (ISG-15, CXCL10 and Oas-3 )(66). Other genes were moderately increased (Fig. 1B), like Mx-1, which is also part of the anti-viral response, and STAT1 and STAT2, which belong to the signaling pathway downstream of IFNAR and are up-regulated by Type I IFNs as part of their positive feedback loop. IL-15 and IRF5 showed only a very small, but statistically significant increase (< 2 fold) (Fig. 1C). There were no differences in IFNAR1, IFNAR2, IRF3 and PKR in the Sle1,2,3 vs. B6 BMDC cultures (Fig. 1C): these are important negative results to assure that the differences we are seeing are not due to non-specific alterations of the levels of RNAs but rather are specific for some IFN responsive genes. IFNa, IL-6 and STAT4 showed a trend toward increased levels in Sle1,2,3 BMDCs compared to B6 BMDCs but their differences did not reach statistical significance. These results indicate that Sle1,2,3 mDCs express an Interferon Signature similar to SLE patients. Furthermore, as the Signature occurred in BMDCs differentiated in vitro, severed from any pro-autoimmune environment in vivo, there is likely a causative mechanism intrinsic to the DCs driving this Signature.
FIGURE 1. Constitutive over-expression of IFN responsive genes in Sle1,2,3 (Sle) BMDCs.
We analyzed by real-time qRT-PCR a panel of seventeen genes as representative of the Interferon Signature in BMDCs at day 6-7 of culture in absence of any stimulation. The results are expressed as fold differences in RNA expression as compared to normal B6 BMDCs. (A) The gene expression levels were over 10 fold for some genes; (B) over 5 fold for some and (C) some showed very modest difference or no change. We calculated the statistical significance with the one sample t test. * is <0.05, ** is <0.01, *** is <0.001. Mean and SE are from a minimum of 4 experiments for some genes to a max of 14 expts for other genes. (D) We analyzed by real-time RT-PCR bone marrow precursors at day 0 of culture. The results are expressed as fold differences in gene expression from B6 controls, results are Mean and SE of 3 independent experiments. None of the values was significant at the one sample t test. (E) We cultured bone marrow cells from young age-matched B6 and Sle1,2,3 females in medium containing 7.5% Flt3L supernatant. We sorted the cells at day 8/9 of culture for B220+CD11c+ pDCs and analyzed the RNA by real-time qRT-PCR for Interferon Signature gene expression. Representative of one out of three independent cultures. The normalization from B6 is indicated by black line in the graph.
Bone marrow precursors do not express the Interferon Signature
We also analyzed the bone marrow precursors, depleted of mature T and B cells, macrophages, and dendritic cells, that we put in culture at day 0 to determine whether these cells already express the Interferon Signature. We analyzed the expression of a selected number of IFN responsive genes from the original 17 genes shown in Fig.1 (IFNb, IRF7, ISG15, Mx-1 and CXCL10) and found that Sle1,2,3 precursors expressed these genes at the same levels than the B6 precursors (Fig. 1D), suggesting that the IFN abnormalities develop together with DC differentiation.
Sle1,2,3 bone marrow-derived pDCs express the Interferon Signature
PDCs were originally called “natural IFN-producing cells” because they are the most potent producers of Type I IFNs I (59). In the context of SLE, pDC can activate and secrete IFNa upon FcR-mediated uptake of autoantibodies complexed to chromatin and RNPs (48). Here we asked whether pDCs from young Sle1,2,3 mice express an Interferon Signature. We generated bone marrow-derived pDCs in medium containing Flt3L from young B6 and Sle1,2,3 mice. The cultures yielded similar percentages of pDCs from both strains (data not shown). The sorted pDCs (B220+ CD11c+) from these cultures showed increased expression of IRF7, ISG15 and CXCL10 genes (Fig 1E), indicating that pDCs from Sle1,2,3 mice express the Interferon Signature like the mDCs do in absence of autoantibodies.
Constitutive activation of the IFN response in Sle1,2,3 BMDCs
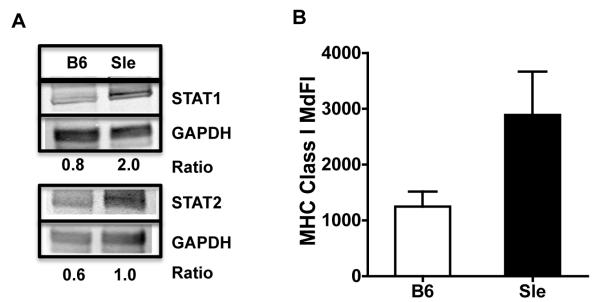
To confirm at the protein level some of the results found with real time qRT-PCR, we measured by Western Blot the constitutive expression of STAT1 and STAT2, the pivotal signaling molecules that form IRF9, the transcription factor complex that is responsible for the transcription of IFN responsive genes. We found that the protein levels of STAT1 and STAT2 were higher in the Sle1,2,3 BMDCs than in the B6 BMDCs as measured by Western Blot (Fig. 2A), indicating that Sle1,2,3 BMDCs have the IFN signaling pathway pre-activated in the absence of any experimental stimulation.
FIGURE 2. Constitutive over-expression of IFN responsive proteins in Sle1,2,3 BMDCs.
(A) We analyzed the total cell lysates from BMDCs by Western blot for STAT1 and STAT2 proteins using GAPDH as loading control; numbers represent the intensity values normalized with GAPDH; representative of one out of 4 experiments. (B) We analyzed by flow cytometry the Median Fluorescence Intensity (MdFI) of MHC Class I expression on CD11c+ cells; Results are Mean and SE from 6 experiments.
Since Type I IFNs up-regulate MHC Class I expression (67), we tested the constitutive expression of MHC Class I by Flow Cytometry as an indicator of an active response to Type I IFN (61) and found that the expression of MHC Class I was higher in the Sle1,2,3 BMDCs compared to B6 BMDCs (Fig. 2B). The fact that the expression of the costimulatory molecules CD80 and CD86, which depends on many types of stimuli, was instead similar in Sle1,2,3 vs. B6 BMDCs (data not shown) suggests an IFN-specific stimulation of the lupus BMDCs.
In summary, these results indicate that BMDCs from young Sle1,2,3 mice have the transcriptional and translational Interferon response constitutively activated in absence of any apparent stimulation.
The Interferon Signature in BMDCs from Sle1,2,3 mice does not depend on their autoantibody levels
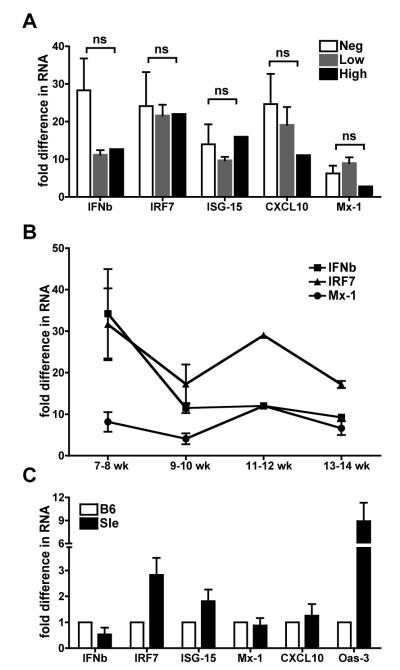
As shown in Fig. 1, we have found an Interferon Signature in BMDCs that were generated from bone marrows of young Sle1,2,3 mice before or at the beginning of the development of the autoimmune process. To determine whether there was any correlation between the levels of Interferon Signature and the titers of autoantibodies (68) in our Sle1,2,3 mice, we grouped the results from the Interferon Signature according to the level of anti-DNA and anti-chromatin autoantibodies measured in the serum by specific ELISA. We found that the Interferon Signature was high in BMDCs generated from bone marrow precursors of mice that were negative for lupus-specific autoantibodies (anti-DNA autoAbs OD <0.3; anti-chromatin autoAbs OD< 0.1) (Fig. 3A) and did not change significantly in the BMDCs that were generated from mice with low positivity (anti-DNA autoAbs OD >0.3 <1; anti-chromatin autoAbs OD > 0.1 < 0.5) or high positivity (anti-DNA autoAbs OD >1;anti-chromatin autoAbs OD > 0.5). The levels of IFNb seemed to decrease with the onset of the disease but the difference was not significant and it might be due to larger variations in the group of mice negative for autoantibodies. Furthermore, we plotted the results according to the age of the mice from which we harvested the bone marrow and we found that the Interferon Signature did not change from 7 weeks up to 14 weeks of age, a time when the first autoantibodies start to appear in the serum (Fig. 3B). Similar analyses of other IFN-responsive genes, like IRF7, ISG-15, Mx-1 etc. led to similar conclusions (data not shown). Our study reveals that the Interferon Signature in BMDCs precedes the onset of the autoimmune process, it does not depend on the presence of autoantibodies and is not affected by autoantibody levels.
