Plasmodium falciparum, the most deadly human malaria parasite, poses a major threat to human health worldwide, with over 500 million clinical cases and between one and two million deaths annually.[1] Natural products and their synthetic derivatives have provided a significant number of successful antimalarial treatments to date, representing approximately 65 % of current drugs.[2] Quinine, discovered from cinchona tree bark, has been used to treat malaria for centuries and was the primary antimalarial drug until it was replaced by chloroquine, a synthetic derivative, in the 1940s.[3] Chloroquine became the mainstay antimalarial until resistant strains began to appear nearly a decade after its introduction. Artemisinin, isolated from the plant Artemisia annua used in traditional Chinese medicine, ushered in a new wave of antimalarials and became the most potent and rapid-acting drug available.[4] Several synthetic artemisinin derivatives have since been developed, and artemisinin-based combination therapies are currently in use throughout the world to treat this parasitic disease. However, artemisinin-resistant strains have recently been reported,[5] and new antiparasitic drugs are urgently needed to combat these strains.
Therapeutic development for malaria is complex due to the intricate host–guest interactions during the plasmodial life cycle. During its maturation, the malaria parasite is transferred to human hosts via mosquitoes, and after a transitory stage in the liver, the parasite resides in the hosts’ red blood cells. While residing in red blood cells, the parasite catabolizes hemoglobin within its food vacuole for its nutritional benefit. This hemoglobin catabolism can be lethal, as it releases free heme molecules which are toxic to the parasite.[6] However, the parasite overcomes heme toxicity through the crystallization of free heme into nontoxic hemozoin, a non-enzymatic process which is crucial to parasite survival and thus an excellent antimalarial drug target.[7] Chloroquine has been shown to inhibit heme crystallization,[8] and structurally related antimalarial drugs such as quinine, amodiaquine, and mefloquine have also been proposed to disrupt hemozoin formation. Resistance to chloroquine has been shown to occur through multiple mutations in a P. falciparum transmembrane protein that, when mutated, pumps chloroquine out of the food vacuole, significantly decreasing the concentration of the drug.[9] The primary mode of action (MOA) for artemisinin remains a matter of debate, although several hypotheses have been reported, including alkylation of heme by carbon-centered free radicals,[10] interference with proteins such as the sarcoplasmic/endoplasmic calcium ATPase (SERCA),[11] as well as disruption of normal mitochondrial functions.[12]
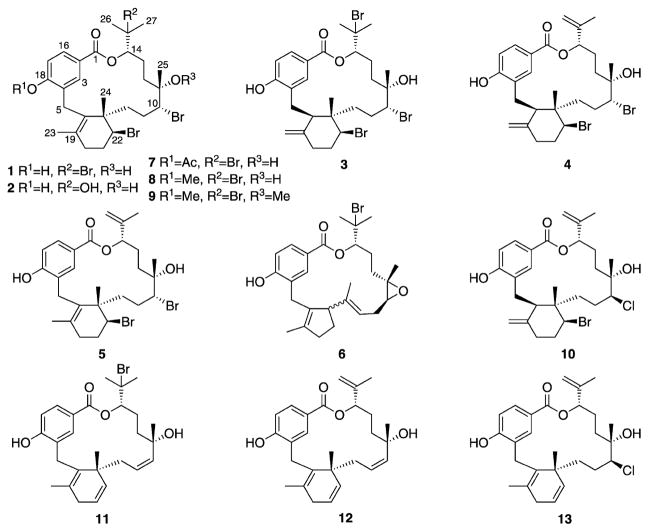
The identification of molecular targets for bioactive natural products is essential for understanding their MOAs, developing new drug leads, and anticipating the evolution of resistance. We previously discovered a unique group of brominated macrocyclic meroditerpenes from the Fijian red macroalga Callophycus serratus[13] and showed that this class of natural products is toxic to P. falciparum in vitro.[14] The most abundant natural product of this class with sub-micromolar inhibition of P. falciparum proliferation, bromophycolide A (1; Figure 1), induces a strong arrest of the parasite erythrocytic cycle,[15] and was therefore used to develop a probe for both cell imaging and molecular target identification studies to gain insight into the potential MOA of 1 in P. falciparum.
Figure 1.
Bromophycolides A (1), C (2), D (3), E (4), M (5), and debromophycolide A (6) isolated from Callophycus serratus[13, 14] and semi-synthetic derivatives of bromophycolide A (7–13).
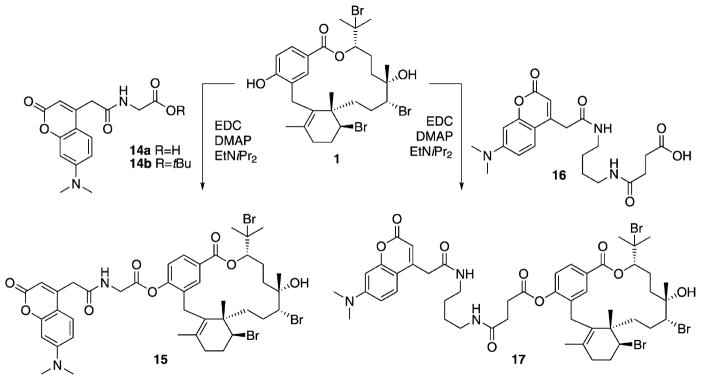
To maintain antimalarial activity while not compromising the integrity of target binding, attempts initially focused on connecting a linker through the tertiary alcohol at C11, because changes at this position resulted in only minor differences in activity, based on structure–activity relationship (SAR) trends gleaned from the known C. serratus natural products. Unfortunately, all synthetic efforts to modify functionality at C11 failed, likely due to a combination of poor reactivity of tertiary alcohols and steric hindrance. From a synthetic perspective, the most chemically reactive and accessible position in 1 is the phenolic hydroxy group at C18. Because the C18 phenol was conserved in all C. serratus metabolites and thus SAR data was lacking, we acetylated the phenol in 1 to test for changes in activity. Fortuitously, the acetylated derivative 7 resulted in an increase in antimalarial activity (IC50 = 241 nM, Tables 1 and 2). Therefore, two immunoaffinity fluorescent (IAF) tags[16] with varying chain lengths were each synthetically appended to 1 through an acetyl linkage (Scheme 1). Probe 15 exhibited an IC50 value of 271 nM, similar to that of 7. Furthermore, both control tags 14a and 14b were completely inactive against P. falciparum, exhibiting IC50 values > 100 μM, confirming that the activity observed in probe 15 was a result of the natural product and not the IAF tag.
Table 1.
Activities of isolated bromophycolides A (1), C (2), D (3), E (4), M (5), debromophycolide A (6); semi-synthetic derivatives of bromophycolide A (7–13); and fluorescent probes 15 and 17.
| Compound | P. falciparum (3D7) IC50 [μM] |
|---|---|
| 1 | 0.50 |
| 2 | 56 |
| 3 | 0.35 |
| 4 | 0.82 |
| 5 | 0.55 |
| 6 | >100 |
| 7 | 0.24 |
| 8 | 15 |
| 9 | 21 |
| 10 | 3.1 |
| 11 | 4.6 |
| 12 | 3.5 |
| 13 | 2.7 |
| 15 | 0.27 |
| 17 | 0.53 |
Table 2.
Efficacies of bromophycolide A (1), 18-acetylbromophycolide A (7), and amodiaquine (AMQ) against three P. falciparum strains, along with their activity in inhibiting heme crystallization, and cytotoxicity against uninfected cell lines.
| Compound | Antiplasmodial activity IC50 [nM] | Heme crystallization IC50 [mol equiv][c] | Cytotoxicity IC50 [μM] | ||||
|---|---|---|---|---|---|---|---|
| 3D7[a] | Dd2[b] | HB3[a] | Vero | J774 | HepG2 | ||
| 1 | 499 ± 51 | 377 ± 92 | 493 ± 44 | 2.5 ± 0.13 | 34.6 | 19.0 | 21.3 |
| 7 | 241 ± 52 | 304 ± 8.4 | 431 ± 25 | 2.0 ± 0.15 | 74.0 | 24.7 | 38.5 |
| 8 | 14 800 ± 1200 | 4880 ± 1100 | 6050 ± 440 | >6.0 | – | – | – |
| AMQ | 7.8[d] | – | 8.5[d] | 1.2 ± 0.19 | – | – | – |
3D7 and HB3 are chloroquine-sensitive parasites.
Dd2 is a chloroquine-resistant parasite.