FIGURE 3. Type I Interferon Signature in young Sle1,2,3 mice does not depend on the autoantibody levels.
A. We compared the levels of expression of the Interferon Signature in BMDCs according to the levels of autoantibodies present in the mice which the BMDCs were generated from. We measured the levels of anti-DNA and anti-chromatin antibodies in the sera of Sle1,2,3 mice at the time of the harvest of bone marrow to grow BMDCs. The mice were categorized as autoantibody negative (anti-DNA autoAbs OD <0.3; anti-chromatin autoAbs OD < 0.1), low positive (anti-DNA autoAbs OD >0.3 <1; anti-chromatin autoAbs OD > 0.1 < 0.5) or high positive (anti-DNA autoAbs OD >1;anti-chromatin autoAbs OD > 0.5). We show fold differences in the gene expression of IFNb, IRF7, ISG-15, Mx-1 and CXCL10, normalized against the B6 control, and comparing the levels among the three groups of Sle1,2,3 BMDCs. The results were not statistically significant using Mann Whitney test. B. We plotted the same results of the Interferon Signature of BMDCs according to the age of the mice, which the BMDCs were generated from. C. We flushed bone marrow cells from 3 B6 and 3 Sle1,2,3 pups (7-day-old mice) and cultured them in complete medium with GM-CSF for 7 days in independent cultures; then we harvested the cells and analyzed their constitutive Interferon Signature for 6 genes by real time qRT-PCR. We normalized the cycle threshold numbers of the 5 genes with cyclophilin (housekeeping gene) for each mouse, we averaged the results of the three B6 mice and used it to normalize the results from 3 Sle1,2,3 mice.
The Interferon Signature is present in BMDCs from one week old Sle1,2,3 pups
To support the evidence for a constitutive Interferon Signature that precedes the onset of the disease, we grew BMDCs from 7-day-old-pups. This age corresponds in immunological terms to the time of birth in humans, in which the adaptive immune cells are still very immature and they are beginning to colonize the spleen and lymph nodes. We determined the gender of the pups by PCR genotyping for Sry male gene (62) and used only female pups as we have used female adult mice for the rest of the experiments shown herein. Because at 7 days of age there are not many mature T and B cells in the bone marrow, and also because of the paucity of cells available from one single mouse, we did not perform any cell depletion and we plated the total population of bone marrow cells from individual pups at one million per ml per well, as described in the Materials and Methods. When we analyzed the BMDCs on day 7 of culture in vitro, we found that >98% were myeloid CD11c+CD11b+ DCs in both the strains (data not shown). The analysis by real time qRT-PCR of the same 17 genes shown in Fig. 1, revealed the presence of the beginning of an Interferon Signature (Fig. 3C and data not shown). Indeed, IRF7, the master regulator of the response to Type I IFNs, showed a 3 fold increase in the BMDCs from Sle1,2,3 pups compared to BMDCs from B6 pups; Oas-3, one of the most up-regulated genes in the constitutive Interferon Signature in adult mice, was highly expressed in Sle1,2,3 (9 fold difference) compared to B6 pups; the rest of the 17 genes did not show any important difference from the controls (Fig. 3C and data not shown). These results indicate that an Interferon Signature is already present in BMDCs generated from lupus-prone mice very early in life, and definitively precedes the onset of any sign of disease. Since the Interferon Signature is not as complete, in terms of number of genes up-regulated and levels of expression, as in the 7-14 weeks old mice, these results suggest the existence of a missing step occurring during the differentiation of DCs both in vitro and in vivo that is required for a full development of the DC Interferon response.
Sle1,2,3 BMDCs hyper-respond to stimulation with IFNa
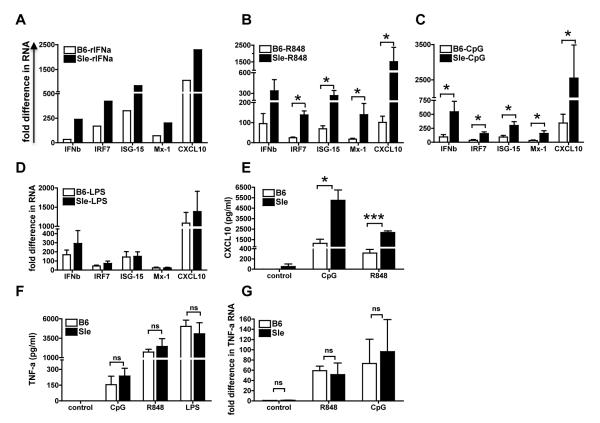
Type I IFNs are a family of cytokines that can be produced by almost any cell in the body and share the same receptor (IFNAR)(7). mDCs produce mostly IFNb while pDCs produce mostly IFNa (1) and these two cytokines are important messengers in the cross-talk between mDCs and pDCs. To reproduce in vitro the in vivo situation in which mDCs are stimulated by the IFNs produced by pDCs, we determined how mDCs responded to exogenous IFN-a. We stimulated BMDCs from young pre-diseased Sle1,2,3 mice with murine recombinant IFNa (2500U/ml) and tested the same selected number of IFN responsive genes as shown in Fig. 3A (IFNb, IRF7, ISG-15, Mx-1 and CXCL10): we found that Sle1,2,3 BMDCs had an increased response to Type I IFN stimulation and expressed the five IFN responsive genes at higher levels than B6 BMDCs (Fig. 4A). Since it has been shown in cell types other than DCs that Type I IFNs induce a positive feedback loop and strengthen the response to a second challenge with Type I IFNs (70), the over-response of Sle1,2,3 BMDCs to IFNa functionally confirms the results that Sle1,2,3 BMDCs have a constitutive active IFN response and suggest that a cross-talk between myeloid and pDCs could amplify such activation.
FIGURE 4. Sle1,2,3 BMDCs hyper-respond to IFNa and TLR7-9 stimulation.
We stimulated B6 and Sle1,2,3 BMDCs at day 6-7 of culture with any of the following stimuli: (A) 2500U/ml mouse recombinant IFNa, (B) 1ug/ml R848 (TLR7 ligand), (C) 10ug/ml CpG 1826 (TLR9 ligand), (D) 100ng/ml LPS (TLR4 ligand). We analyzed the Interferon Signature by real-time qRT-PCR after 5 hours of stimulation. Results are shown as average fold difference in RNA expression from unstimulated B6 control; A,B,C,D show Mean and SE from 3-4 independent experiments. We measured CXCL10 (E) and TNF-a (F) in BMDC culture supernatants 24h after stimulation with TLR ligands and TNF-a gene expression (G) after 5h with TLR ligands; Mean and SE from 3 independent experiments.
Sle1,2,3 BMDCs hyper-respond to TLR7 and TLR9 ligands
The role for TLRs in stimulating DCs in lupus was suggested by the finding that DCs secrete inflammatory cytokines via a TLR9- or TLR7-dependent mechanism upon stimulation with nucleic acid-containing immune complexes (41, 45, 47, 48, 71). Santiago-Raber at al. have also shown that TLR7 and TLR9 are important regulators of disease severity and B cell reactivity in B6.Nba2.Yaa lupus prone mice (43, 72). These results validated the notion that TLRs, presumably recognizing endogenous ligands, play a key role in systemic autoimmunity. However, the constitutive sensitivity of lupus DCs to these stimulators remains undetermined. We found that the response of BMDCs from young pre-diseased Sle1,2,3 mice to TLR7 (R848) and TLR9 (CpG-B 1826) ligands was significantly higher compared to the response of B6 BMDCs (Fig. 4 B, C). This hyper-reactivity was specific for TLR7 and TLR9 because the response to LPS, ligand of TLR4, was not different in B6 and Sle1,2,3 BMDCs (Fig. 4D).
The chemokine Interferon-inducible protein 10 (IP-10)/CXCL10 is over-expressed in lupus patients and correlates with disease activity (69, 73). We have found that Sle1,2,3 BMDCs constitutively have a very high gene expression of CXCL10 (Fig. 2A) that was greatly increased upon TLR7 and 9 stimulation compared to the unstimulated Sle1,2,3 BMDCs and to the B6 BMDCs upon stimulation (Fig. 4 B-C). We also measured CXCL10 protein levels by ELISA and found that Sle1,2,3 BMDCs secreted significantly more CXCL10 in the culture supernatants than B6 BMDCs in response to R848 and CpG stimulation (Fig. 4E). These results confirm that Sle1,2,3 BMDCs hyper-activate upon TLR7/9 stimulation and suggest that DCs can be an important source of CXCL10 in lupus.