IC50 values[19] are reported as molar equivalents of bromophycolide to heme.
Values for AMQ are reported from Hawley et al.[30]
A true IC50 value could not be determined due to solubility limitation above 6 mol equiv.
Scheme 1.
Probes 15 and 17 were prepared by coupling acids 14a and 16, respectively, with bromophycolide A (1). EDC =N′-(3-dimethylaminopropyl)-N-ethylcarbodiimide; DMAP =4-dimethylaminopyridine; EtNiPr2 = N,N-diisopropylethylamine; DMF =N,N-dimethylformamide.
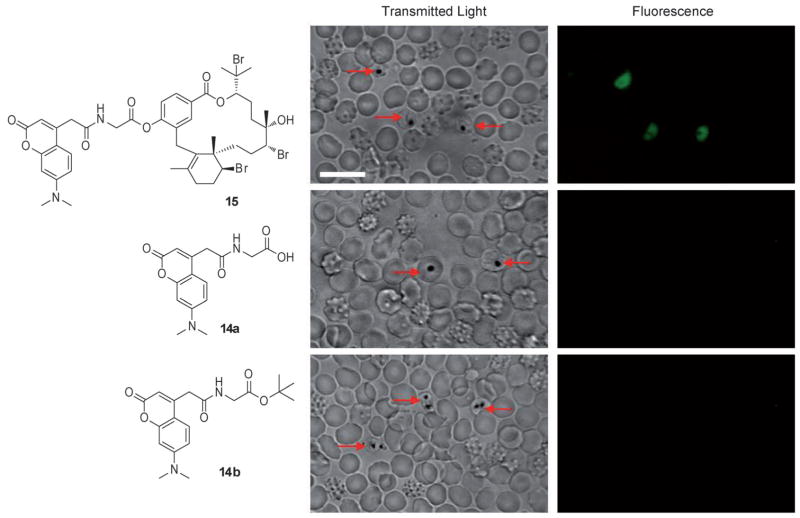
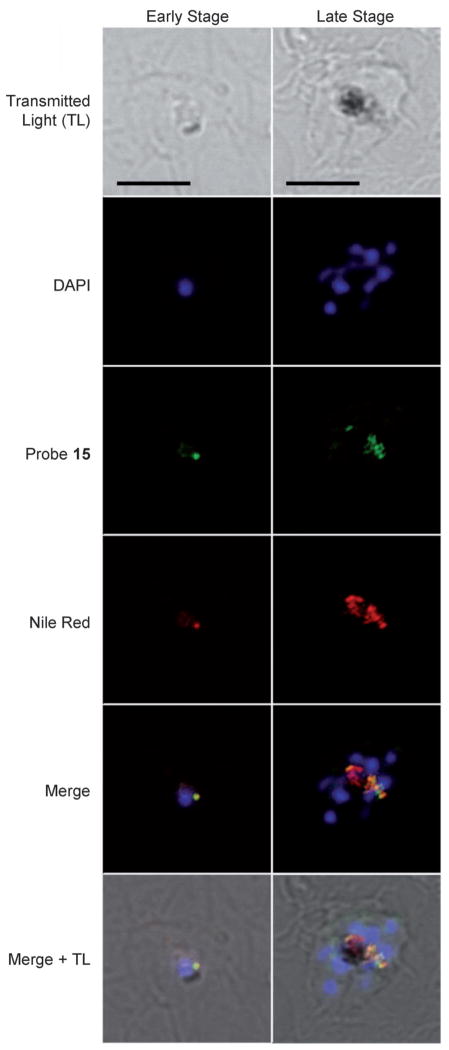
With active probes in hand, we turned to confocal microscopy to study the intracellular distribution of probe 15 in P. falciparum-infected erythrocytes. Fluorescence was observed only in P. falciparum-infected erythrocytes, which were distinguished by the presence of hemozoin (dark crystals, Figure 2). No fluorescence was observed if P. falciparum-infected erythrocytes were incubated with either control tag 14a or 14b, offering further evidence that 15 provided an effective mimic of natural product 1 (Figure 2). To determine its specific intracellular target, the histological localization of probe 15 was compared with that of fluorescent organelle and lipid probes by co-incubation and imaging using multi-channel fluorescence confocal microscopy. Incubation of infected erythrocytes with both 15 and Nile Red, a neutral lipid stain, repeatedly showed co-localization of the two stains, suggesting that 15 was associating with intracellular neutral lipids (Figure 3).[17] Previous studies by Pisciotta et al. established that heme crystallization occurs inside neutral lipid nanospheres, which encompass hemozoin in P. falciparum food vacuoles.[18] Therefore, molecular imaging studies with 15 and infected erythrocytes provided the first suggestion that bromophycolides could be targeting heme crystallization.