TNF-a production is normal in Sle1,2,3 BMDCs
Tumor necrosis factor (TNF)-a is a pleiotropic cytokine that has both proinflammatory and immunoregulatory effects. Its involvement in lupus is ambiguous as it was reported to be increased in SLE patients and correlated with disease activity, but anti-TNF blockers occasionally trigger a latent disease (74)(75). The lupus prone mice (NZBxNZW)F1 and Sle1,2,3 mice are low producers and respond less than wild type mice to TNF-a (76) but the same cytokine has been found to accelerate nephritis (77). It has been also suggested that TNF-a could be beneficial in lupus because it is able to antagonize Type I IFNs (78). We investigated the ability of Sle1,2,3 BMDCs to produce TNF-a. There were no detectable constitutive levels of TNF-a in the culture supernatants of Sle1,2,3 as well as B6 BMDCs (Fig. 4F), while the levels of TNF-a after TLR stimulation were highly up-regulated by the three TLR ligands used, R848, CpGs and LPS, with no differences between B6 and Sle1,2,3 BMDCs (Fig. 4F). Gene expression analysis confirmed these results at RNA level (Fig. 4G). These data indicate that Sle1,2,3 BMDCs do not have a constitutive activation of the signaling pathway leading to TNF-a production (NF-kB-dependent), as they have for Type I IFNs (STAT1-2 dependent). These data also suggest that the Interferon Signature in Sle1,2,3 DCs is the result of a specific activation of the Type I IFN pathway rather than a sign of a generalized hyper-activation of the Sle1,2,3 DCs.
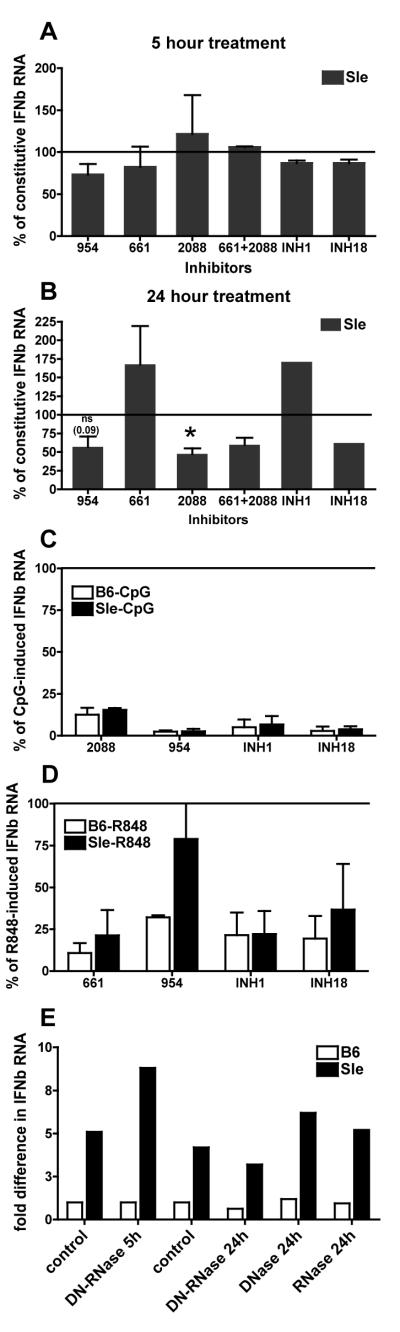
Inhibitors of nucleic acids or nucleic acid sensing receptors partially block the constitutive Interferon Signature of Sle1,2,3 BMDCs
The finding that Sle1,2,3 BMDCs over-produce IFN-responsive genes upon TLR7 and TLR9 stimulation (Fig. 4B-C) prompted us to investigate whether the constitutive Interferon Signature is due to an over-response to DNA/RNA fragments present in the culture supernatants and possibly released in vitro by apoptotic cells naturally occurring in the culture. These nucleic acids would trigger TLR7 and TLR9 to which Sle1,2,3 BMDCs respond by over-producing IFN-responsive genes (Fig. 4B-C). We used inhibitory oligonucleotides that have been reported in literature to block the response to TLR7 and 9 stimulation (46, 63): the oligonucleotide 661 specifically inhibits TLR7 induced responses and the oligonucleotide 2088 inhibits TLR9 induced responses, while 954, INH1 and INH18 oligonucleotides have been reported to suppress both TLR7 and TLR9 induced responses (48, 63). We found that none of these inhibitors decreased the constitutive Interferon Signature by any significant measure after 5 hours of incubation (Fig. 5A); after 24 hours of incubation 2088, 954 and INH18 reduced the Interferon Signature by roughly 50%, while 661 and INH1 increased the constitutive Interferon Signature in Sle1,2,3 BMDCs (Fig. 5B). Extending the time of incubation with the inhibitors to 48 hours did not change the extent or pattern of the suppression observed at 24 hours (data not shown). At the same doses, all the inhibitors, with the exclusion of one, efficiently inhibited the responses of both B6 and Sle1,2,3 BMDCs to the exogenous ligands of TLR7 or TLR9, R848 and CpG, when added respectively to the culture (Fig. 5C, D). We have found that the oligo 954 inhibited the induction of IFNb by CpGs but much less so by R848, with an even less inhibition in Sle1,2,3 than in B6, as previously suggested in other studies (63). The results of the five inhibitors together suggest that the constitutive Interferon Signature of Sle1,2,3 BMDCs, which is partially decreased by TLR9 inhibitors but not by TLR7 inhibitors, may be in part induced by TLR9 triggering.
FIGURE 5. Inhibitors of nucleic acids or nucleic acid sensing receptors partially block the constitutive Interferon Signature of Sle1,2,3 BMDCs.
We treated Sle1,2,3 and B6 BMDC cultures with the oligonucleotides 954 (20ug/ml), 661(20ug/ml), 2088 (20ug/ml), INH1(10ug/ml) or INH18 (10ug/ml) for 5h (A) and 24h (B) and then harvested them and measured by real time qRT-PCR the expression of IFN-responsive genes; the results are shown as Mean and SE of the percentages of the constitutive expression of the IFNb RNA in Sle1,2,3 BMDCs, calculated from 2-4 experiments; the Sle1,2,3 constitutive expression taken as 100% is shown as the straight line. We used ligand-induced stimulation as positive controls; we pretreated BMDCs with the same inhibitors for 30 minutes and then we added R848 (C) or CpG (D) and harvested BMDCs after a total of 5h; the results are shown as Mean and SE of the percentages of the expression of IFNb RNA in Sle1,2,3 BMDCs after the indicated stimulations, calculated from 2-4 experiments. E. We added DNase I (6.8 units/ml) or RNase (25ug/ml), alone or in combination and harvested BMDCs 5h or 24h later. Representative of one out of two independent experiments are shown.
To confirm this interpretation, we added DNase or RNase, alone or in combination, to the culture supernatant of the BMDCs to degrade DNA and RNA, and we found that these treatments did not affect the Interferon Signature of Sle1,2,3 BMDCs after 5 hours of stimulation, while DNase and RNAse combined reduced the Interferon Signature only by 30% after 24 hours of incubation (Fig. 5E). These results indicate that the Interferon Signature of Sle1,2,3 BMDCs is only partially due to an over-response of TLR7 and TLR9 to a hypothetical chronic stimulation by endogenous nucleic acids. Since at least half of the Interferon Signature remained unaffected by TLR7/9 inhibitors, these results suggest a second TLR-independent mechanism mediating the induction of the Interferon Signature in Sle1,2,3 BMDCs.
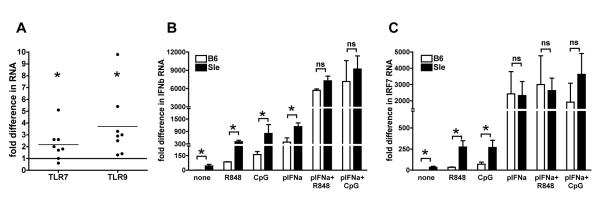
Pre-treatment with Type I IFN increases the response of BMDCs to TLR7 and TLR9 ligands
Although the TLR7/9 inhibitors had limited effects on the Interferon Signature, it is still important to understand why the Sle1,2,3 BMDCs over-respond to these TLR stimuli because it could be an important mechanism for fueling the excessive production of Type I IFNs in lupus (43). First, we hypothesized that Sle1,2,3 BMDCs express more TLR7 and TLR9. We analyzed the expression of TLR7 and TLR9 genes in BMDCs by real-time qRT-PCR and found a 2-3 fold increase in the expression of both genes in the Sle1,2,3 as compared to the B6 BMDCs (Fig. 6A). Furthermore, Type I IFNs have been shown to increase TLR induced responses in macrophages (79): therefore, we investigated whether exposure to Type I IFNs during the culture, as it occurs in Sle1,2,3 BMDCs due to their own Interferon Signature, could increase the response to TLR stimulation in wild type BMDCs. Pre-treatment of B6 BMDCs for 12 hours with rIFNa dramatically increased the expression of IFNb upon TLR7 and TLR9 stimulation as it did in Sle1,2,3 BMDCs and eliminated the big difference in the BMDCs’ response from the two strains (Fig. 6B). Also TLR-induced IRF7 was strongly upregulated by pre-treatment with rIFNa in BMDCs from both strains of mice (Fig. 6C), indicating that the TLR-dependent induction of the master regulator of Type I IFN responses is also sensitive to IFNs. These results indicate that Type I IFNs increase the response of mDCs to TLR7/9 stimulation and suggest that the chronic exposure to Type I IFNs, due to the constitutive Interferon Signature, could be responsible for the over-response to TLR7/9 stimulation in Sle1,2,3 BMDCs
FIGURE 6. Exposure to Type I Interferon increases the response of DCs to TLR7 and TLR9 stimulation.