Figure 2.
Confocal microscopy images of live mixed-stage P. falciparum-infected erythrocytes incubated with either 10 μM bromophycolide A probe 15, 10 μM control tags 14a or 14b in complete medium. Hemozoin crystals were identified in the transmitted light image by dark crystals. Probe 15 localized only in P. falciparum-infected erythrocytes, as observed in the fluorescent image. No fluorescence was observed when P. falciparum-infected erythrocytes were incubated with either 14a or 14b. Red arrows indicate P. falciparum-infected erythrocytes. Echinocytes (altered red blood cells) are identified by the formation of small knoblike spicules on the membrane surface that cause the cell to transform from a biconcave disk into a seaurchin-like shape, an effect often observed with live cell imaging. Scale bar: 10 μm.
Figure 3.
Confocal images of fixed parasites incubated with 2 μM probe 15 (green), 30 nM Nile Red (red), and 70 nM DAPI (blue). Co-localization of 15 and Nile Red (neutral lipids) is indicated by the yellow merged image. Interestingly, whereas probe 15 emission spectra showed a maximum at λ 450 ± 10 nm (blue channel) in both complete medium and methanol, 15 fluoresced in the green channel after parasite fixation with paraformaldehyde. The latter effect could result from solvatochromic effects while localized within the lipid environment. Scale bars: 5 μm.
Because heme crystallization is a non-enzymatic process, parasite-free colorimetric high-throughput screens for heme crystallization inhibitors have been developed[19] and refined.[20] Natural product 1 and synthetic derivatives 7 and 8 were tested for heme crystallization inhibition and compared with the potent antimalarial drug amodiaquine as positive control. Bromophycolide A (1) inhibited heme crystallization with an IC50 value of 2.5 molar equivalents of compound to heme, and acetylated 7 was slightly more effective than 1, with an IC50 of 2.0 molar equivalents, consistent with growth inhibition data (Table 2); the IC50 for amodiaquine (1.2 molar equivalents), a fold decrease in antimalarial IC50 values, was found to be ineffective at inhibiting heme crystallization at relevant concentrations (Table 2). Collectively, these data provided compelling evidence for heme crystallization inhibition as a cellular target of the bromophycolides.
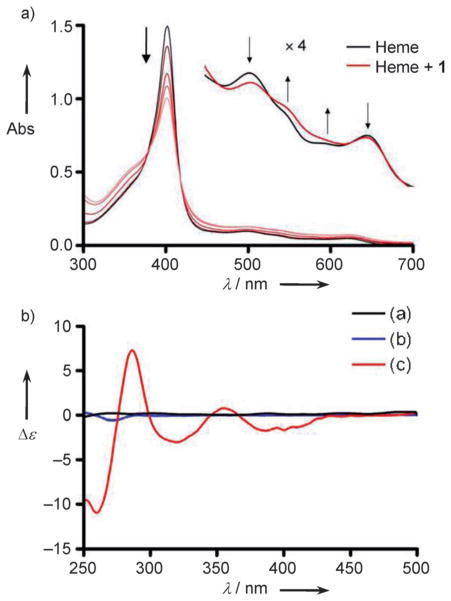
To further explore possible heme–bromophycolide chemical modes of interaction, the absorbance spectra of ferriprotoporphyrin IX (FeIIIPPIX), commercially available heme, was recorded in 20 mM HEPES (pH 7.4) containing 40 % DMSO, conditions that were previously demonstrated to evaluate monomeric FeIIIPPIX.[21] FeIIIPPIX gives a characteristic absorption spectrum with an intense Soret band positive control, was consistent with previously reported at λ 402 nm, the intensity of which was decreased by 45 % values.[19] Furthermore, methylated 8, which exhibited 20–50- upon titration of 1, suggesting that 1 forms a saturable complex with FeIIIPPIX (Figure 4a). Isosbestic points offer further evidence of FeIIIPPIX–1 complexation, and addition of 1 also caused notable changes in the Q-bands (Figure 4a, inset).