A. We analyzed TLR7 and TLR9 RNA expression in BMDCs from young female age-matched B6 and Sle1,2,3 mice by qRT-PCR. Results are fold difference in gene expression from B6 control; Mean and SE from 4 independent experiments. We calculated the statistical significance with the one sample t test. B-C. We treated B6 and Sle1,2,3 BMDCs overnight with IFNa (2500U/ml), then stimulated them with either R848 (1ug/ml) or CpG (10ug/ml) for 5h and then processed RNA to measure IFNb (B) and IRF7 (C) gene expression by qRT-PCR as described above. Results are Mean and SE from 3 independent experiments; p-pretreated.
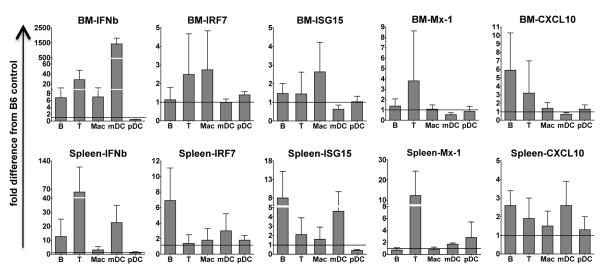
Sle1,2,3 mice express the Interferon Signature in vivo
We investigated whether the Interferon Signature that we have observed in lupus BMDCs in vitro is expressed by Sle1,2,3 mice in vivo and also whether it is specific to DCs or it is expressed by other immune populations. We sorted five immune subsets (B cells, T cells, macrophages, mDCs and pDCs) from bone marrow and spleen of young pre-diseased Sle1,2,3 mice and gender and age matched B6 controls. Although none of the results reached statistical significance due to large variabilities between individual mice, especially in the wild type mice, we found important differences between Sle1,2,3 and wild type B6 mice. Out of the five IFN-responsive genes that we analyzed (IFNb, IRF7, ISG15, Mx-1 and CXCL10), we found the most evident differences in the expression of IFNb. Indeed, mDCs from B6 mice expressed very low levels of IFNb while mDCs both from bone marrow and spleen of Sle1,2,3 mice showed increased expression of IFNb, with a more variable expression in the spleen, while the bone marrow mDCs showed much higher and more consistent increase in the expression of IFNb (ranging from 744 to 2084 fold increase compare to B6 mDCs, p<0.06) (Fig. 7). The expression of IRF7, ISG-15, MX-1 and CXCL10 was variable, with a trend for higher levels of ISG-15 and CXCL10 in the Sle1,2,3 mDCs from the spleen. Surprisingly, Sle1,2,3 pDCs did not show any hint of Interferon Signature, while B and T cell subsets showed increased IFNb gene expression both in the bone marrow and in the spleen. B cells had elevated IRF7 and ISG-15 expression in the spleen and higher CXCL10 in both spleen and bone marrow; T cells showed increased Mx-1 expression in the spleen. The macrophage subset showed a very weak Interferon Signature both in spleen and bone marrow (Fig. 7). In summary, although much variability between individual mice, we have found that ex vivo mDCs from spleen and bone marrow of young pre-diseased Sle1,2,3 mice express an Interferon Signature, that is also in part shared by other immune cells such as B and T cells, while macrophages are less involved and pDCs seem to be refractory to the hyper-expression of IFN responsive genes.
FIGURE 7. Increased Interferon Signature is found in ex vivo bone marrow and spleen of young Sle1,2,3 mice.
We processed bone marrow and spleen of young pre-diseased Sle1,2,3 female mice and age matched B6 for sorting by Flow cytometry to analyze the Interferon gene Signature by qRT-PCR in the different immune populations, as described in Materials and Methods. Bone marrow (top panel) and Spleen (bottom panel) cells were sorted as: B cells (CD19 positive), T cells (CD3 positive), Macrophages (CD19-CD3-CD11b+CD11c-), mDC (CD19-CD3-CD11c+CD11b+) and pDC (CD19-CD3-B220+CD11c+). Bar graphs are mean and SEM of at least 3 sets of mice. The normalization from B6 is indicated by black line in the graph.
Discussion
We have shown that Sle1,2,3 lupus prone mice have an Interferon Signature similar to that discovered in SLE patients. Since this Signature was expressed by bone marrow-derived mDCs in vitro, severed from the rest of the pro-autoimmune immune system, we propose that the Interferon Signature is intrinsic to the DCs. In vivo, T and B cells expressed the Interferon Signature like the mDCs, suggesting that it may be intrinsic to the lymphocytes as well; alternatively, lymphocytes could express the Interferon Signature in response to the Type I IFNs produced by the adjacent mDCs.
Since we have found the Interferon Signature in young Sle1,2,3 mice that were negative for autoantibodies, we propose that Sle1,2,3 DCs have a primary defect that predates the disease and participates to the initiation of the autoimmune process. Compared to the strong, statistically significant over-expression of IFN-responsive genes by the Sle1,2,3 mDCs in vitro, the Interferon Signature in vivo was incomplete. These results can be explained in the context of the “two hit model” for the role of IFNs in lupus pathogenesis: this model proposes that a genetically determined threshold of Type I IFN production is a primary risk factor in human disease (80) that is then triggered/amplified by lupus autoantibodies that form immune complexes with DNA and riboucleoproteins and activate TLR7 and TLR9 in DCs (63)(80). The incomplete Interferon Signature in vivo in Sle1,2,3 mice could require the second hit of autoantibodies to reach the levels seen in SLE patients. Our results suggest that the pathogenesis of lupus in our Sle1,2,3 model shows parallels with the human disease. Longitudinal analysis of the in vivo expression of the Interferon Signature with the development of autoantibodies and disease pathology in Sle1,2,3 mice will test this hypothesis. Since DCs in vitro did not require autoantibodies to express a strong Signature, we hypothesize that in vivo there are inhibitory signals provided by cells other than DCs and therefore absent in DCs generated in vitro.
Trisomy of the Type I IFN cluster genes on chromosome 9p was associated with lupus-like autoimmunity in few families of SLE patients (81). In Sle1,2,3 mice the cluster of IFN genes is located in the susceptibility locus Sle2, with the alleles for IFNs derived specifically from the NZW parental strain (52, 82). It was previously demonstrated that the single congenic B6.Sle2 produces lower levels of Type I IFN (82), and it has an autoimmune phenotype mostly affecting the B cell compartment without full disease development (83). These results suggest that the polymorphisms in IFN genes are not responsible for the Interferon Signature in the triple congenic Sle1,2,3 mice that it could be rather the result of epistatic contributions from the other Sle loci.
Polymorphisms in several genes involved in the IFN pathway have been associated with SLE susceptibility (34-36)(33), including IRF7, a master regulator of the IFN inducible pathway (84)(85), that was constitutively highly expressed in our Sle1,2,3 BMDCs as compared to the B6 BMDCs, and in FLt3L induced Sle1,2,3 pDCs. IRF7 was over-expressed even in BMDCs generated from 7 days old pups suggesting that a dysregulation in IRF7 expression maybe key in the initiation of the pathogenic events that follow. The mouse IRF7 gene is located on chromosome 7 (GenBank Accession no. U73037 http://www.ncbi.nlm.nih.gov/genbank/) that also harbors the Sle3 susceptibility locus but IRF7 is outside the Sle3 locus, at the opposite end of the chromosome, indicating that the IRF7 allele is from the non-autoimmune B6 genome and its higher expression is not directly due to a lupus-related polymorphism in these mice. Since pre-treatment with IFNa strongly up-regulated IRF7 both in Sle1,2,3 and in B6 BMDCs, (Fig. 6C) but IRF7 was also upregulated in BMDC from lupus prone pups (Fig. 3C), in which there is little or no over-expression of IFNab, we do not know yet whether IRF7 is the cause or an effect of the Interferon Signature.
The Interferon Signature was absent in lineage negative bone marrow precursors (Fig. 1D) and was only partial in DCs from one-week-old pups (Fig. 3C) in which the innate immune system is still acquiring its ability to differentiate into effector cells. These results suggest that the Interferon Signature in these mice depends on a defect that expands as the DCs differentiate and that a full differentiation to DCs is necessary to develop the complete Interferon Signature.