Figure 4.
Spectroscopic changes observed when FeIIIPPIX (heme) is incubated with various concentrations of bromophycolide A (1). a) Changes in the Soret band (λ 402 nm) and Q-bands of heme. The Soret band absorbance (Abs) decreases as concentrations ranging from 1:1 to 16:1 mol equiv 1–heme increases, as depicted with increasingly light shades of red. Inset: changes in the Q-bands of heme upon addition of 1. The arrows indicate the direction of change after addition of 1, and the isosbestic points support the presence of a FeIIIPPIX–1 complex. b) CD spectra of heme and 1. Heme is an achiral molecule and lacks a CD signal (black line, 12 μM). Chiral natural product 1 gives a weak Cotton effect at λ 270 nm (blue line, 72 μM), and addition of 1 to a solution of heme results in induced Cotton effects around 400 and 350 nm (red line, 12 μM hemin +72 μM 1). All spectra have been corrected for dilution by subtracting a reference cell. Conditions: 23 °C, 40 % DMSO, 20 mM HEPES buffer, pH 7.4.
Using a 1:1 complexation model between FeIIIPPIX and natural product 1,[21] the association constant for the FeIIIPPIX–1 complex was calculated to have a log K value of 4.52 ± 0.18, suggesting favorable interactions between these molecules (Supporting Information figure S2). This FeIIIPPIX–1 association constant is similar to other known heme crystallization inhibitors, such as chloroquine and quinine, the respective log K values of which are 5.52 ± 0.03 and 4.10 ± 0.02.[21] Titration of acetylated derivative 7 into a heme solution also resulted in a decrease in absorbance of the Soret band (25 %), whereas titration of methylated 8 resulted in virtually no (2%) decrease in the Soret band absorbance (Supporting Information figure S3). Because these binding studies are indicative of the chemical interactions of the bromophycolides with heme, the binding concentration at half-maximum (25 μM for 1 in the spectro-photometric titration experiment, Supporting Information figure S2) does not relate directly to the in vitro biological inhibition potency (0.50 μM for 1, Table 1).
Circular dichroism (CD) spectroscopic analyses indicated induced Cotton effects at λ 400 and 350 nm under similar conditions used for titration experiments, further supporting a measurable association between heme and natural product 1. Achiral FeIIIPPIX alone does not produce a CD signal, and chiral 1 alone exhibits a weak Cotton effect (Figure 4b). The induced Cotton effects of 1 with heme are consistent with previously reported interactions of tyrosine, and thus a phenolic hydroxy group, with heme in cytochrome c.[22] The spectrophotometric and CD spectroscopic data support hydroxy group coordination of 1 to the iron center of heme, as a hydroxy group ligation to heme resulted in both a decrease in the Soret band, changes in the Q-bands (Figure 4a), and a strongly induced Cotton effect (Figure 4b). Because the phenolic hydroxy group is absent in both acetylated derivative 7 and methylated derivative 8, strong Cotton effects are not observed, thus supporting our hypothesis that natural product 1 coordinates to heme through the phenolic hydroxy group (Supporting Information figure S4). The induced Cotton effects of the heme:7 titration could be a result of π–π interactions, which often result in weaker Cotton effects than a ligation.[23] However, acetylated 7 is more hydrophobic than natural product 1, thus a weaker CD spectrum of heme:7 may also be a result of lower solubility of compound 7 in the buffer solution. Methylated derivative 8 does not exhibit induced Cotton effects when titrated into a heme solution, consistent with the UV/Vis titration studies, as well as biological data (Supporting Information figure S4).