The finding that Sle1,2,3 BMDCs express constitutively high levels of STAT1 and STAT2 (Fig. 2A), together with the high expression of IRF7, indicates that Sle1,2,3 mDCs have already upregulated the molecular apparatus required to respond to Type I IFNs and to stimuli that are dependent on Type I IFNs such as TLR ligands. This leads them to express Interferon responsive genes at high levels. This molecular setting is normally not present in mDCs while it is characteristic of pDCs (84), suggesting the highly speculative hypothesis that in Sle1,2,3 mice mDCs behave more like pDCs in their heightened Interferon production. The analysis of the pDCs from the young Sle1,2,3 mice yielded puzzling results. Indeed, pDCs generated in vitro showed an Interferon Signature while pDCs sorted ex vivo did not. It will be important to determine the cause of this dichotomy in vitro-in vivo in order to understand the role of pDCs in sustaining the Interferon Signature before the onset of the disease. Compared to the human immune system in which the TLR7 and TLR9 are exclusively present in the pDCs, these TLRs are also expressed in mDCs in mice and respond well to the respective ligands: this difference may shift the predominant control pathways of TLR activation. It is also possible that other types of IFNs other than IFNa/b play a role as described in humans (86); their study could shed more light in lupus pathology.
The hyper-activation of the molecular machinery to produce Type I IFNs can explain the hyper-response that we found in Sle1,2,3 BMDCs to the stimulation with recombinant IFNa (Fig. 4A). The phenomenon that Interferon induces a positive feedback loop that potentiates subsequent responses to Interferon was already known in other types of cells (70) and we have confirmed it in BMDCs, both from control B6 and Sle1,2,3 mice, in which it is heightened.
We propose that the hyper-activation of the Interferon machinery also mediates the hyper-response to R848 and CpGs as the result of the positive feedback loop of the IFN response induced by the exposure to the Interferon Signature. Indeed, the pretreatment with Type I IFNs strongly increased the expression of IFNb and IRF7 both in B6 and Sle1,2,3 BMDCs, indicating that Type I IFNs increase the response of mDCs to TLR7/9 stimulation. Furthermore, the results that B6 and Sle1,2,3 BMDCs responded equally to TLR7/9 stimulation after pre-treatment with high doses of rIFNa, suggest that we had saturated the response of the BMDCs of both strains to TLR7/9 and that Sle1,2,3 BMDCs do not have an intrinsic hyper-response to TLR7/9 but rather they are pre-primed to higher IFN responses (Fig.6B-C). The DC sensitization by the Interferon Signature to over-respond to TLR7 and TLR9 would create a vicious circle because higher responses to TLR stimulation amplify the Interferon Signature (Fig. 4B-C). Moreover, the result that TNF-a expression is similar in Sle1,2,3 and B6 BMDCs upon TLR stimulation suggests that the hyper-response to R848 and CpGs is not actually a direct hyper-reactivity of the TLR7/9 pathway but rather a hyper-activated response to the autocrine IFNb induced by TLR stimulation. It remains to be explained why this mechanism is not acting upon LPS stimulation. It has been previously proposed that Type I IFNs and TNF-a antagonize their reciprocal expression (78). Our results that Sle1,2,3 BMDCs and B6 BMDCs produce the same amounts of TNF-a although they are exposed to different amounts of Type I IFNs suggest that the Type I IFNs do not inhibit TNF-a production in myeloid dendritic cells. It remains to be demonstrated whether the opposite is true, and these results would justify the anecdotal triggerings of SLE upon TNF blockers(74).
Accumulating evidence supports the role for TLR7 and TLR9 in the development of murine lupus (36, 40, 41, 43, 45-48, 71). TLR7 and TLR9 are abundant in the pDCs, but also mouse mDCs express and respond to TLR7 and TLR9 and we found that these responses were significantly increased in the Sle1,2,3 BMDCs (Fig. 4B-C), in part because of the increased expression of the TLR7 and TLR9 themselves (Fig. 6A). Recently, TLR blockade has provided promising results in animal models using TLR7 and/or TLR9 specific antagonists (46, 63, 87). Since these antagonists, as well as the treatment with DNase and RNAse, could only partially inhibit the Interferon Signature (Fig. 5E), we conclude that, although Sle1,2,3 BMDCs are sensitized to over-react to the TLR7 and 9 ligands specifically, the Interferon Signature is mediated by TLR7/9 dependent and independent mechanisms.
In conclusion, we present evidence that Sle1,2,3 mice are a useful model to study the role of Type I IFNs in lupus pathogenesis and understand the dynamics between genetic threshold and immunologic and environmental stimuli in determining the Interferon Signature. These mice will also be useful to test novel therapeutic approaches aimed to target the Interferon Signature and determine the impact on the onset and severity of the disease.
Supplementary Material
Acknowledgments
We thank Drs. Phillip L. Cohen, Doina Ganea and Marc Monestier for reading the manuscript. We thank Dr. Xiaoxuan Fan at the Flow Cytometry Facility, Temple University, for helping with the sorting experiments.
This work was supported by the National Institutes of Health, National Institute of Allergy and Infectious Diseases Grant RO1-AI076423 (to S.G.) and the Arthritis Foundation (Postdoctoral Fellowship to U.S.) .
References
- 1.Theofilopoulos AN, Baccala R, Beutler B, Kono DH. Type I interferons (alpha/beta) in immunity and autoimmunity. Annu Rev Immunol. 2005;23:307–336. doi: 10.1146/annurev.immunol.23.021704.115843. [DOI] [PubMed] [Google Scholar]
- 2.Banchereau J, Pascual V. Type I interferon in systemic lupus erythematosus and other autoimmune diseases. Immunity. 2006;25:383–392. doi: 10.1016/j.immuni.2006.08.010. [DOI] [PubMed] [Google Scholar]
- 3.Gallucci S, Lolkema M, Matzinger P. Natural adjuvants: endogenous activators of dendritic cells. Nat Med. 1999;5:1249–1255. doi: 10.1038/15200. [DOI] [PubMed] [Google Scholar]
- 4.Finkelman FD, Svetic A, Gresser I, Snapper C, Holmes J, Trotta PP, Katona IM, Gause WC. Regulation by interferon alpha of immunoglobulin isotype selection and lymphokine production in mice. J Exp Med. 1991;174:1179–1188. doi: 10.1084/jem.174.5.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Le Bon A, Schiavoni G, D’Agostino G, Gresser I, Belardelli F, Tough DF. Type i interferons potently enhance humoral immunity and can promote isotype switching by stimulating dendritic cells in vivo. Immunity. 2001;14:461–470. doi: 10.1016/s1074-7613(01)00126-1. [DOI] [PubMed] [Google Scholar]
- 6.Le Bon A, Etchart N, Rossmann C, Ashton M, Hou S, Gewert D, Borrow P, Tough DF. Cross-priming of CD8+ T cells stimulated by virus-induced type I interferon. Nat Immunol. 2003;4:1009–1015. doi: 10.1038/ni978. [DOI] [PubMed] [Google Scholar]
- 7.Taniguchi T, Takaoka A. The interferon-alpha/beta system in antiviral responses: a multimodal machinery of gene regulation by the IRF family of transcription factors. Curr Opin Immunol. 2002;14:111–116. doi: 10.1016/s0952-7915(01)00305-3. [DOI] [PubMed] [Google Scholar]
- 8.Hooks JJ, Moutsopoulos HM, Geis SA, Stahl NI, Decker JL, Notkins AL. Immune interferon in the circulation of patients with autoimmune disease. N Engl J Med. 1979;301:5–8. doi: 10.1056/NEJM197907053010102. [DOI] [PubMed] [Google Scholar]
- 9.Ytterberg SR, Schnitzer TJ. Serum interferon levels in patients with systemic lupus erythematosus. Arthritis Rheum. 1982;25:401–406. doi: 10.1002/art.1780250407. [DOI] [PubMed] [Google Scholar]
- 10.Weckerle CE, Franek BS, Kelly JA, Kumabe M, Mikolaitis RA, Green SL, Utset TO, Jolly M, James JA, Harley JB, Niewold TB. Network analysis of associations between serum interferon-alpha activity, autoantibodies, and clinical features in systemic lupus erythematosus. Arthritis Rheum. 2010;63:1044–1053. doi: 10.1002/art.30187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crow MK, Kirou KA, Wohlgemuth J. Microarray analysis of interferon-regulated genes in SLE. Autoimmunity. 2003;36:481–490. doi: 10.1080/08916930310001625952. [DOI] [PubMed] [Google Scholar]
- 12.Bennett L, Palucka AK, Arce E, Cantrell V, Borvak J, Banchereau J, Pascual V. Interferon and granulopoiesis signatures in systemic lupus erythematosus blood. J Exp Med. 2003;197:711–723. doi: 10.1084/jem.20021553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baechler EC, Batliwalla FM, Karypis G, Gaffney PM, Ortmann WA, Espe KJ, Shark KB, Grande WJ, Hughes KM, Kapur V, Gregersen PK, Behrens TW. Interferon-inducible gene expression signature in peripheral blood cells of patients with severe lupus. Proc Natl Acad Sci U S A. 2003;100:2610–2615. doi: 10.1073/pnas.0337679100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baechler EC, Gregersen PK, Behrens TW. The emerging role of interferon in human systemic lupus erythematosus. Curr Opin Immunol. 2004;16:801–807. doi: 10.1016/j.coi.2004.09.014. [DOI] [PubMed] [Google Scholar]
- 15.Ronnblom L, Eloranta ML, Alm GV. The type I interferon system in systemic lupus erythematosus. Arthritis Rheum. 2006;54:408–420. doi: 10.1002/art.21571. [DOI] [PubMed] [Google Scholar]
- 16.Mathian A, Weinberg A, Gallegos M, Banchereau J, Koutouzov S. IFN-alpha induces early lethal lupus in preautoimmune (New Zealand Black x New Zealand White) F1 but not in BALB/c mice. J Immunol. 2005;174:2499–2506. doi: 10.4049/jimmunol.174.5.2499. [DOI] [PubMed] [Google Scholar]
- 17.Agrawal H, Jacob N, Carreras E, Bajana S, Putterman C, Turner S, Neas B, Mathian A, Koss MN, Stohl W, Kovats S, Jacob CO. Deficiency of type I IFN receptor in lupus-prone New Zealand mixed 2328 mice decreases dendritic cell numbers and activation and protects from disease. J Immunol. 2009;183:6021–6029. doi: 10.4049/jimmunol.0803872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jorgensen TN, Thurman J, Izui S, Falta MT, Metzger TE, Flannery SA, Kappler J, Marrack P, Kotzin BL. Genetic susceptibility to polyI:C-induced IFNalpha/beta-dependent accelerated disease in lupus-prone mice. Genes Immun. 2006;7:555–567. doi: 10.1038/sj.gene.6364329. [DOI] [PubMed] [Google Scholar]
- 19.Steinberg AD, Baron S, Talal N. The pathogenesis of autoimmunity in New Zealand mice, I. Induction of antinucleic acid antibodies by polyinosinic-polycytidylic acid. Proc Natl Acad Sci U S A. 1969;63:1102–1107. doi: 10.1073/pnas.63.4.1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Niewold TB, Swedler WI. Systemic lupus erythematosus arising during interferon-alpha therapy for cryoglobulinemic vasculitis associated with hepatitis C. Clin Rheumatol. 2005;24:178–181. doi: 10.1007/s10067-004-1024-2. [DOI] [PubMed] [Google Scholar]
- 21.Santiago-Raber ML, Baccala R, Haraldsson KM, Choubey D, Stewart TA, Kono DH, Theofilopoulos AN. Type-I interferon receptor deficiency reduces lupus-like disease in NZB mice. J Exp Med. 2003;197:777–788. doi: 10.1084/jem.20021996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Braun D, Geraldes P, Demengeot J. Type I Interferon controls the onset and severity of autoimmune manifestations in lpr mice. J Autoimmun. 2003;20:15–25. doi: 10.1016/s0896-8411(02)00109-9. [DOI] [PubMed] [Google Scholar]
- 23.Jorgensen TN, Roper E, Thurman JM, Marrack P, Kotzin BL. Type I interferon signaling is involved in the spontaneous development of lupus-like disease in B6.Nba2 and (B6.Nba2 × NZW)F(1) mice. Genes Immun. 2007;8:653–662. doi: 10.1038/sj.gene.6364430. [DOI] [PubMed] [Google Scholar]
- 24.Hron JD, Peng SL. Type I IFN protects against murine lupus. J Immunol. 2004;173:2134–2142. doi: 10.4049/jimmunol.173.3.2134. [DOI] [PubMed] [Google Scholar]
- 25.Liu J, Karypis G, Hippen KL, Vegoe AL, Ruiz P, Gilkeson GS, Behrens TW. Genomic view of systemic autoimmunity in MRLlpr mice. Genes Immun. 2006;7:156–168. doi: 10.1038/sj.gene.6364286. [DOI] [PubMed] [Google Scholar]
- 26.Nacionales DC, Kelly-Scumpia KM, Lee PY, Weinstein JS, Lyons R, Sobel E, Satoh M, Reeves WH. Deficiency of the type I interferon receptor protects mice from experimental lupus. Arthritis Rheum. 2007;56:3770–3783. doi: 10.1002/art.23023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nacionales DC, Kelly KM, Lee PY, Zhuang H, Li Y, Weinstein JS, Sobel E, Kuroda Y, Akaogi J, Satoh M, Reeves WH. Type I interferon production by tertiary lymphoid tissue developing in response to 2,6,10,14-tetramethyl-pentadecane (pristane) Am J Pathol. 2006;168:1227–1240. doi: 10.2353/ajpath.2006.050125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Craft JE. Dissecting the immune cell mayhem that drives lupus pathogenesis. Sci Transl Med. 2011;3:73ps79. doi: 10.1126/scitranslmed.3002138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Niewold TB. Interferon alpha as a primary pathogenic factor in human lupus. J Interferon Cytokine Res. 2011;31:887–892. doi: 10.1089/jir.2011.0071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harley IT, Kaufman KM, Langefeld CD, Harley JB, Kelly JA. Genetic susceptibility to SLE: new insights from fine mapping and genome-wide association studies. Nat Rev Genet. 2009;10:285–290. doi: 10.1038/nrg2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kariuki SN, Niewold TB. Genetic regulation of serum cytokines in systemic lupus erythematosus. Transl Res. 2009;155:109–117. doi: 10.1016/j.trsl.2009.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Santana-de Anda K, Gomez-Martin D, Diaz-Zamudio M, Alcocer-Varela J. Interferon regulatory factors: beyond the antiviral response and their link to the development of autoimmune pathology. Autoimmun Rev. 2011;11:98–103. doi: 10.1016/j.autrev.2011.08.006. [DOI] [PubMed] [Google Scholar]
- 33.Salloum R, Niewold TB. Interferon regulatory factors in human lupus pathogenesis. Transl Res. 2011;157:326–331. doi: 10.1016/j.trsl.2011.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harley JB, Alarcon-Riquelme ME, Criswell LA, Jacob CO, Kimberly RP, Moser KL, Tsao BP, Vyse TJ, Langefeld CD, Nath SK, Guthridge JM, Cobb BL, Mirel DB, Marion MC, Williams AH, Divers J, Wang W, Frank SG, Namjou B, Gabriel SB, Lee AT, Gregersen PK, Behrens TW, Taylor KE, Fernando M, Zidovetzki R, Gaffney PM, Edberg JC, Rioux JD, Ojwang JO, James JA, Merrill JT, Gilkeson GS, Seldin MF, Yin H, Baechler EC, Li QZ, Wakeland EK, Bruner GR, Kaufman KM, Kelly JA. Genome-wide association scan in women with systemic lupus erythematosus identifies susceptibility variants in ITGAM, PXK, KIAA1542 and other loci. Nat Genet. 2008;40:204–210. doi: 10.1038/ng.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gateva V, Sandling JK, Hom G, Taylor KE, Chung SA, Sun X, Ortmann W, Kosoy R, Ferreira RC, Nordmark G, Gunnarsson I, Svenungsson E, Padyukov L, Sturfelt G, Jonsen A, Bengtsson AA, Rantapaa-Dahlqvist S, Baechler EC, Brown EE, Alarcon GS, Edberg JC, Ramsey-Goldman R, McGwin G, Jr., Reveille JD, Vila LM, Kimberly RP, Manzi S, Petri MA, Lee A, Gregersen PK, Seldin MF, Ronnblom L, Criswell LA, Syvanen AC, Behrens TW, Graham RR. A large-scale replication study identifies TNIP1, PRDM1, JAZF1, UHRF1BP1 and IL10 as risk loci for systemic lupus erythematosus. Nat Genet. 