With one possible mode of action established for 1, potential protein targets within the malaria parasite were also explored. Taking advantage of the dual utility of probes 15 and 17 as cellular visualization agents and for affinity chromatography, P. falciparum lysates were screened for bromophycolide binding proteins. A selective monoclonal antibody (mAb) against the IAF tag[24] was available for immunoprecipitation (IP) with P. falciparum lysates incubated with probes 15 and 17. Probe 17 contains a longer linker relative to probe 15 (Scheme 1), and the increased spacing between the target compound and tag is sometimes advantageous for protein affinity experiments. Repeated IP experiments failed to result in isolation of any bromophycolide binding proteins. This anti-IAF-mAb affinity method has been successful with IP of protein targets in multiple systems in our research groups, from cancerous cells[24] to micro-invertebrates.[25] Although a negative IP experiment cannot prove the lack of a protein target, these data, along with the observation that probe 15 localized in a low protein environment (i.e., neutral lipid nanospheres), provide a strong suggestion that a prominent mode of action of 1 involves the inhibition of heme crystallization. This hypothesis is further supported by recent studies which indicate that samples of heme isolated from P. falciparum trophozoites, a feeding stage of the parasite, had minimal protein content, but were rich in neutral lipids.[18]
To date, we have isolated 33 unique C. serratus natural products, the IC50 values of which in P. falciparum ranged from 0.3 to >100 μM, with anticancer IC50 values ranging from 5 to 80 μM, providing a small library to analyze SAR trends (representative natural products 1–6, Figure 1).[13, 14, 26] Several semi-synthetic derivatives of 1 were also prepared (7–13) to further enhance SAR studies and to provide insight for potential future designs of bromophycolide-inspired compounds. In examining the natural product SAR trends, the most striking observation is related to the diterpene head. Whereas replacing the 1-bromoisopropyl at C14 with an isopropenyl functionality caused little change in antimalarial activity, substitution of bromine at C15 with a hydroxy group resulted in a dramatic loss of activity (Table 1), suggesting that a hydrogen bond donating group is poorly suited at this position (Supporting Information figure S1).
Moving to functional groups in the aliphatic macrocycle, synthetic modifications at C10 and C11 did not substantially affect activity. Replacing the bromine at C10 with chlorine resulted in a slight decrease in activity, although it should be noted that the configuration at C10 was inverted (in 4 versus 10). Regioisomerization of the double bond in the cyclohexene ring in both natural products and synthetic derivatives demonstrated only minor alterations in activity. Interestingly, elimination of all bromines from 1 (along with some other significant changes to the skeleton) only moderately decreased the IC50 value from 0.50 to 3.5 μM (1 versus 12), whereas the debrominated and rearranged natural product 6 was completely inactive. The final portion of 1 to investigate was the p-hydroxybenzoate group. As previously discussed, acylation of the phenol in 1 to 7 resulted in a slight increase in activity. Surprisingly, methylation of the phenol in 1 to 8 led to a 20-fold decrease in activity. Collectively, these data suggest that substitutions at positions C15 and C18 have the most striking effects on antimalarial activity.
The discovery and development of heme crystallization inhibitors remains an excellent antimalarial strategy, as heme crystallization is a physicochemical process that is independent of export mechanisms within the parasite.[27] Drug resistance most commonly involves mutations in a drug’s target protein or proteins that regulate efflux in order to remove the drug,[28] as the latter was shown for chloroquine resistance. Given that both 1 and 7 effectively inhibited chloroquine-resistant parasites (Dd2) with IC50 values of 377 and 304 nM, respectively (Table 2), it is clear that the efflux systems in chloroquine-resistant parasites do not recognize bromophycolides. Furthermore, 1 and 7 exhibited a respective 50- and 300-fold increase in activity for P. falciparum parasites over mammalian (Vero) cells (Table 2), providing support for these molecules as viable antimalarial leads. The molecular structures of the bromophycolides are notably different from other natural product heme crystallization inhibitors such as the xanthones and terpene isonitriles from marine sponges,[29] and well-known natural products quinine and artemisinin, thus suggesting a new class of therapeutic motifs. As historically evident, natural products continue to provide a rich resource for the discovery of unique antimalarial drugs, and these studies now suggest a new class of marine meroditerpenes as discreet inhibitors of P. falciparum.
Supplementary Material
Acknowledgments
This work was supported by the U.S. National Institutes of Health’s International Cooperative Biodiversity Groups program (Grant No. U01 TW007401). The authors thank MR4 for providing malaria parasites contributed by D. Carucci, A. Craig, and T. Wellems. We thank D. Hostetler, F. Fernandez, M. C. Sullards, and D. Bostwick for mass spectroscopic analyses; L. Gelbaum for NMR assistance; and E. Zhao and B. Hammer for use of their microplate reader.
Footnotes
Supporting information for this article is available on the WWW under http://dx.doi.org/10.1002/cmdc.201100252.