2009;41:1228–1233. doi: 10.1038/ng.468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Salloum R, Franek BS, Kariuki SN, Rhee L, Mikolaitis RA, Jolly M, Utset TO, Niewold TB. Genetic variation at the IRF7/PHRF1 locus is associated with autoantibody profile and serum interferon-alpha activity in lupus patients. Arthritis Rheum. 2010;62:553–561. doi: 10.1002/art.27182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jacob CO, Zhu J, Armstrong DL, Yan M, Han J, Zhou XJ, Thomas JA, Reiff A, Myones BL, Ojwang JO, Kaufman KM, Klein-Gitelman M, McCurdy D, Wagner-Weiner L, Silverman E, Ziegler J, Kelly JA, Merrill JT, Harley JB, Ramsey-Goldman R, Vila LM, Bae SC, Vyse TJ, Gilkeson GS, Gaffney PM, Moser KL, Langefeld CD, Zidovetzki R, Mohan C. Identification of IRAK1 as a risk gene with critical role in the pathogenesis of systemic lupus erythematosus. Proc Natl Acad Sci U S A. 2009;106:6256–6261. doi: 10.1073/pnas.0901181106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jacob CO, Zang S, Li L, Ciobanu V, Quismorio F, Mizutani A, Satoh M, Koss M. Pivotal role of Stat4 and Stat6 in the pathogenesis of the lupus-like disease in the New Zealand mixed 2328 mice. J Immunol. 2003;171:1564–1571. doi: 10.4049/jimmunol.171.3.1564. [DOI] [PubMed] [Google Scholar]
- 39.Rozzo SJ, Allard JD, Choubey D, Vyse TJ, Izui S, Peltz G, Kotzin BL. Evidence for an interferon-inducible gene, Ifi202, in the susceptibility to systemic lupus. Immunity. 2001;15:435–443. doi: 10.1016/s1074-7613(01)00196-0. [DOI] [PubMed] [Google Scholar]
- 40.Marshak-Rothstein A, Rifkin IR. Immunologically active autoantigens: the role of toll-like receptors in the development of chronic inflammatory disease. Annu Rev Immunol. 2007;25:419–441. doi: 10.1146/annurev.immunol.22.012703.104514. [DOI] [PubMed] [Google Scholar]
- 41.Means TK, Latz E, Hayashi F, Murali MR, Golenbock DT, Luster AD. Human lupus autoantibody-DNA complexes activate DCs through cooperation of CD32 and TLR9. J Clin Invest. 2005;115:407–417. doi: 10.1172/JCI23025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Denman AM. Systemic lupus erythematosus--is a viral aetiology a credible hypothesis? J Infect. 2000;40:229–233. doi: 10.1053/jinf.2000.0670. [DOI] [PubMed] [Google Scholar]
- 43.Santiago-Raber ML, Baudino L, Izui S. Emerging roles of TLR7 and TLR9 in murine SLE. J Autoimmun. 2009;33:231–238. doi: 10.1016/j.jaut.2009.10.001. [DOI] [PubMed] [Google Scholar]
- 44.Lovgren T, Eloranta ML, Bave U, Alm GV, Ronnblom L. Induction of interferon-alpha production in plasmacytoid dendritic cells by immune complexes containing nucleic acid released by necrotic or late apoptotic cells and lupus IgG. Arthritis Rheum. 2004;50:1861–1872. doi: 10.1002/art.20254. [DOI] [PubMed] [Google Scholar]
- 45.Pisitkun P, Deane JA, Difilippantonio MJ, Tarasenko T, Satterthwaite AB, Bolland S. Autoreactive B cell responses to RNA-related antigens due to TLR7 gene duplication. Science. 2006;312:1669–1672. doi: 10.1126/science.1124978. [DOI] [PubMed] [Google Scholar]
- 46.Barrat FJ, Meeker T, Chan JH, Guiducci C, Coffman RL. Treatment of lupus-prone mice with a dual inhibitor of TLR7 and TLR9 leads to reduction of autoantibody production and amelioration of disease symptoms. Eur J Immunol. 2007;37:3582–3586. doi: 10.1002/eji.200737815. [DOI] [PubMed] [Google Scholar]
- 47.Christensen SR, Shupe J, Nickerson K, Kashgarian M, Flavell RA, Shlomchik MJ. Toll-like receptor 7 and TLR9 dictate autoantibody specificity and have opposing inflammatory and regulatory roles in a murine model of lupus. Immunity. 2006;25:417–428. doi: 10.1016/j.immuni.2006.07.013. [DOI] [PubMed] [Google Scholar]
- 48.Barrat FJ, Meeker T, Gregorio J, Chan JH, Uematsu S, Akira S, Chang B, Duramad O, Coffman RL. Nucleic acids of mammalian origin can act as endogenous ligands for Toll-like receptors and may promote systemic lupus erythematosus. J Exp Med. 2005;202:1131–1139. doi: 10.1084/jem.20050914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brinkmann V, Reichard U, Goosmann C, Fauler B, Uhlemann Y, Weiss DS, Weinrauch Y, Zychlinsky A. Neutrophil extracellular traps kill bacteria. Science. 2004;303:1532–1535. doi: 10.1126/science.1092385. [DOI] [PubMed] [Google Scholar]
- 50.Lande R, Ganguly D, Facchinetti V, Frasca L, Conrad C, Gregorio J, Meller S, Chamilos G, Sebasigari R, Riccieri V, Bassett R, Amuro H, Fukuhara S, Ito T, Liu YJ, Gilliet M. Neutrophils activate plasmacytoid dendritic cells by releasing self-DNA-peptide complexes in systemic lupus erythematosus. Sci Transl Med. 2011;3:73ra19. doi: 10.1126/scitranslmed.3001180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Garcia-Romo GS, Caielli S, Vega B, Connolly J, Allantaz F, Xu Z, Punaro M, Baisch J, Guiducci C, Coffman RL, Barrat FJ, Banchereau J, Pascual V. Netting neutrophils are major inducers of type I IFN production in pediatric systemic lupus erythematosus. Sci Transl Med. 2011;3:73ra20. doi: 10.1126/scitranslmed.3001201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Morel L, Mohan C, Yu Y, Croker BP, Tian N, Deng A, Wakeland EK. Functional dissection of systemic lupus erythematosus using congenic mouse strains. J Immunol. 1997;158:6019–6028. [PubMed] [Google Scholar]
- 53.Mohan C, Morel L, Yang P, Wakeland EK. Genetic dissection of systemic lupus erythematosus pathogenesis: Sle2 on murine chromosome 4 leads to B cell hyperactivity. J Immunol. 1997;159:454–465. [PubMed] [Google Scholar]
- 54.Morel L, Croker BP, Blenman KR, Mohan C, Huang G, Gilkeson G, Wakeland EK. Genetic reconstitution of systemic lupus erythematosus immunopathology with polycongenic murine strains. Proc Natl Acad Sci U S A. 2000;97:6670–6675. doi: 10.1073/pnas.97.12.6670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pascual V, Banchereau J, Palucka AK. The central role of dendritic cells and interferon-alpha in SLE. Curr Opin Rheumatol. 2003;15:548–556. doi: 10.1097/00002281-200309000-00005. [DOI] [PubMed] [Google Scholar]
- 56.Elkon KB, Stone VV. Type I interferon and systemic lupus erythematosus. J Interferon Cytokine Res. 2011;31:803–812. doi: 10.1089/jir.2011.0045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Colonna L, Dinnall JA, Shivers DK, Frisoni L, Caricchio R, Gallucci S. Abnormal costimulatory phenotype and function of dendritic cells before and after the onset of severe murine lupus. Arthritis Res Ther. 2006;8:R49. doi: 10.1186/ar1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bondanza A, Zimmermann VS, Dell’Antonio G, Dal Cin E, Capobianco A, Sabbadini MG, Manfredi AA, Rovere-Querini P. Cutting edge: dissociation between autoimmune response and clinical disease after vaccination with dendritic cells. J Immunol. 2003;170:24–27. doi: 10.4049/jimmunol.170.1.24. [DOI] [PubMed] [Google Scholar]
- 59.Liu YJ. IPC: professional type 1 interferon-producing cells and plasmacytoid dendritic cell precursors. Annu Rev Immunol. 2005;23:275–306. doi: 10.1146/annurev.immunol.23.021704.115633. [DOI] [PubMed] [Google Scholar]
- 60.Jog NR, Dinnall JA, Gallucci S, Madaio MP, Caricchio R. Poly(ADP-ribose) polymerase-1 regulates the progression of autoimmune nephritis in males by inducing necrotic cell death and modulating inflammation. J Immunol. 2009;182:7297–7306. doi: 10.4049/jimmunol.0803565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sriram U, Biswas C, Behrens EM, Dinnall JA, Shivers DK, Monestier M, Argon Y, Gallucci S. IL-4 suppresses dendritic cell response to type I interferons. J Immunol. 2007;179:6446–6455. doi: 10.4049/jimmunol.179.10.6446. [DOI] [PubMed] [Google Scholar]
- 62.Lambert JF, Benoit BO, Colvin GA, Carlson J, Delville Y, Quesenberry PJ. Quick sex determination of mouse fetuses. J Neurosci Methods. 2000;95:127–132. doi: 10.1016/s0165-0270(99)00157-0. [DOI] [PubMed] [Google Scholar]
- 63.Lenert P, Yasuda K, Busconi L, Nelson P, Fleenor C, Ratnabalasuriar RS, Nagy PL, Ashman RF, Rifkin IR, Marshak-Rothstein A. DNA-like class R inhibitory oligonucleotides (INH-ODNs) preferentially block autoantigen-induced B-cell and dendritic cell activation in vitro and autoantibody production in lupus-prone MRL-Fas(lpr/lpr) mice in vivo. Arthritis Res Ther. 2009;11:R79. doi: 10.1186/ar2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Brasel K, De Smedt T, Smith JL, Maliszewski CR. Generation of murine dendritic cells from flt3-ligand-supplemented bone marrow cultures. Blood. 2000;96:3029–3039. [PubMed] [Google Scholar]
- 65.Nie YJ, Mok MY, Chan GC, Chan AW, Jin OU, Kavikondala S, Lie AK, Lau CS. Phenotypic and functional abnormalities of bone marrow-derived dendritic cells in systemic lupus erythematosus. Arthritis Res Ther. 2010;12:R91. doi: 10.1186/ar3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lenschow DJ, Lai C, Frias-Staheli N, Giannakopoulos NV, Lutz A, Wolff T, Osiak A, Levine B, Schmidt RE, Garcia-Sastre A, Leib DA, Pekosz A, Knobeloch KP, Horak I, Virgin H. W. t. IFN-stimulated gene 15 functions as a critical antiviral molecule against influenza, herpes, and Sindbis viruses. Proc Natl Acad Sci U S A. 2007;104:1371–1376. doi: 10.1073/pnas.0607038104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cheng Y, King NJ, Kesson AM. Major histocompatibility complex class I (MHC-I) induction by West Nile virus: involvement of 2 signaling pathways in MHC-I up-regulation. J Infect Dis. 2004;189:658–668. doi: 10.1086/381501. [DOI] [PubMed] [Google Scholar]
- 68.Landolt-Marticorena C, Bonventi G, Lubovich A, Ferguson C, Unnithan T, Su J, Gladman DD, Urowitz M, Fortin PR, Wither J. Lack of association between the interferon-alpha signature and longitudinal changes in disease activity in systemic lupus erythematosus. Ann Rheum Dis. 2009;68:1440–1446. doi: 10.1136/ard.2008.093146. [DOI] [PubMed] [Google Scholar]
- 69.Bauer JW, Petri M, Batliwalla FM, Koeuth T, Wilson J, Slattery C, Panoskaltsis-Mortari A, Gregersen PK, Behrens TW, Baechler EC. Interferon-regulated chemokines as biomarkers of systemic lupus erythematosus disease activity: a validation study. Arthritis Rheum. 2009;60:3098–3107. doi: 10.1002/art.24803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Marie I, Durbin JE, Levy DE. Differential viral induction of distinct interferon-alpha genes by positive feedback through interferon regulatory factor-7. EMBO J. 1998;17:6660–6669. doi: 10.1093/emboj/17.22.6660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Boule MW, Broughton C, Mackay F, Akira S, Marshak-Rothstein A, Rifkin IR. Toll-like receptor 9-dependent and -independent dendritic cell activation by chromatin-immunoglobulin G complexes. J Exp Med. 2004;199:1631–1640. doi: 10.1084/jem.20031942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Santiago-Raber ML, Dunand-Sauthier I, Wu T, Li QZ, Uematsu S, Akira S, Reith W, Mohan C, Kotzin BL, Izui S. Critical role of TLR7 in the acceleration of systemic lupus erythematosus in TLR9-deficient mice. J Autoimmun. 2010;34:339–348. doi: 10.1016/j.jaut.2009.11.001. [DOI] [PubMed] [Google Scholar]
- 73.Feng X, Wu H, Grossman JM, Hanvivadhanakul P, FitzGerald JD, Park GS, Dong X, Chen W, Kim MH, Weng HH, Furst DE, Gorn A, McMahon M, Taylor M, Brahn E, Hahn BH, Tsao BP. Association of increased interferon-inducible gene expression with disease activity and lupus nephritis in patients with systemic lupus erythematosus. Arthritis Rheum. 2006;54:2951–2962. doi: 10.1002/art.22044. [DOI] [PubMed] [Google Scholar]
- 74.Shakoor N, Michalska M, Harris CA, Block JA. Drug-induced systemic lupus erythematosus associated with etanercept therapy. Lancet. 2002;359:579–580. doi: 10.1016/S0140-6736(02)07714-0. [DOI] [PubMed] [Google Scholar]
- 75.Palucka AK, Blanck JP, Bennett L, Pascual V, Banchereau J. Cross-regulation of TNF and IFN-alpha in autoimmune diseases. Proc Natl Acad Sci U S A. 2005;102:3372–3377. doi: 10.1073/pnas.0408506102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Blenman KR, Bahjat FR, Moldawer LL, Morel L. Aberrant signaling in the TNFalpha/TNF receptor 1 pathway of the NZM2410 lupus-prone mouse. Clin Immunol. 2004;110:124–133. doi: 10.1016/j.clim.2003.09.009. [DOI] [PubMed] [Google Scholar]
- 77.Brennan DC, Yui MA, Wuthrich RP, Kelley VE. Tumor necrosis factor and IL-1 in New Zealand Black/White mice. Enhanced gene expression and acceleration of renal injury. J Immunol. 1989;143:3470–3475. [PubMed] [Google Scholar]
- 78.Banchereau J, Pascual V, Palucka AK. Autoimmunity through cytokine-induced dendritic cell activation. Immunity. 2004;20:539–550. doi: 10.1016/s1074-7613(04)00108-6. [DOI] [PubMed] [Google Scholar]
- 79.Siren J, Pirhonen J, Julkunen I, Matikainen S. IFN-alpha regulates TLR-dependent gene expression of IFN-alpha, IFN-beta, IL-28, and IL-29. J Immunol. 2005;174:1932–1937. doi: 10.4049/jimmunol.174.4.1932. [DOI] [PubMed] [Google Scholar]
- 80.Niewold TB, Hua J, Lehman TJ, Harley JB, Crow MK. High serum IFN-alpha activity is a heritable risk factor for systemic lupus erythematosus. Genes Immun. 2007;8:492–502. doi: 10.1038/sj.gene.6364408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhuang H, Kosboth M, Lee P, Rice A, Driscoll DJ, Zori R, Narain S, Lyons R, Satoh M, Sobel E, Reeves WH. Lupus-like disease and high interferon levels corresponding to trisomy of the type I interferon cluster on chromosome 9p. Arthritis Rheum. 2006;54:1573–1579. doi: 10.1002/art.21800. [DOI] [PubMed] [Google Scholar]
- 82.Li J, Liu Y, Xie C, Zhu J, Kreska D, Morel L, Mohan C. Deficiency of type I interferon contributes to Sle2-associated component lupus phenotypes. Arthritis Rheum. 2005;52:3063–3072. doi: 10.1002/art.21307. [DOI] [PubMed] [Google Scholar]
- 83.Xu Z, Duan B, Croker BP, Wakeland EK, Morel L. Genetic dissection of the murine lupus susceptibility locus Sle2: contributions to increased peritoneal B-1a cells and lupus nephritis map to different loci. J Immunol. 2005;175:936–943. doi: 10.4049/jimmunol.175.2.936. [DOI] [PubMed] [Google Scholar]
- 84.Honda K, Yanai H, Negishi H, Asagiri M, Sato M, Mizutani T, Shimada N, Ohba Y, Takaoka A, Yoshida N, Taniguchi T. IRF-7 is the master regulator of type-I interferon-dependent immune responses. Nature. 2005;434:772–777. doi: 10.1038/nature03464. [DOI] [PubMed] [Google Scholar]
- 85.Fu Q, Zhao J, Qian X, Wong JL, Kaufman KM, Yu CY, Mok MY, Harley JB, Guthridge JM, Song YW, Cho SK, Bae SC, Grossman JM, Hahn BH, Arnett FC, Shen N, Tsao BP. Association of a functional IRF7 variant with systemic lupus erythematosus. Arthritis Rheum. 2010 doi: 10.1002/art.30193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Harley IT, Niewold TB, Stormont RM, Kaufman KM, Glenn SB, Franek BS, Kelly JA, Kilpatrick JR, Hutchings D, Divers J, Bruner GR, Edberg JC, McGwin G, Jr., Petri MA, Ramsey-Goldman R, Reveille JD, Vila-Perez LM, Merrill JT, Gilkeson GS, Vyse TJ, Alarcon-Riquelme ME, Cho SK, Jacob CO, Alarcon GS, Moser KL, Gaffney PM, Kimberly RP, Bae SC, Langefeld CD, Harley JB, Guthridge JM, James JA. The role of genetic variation near interferon-kappa in systemic lupus erythematosus. J Biomed Biotechnol. 2010 doi: 10.1155/2010/706825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Dong L, Ito S, Ishii KJ, Klinman DM. Suppressive oligodeoxynucleotides delay the onset of glomerulonephritis and prolong survival in lupus-prone NZB x NZW mice. Arthritis Rheum. 2005;52:651–658. doi: 10.1002/art.20810. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.