Contributor Information
Prof. Dr. Karine Le Roch, Email: karine.leroch@ucr.edu.
Prof. Dr. Julia Kubanek, Email: julia.kubanek@biology.gatech.edu.
References
- 1.Snow RW, Guerra CA, Noor AM, Myint HY, Hay SI. Nature. 2005;434:214–217. doi: 10.1038/nature03342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Newman DJ, Cragg GM. J Nat Prod. 2007;70:461–477. doi: 10.1021/np068054v. [DOI] [PubMed] [Google Scholar]
- 3.Hyde JE. Trends Parasitol. 2005;21:494–498. doi: 10.1016/j.pt.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.a) Klayman DL, Lin AJ, Acton N, Scovill JP, Hoch JM, Milhous WK, Theoharides AD. J Nat Prod. 1984;47:715–717. doi: 10.1021/np50034a027. [DOI] [PubMed] [Google Scholar]; b) Klayman DL. Science. 1985;228:1049–1055. doi: 10.1126/science.3887571. [DOI] [PubMed] [Google Scholar]
- 5.a) Noedl H, Se Y, Schaecher K, Smith BL, Socheat D, Fukuda MM ARC1 Study Consortium. New England J Med. 2008;359:2619–2620. doi: 10.1056/NEJMc0805011. [DOI] [PubMed] [Google Scholar]; b) Dondorp AM, Nosten F, Yi P, Das D, Phyo AP, Tarning J, Lwin KM, Ariey F, Hanpithakpong W, Lee SJ, Ringwald R, Silamut K, Imwong M, Chotivanich K, Lim P, Herdman T, An SS, Yeung S, Singhasivanon P, Day NP, Lindegardh N, Socheat D, White NJ. New England J Med. 2009;361:455–467. doi: 10.1056/NEJMoa0808859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goldberg DE, Slater AFG, Cerami A, Henderson GB. Proc Natl Acad Sci USA. 1990;87:2931–2935. doi: 10.1073/pnas.87.8.2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rosenthal PJ, Meshnick SR. Mol Biochem Parasitol. 1996;83:131–139. doi: 10.1016/s0166-6851(96)02763-6. [DOI] [PubMed] [Google Scholar]
- 8.Chai AC, Chevli R, Fitch CD. Biochemistry. 1980;19:1543–1549. doi: 10.1021/bi00549a600. [DOI] [PubMed] [Google Scholar]
- 9.Fidock DA, Nomura T, Talley AK, Cooper RA, Dzekunov SM, Ferdig MT, Ursos LMB, Sidhu ABS, Naude B, Deitsch KW, Su XZ, Wootton JC, Roepe PD, Wellems TE. Mol Cell. 2000;6:861–871. doi: 10.1016/s1097-2765(05)00077-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.a) Robert A, Meunier B. Chem Soc Rev. 1998;27:273–279. [Google Scholar]; b) Hong YL, Yang YZ, Meshnick SR. Mol Biochem Parasitol. 1994;63:121–128. doi: 10.1016/0166-6851(94)90014-0. [DOI] [PubMed] [Google Scholar]
- 11.Eckstein-Ludwig U, Webb RJ, van Goethem IDA, East JM, Lee AG, Kimura M, O’Neill PM, Bray PG, Ward SA, Krishna S. Nature. 2003;424:957–961. doi: 10.1038/nature01813. [DOI] [PubMed] [Google Scholar]
- 12.Li W, Mo WK, Shen D, Sun LB, Wang J, Lu S, Gitschier JM, Zhou B. PLoS Genet. 2005;1:329–334. doi: 10.1371/journal.pgen.0010036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.a) Kubanek J, Prusak AC, Snell TW, Giese RA, Hardcastle KI, Fairchild CR, Aalbersberg W, Raventos-Suarez C, Hay ME. Org Lett. 2005;7:5261–5264. doi: 10.1021/ol052121f. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Kubanek J, Prusak AC, Snell TW, Giese RA, Fairchild CR, Aalbersberg W, Hay ME. J Nat Prod. 2006;69:731–735. doi: 10.1021/np050463o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.a) Lane AL, Stout EP, Lin AS, Prudhomme J, Le Roch K, Fairchild CR, Franzblau SG, Hay ME, Aalbersberg W, Kubanek J. J Org Chem. 2009;74:2736–2742. doi: 10.1021/jo900008w. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Lin AS, Stout EP, Prudhomme J, Le Roch K, Fairchild CR, Franzblau SG, Aalbersberg W, Hay ME, Kubanek J. J Nat Prod. 2010;73:275–278. doi: 10.1021/np900686w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cervantes S, Stout EP, Prudhomme J, Engel S, Bruton M, Cervantes M, Carter D, Tae-Chang Y, Hay ME, Aalbersberg W, Kubanek J, Le Roch K. 2011 doi: 10.1186/1471-2334-12-1. unpublished results. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alexander MD, Burkart MD, Leonard MS, Portonovo P, Liang B, Ding X, Joullié MM, Gulledge BM, Aggen JB, Chamberlin AR, Sandler J, Fenical W, Cui J, Gharpure SJ, Polosukhin A, Zhang HR, Evans PA, Richardson AD, Harper MK, Ireland CM, Vong BG, Brady TP, Theodorakis EA, La Clair JJ. ChemBioChem. 2006;7:409–416. doi: 10.1002/cbic.200500466. [DOI] [PubMed] [Google Scholar]
- 17.The localization of probe 15 arose due to its natural product and not its pendant dyes, as compounds 14a and 14b did not localize within the neutral lipids. While the potential for dyes to localize within lipids and membranes is always an issue, the 7-dimethylamino-4-coumarin acetamide tag did not suffer from this issue here or within prior reports; see further references [16] and [24, 25].
- 18.Pisciotta JM, Coppens I, Tripathi AK, Scholl PF, Shuman J, Bajad S, Shulaev V, Sullivan DJ. Biochem J. 2007;402:197–204. doi: 10.1042/BJ20060986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ncokazi KK, Egan TJ. Anal Biochem. 2005;338:306–319. doi: 10.1016/j.ab.2004.11.022. [DOI] [PubMed] [Google Scholar]
- 20.Rush MA, Baniecki ML, Mazitschek R, Cortese JF, Wiegand R, Clardy J, Wirth DF. Antimicrob Agents Chemother. 2009;53:2564–2568. doi: 10.1128/AAC.01466-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Egan TJ, Mavuso WW, Ross DC, Marques HM. J Inorg Biochem. 1997;68:137–145. doi: 10.1016/s0162-0134(97)00086-x. [DOI] [PubMed] [Google Scholar]
- 22.a) Frauenhoff MM, Scott RA. Proteins Struct Funct Genet. 1992;14:202–212. doi: 10.1002/prot.340140207. [DOI] [PubMed] [Google Scholar]; b) Pietri R, Granell L, Cruz A, De Jesus W, Lewis A, Leon R, Cadilla CL, Garriga JL. Biochim Biophys Acta. 2005;1747:195–203. doi: 10.1016/j.bbapap.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 23.Fe–OH coordination of bromophycolides with heme may require that the C18-OAc derivative 7 undergoes deacylation by esterases, making 7 a putative prodrug of the hydrophobic natural product 1. This is seemingly unsupported by the cellular studies with probe 15 as shown in Figures 2 and 3, as release of the tags would fail to deliver the observed localization. However, proliferation assays need 72 h of compound/parasite incubation, whereas microscopy studies only require 1–2 h of probe exposure.
- 24.a) Hughes CC, MacMillan JB, Gaudencio SP, Fenical W, La Clair JJ. Angew Chem. 2009;121:739–741. doi: 10.1002/anie.200804107. [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew Chem Int Ed. 2009;48:728–732. doi: 10.1002/anie.200804107. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Hughes CC, Yang YL, Liu WT, Dorrestein PC, La Clair JJ, Fenical W. J Am Chem Soc. 2009;131:12094–12096. doi: 10.1021/ja903149u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stout EP, La Clair JJ, Snell TW, Shearer TL, Kubanek J. Proc Natl Acad Sci USA. 2010;107:11859–11864. doi: 10.1073/pnas.1006074107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.a) Lane AL, Stout EP, Hay ME, Prusak AC, Hardcastle K, Fairchild CR, Franzblau SG, Le Roch K, Prudhomme J, Aalbersberg W, Kubanek J. J Org Chem. 2007;72:7343–7351. doi: 10.1021/jo071210y. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Stout EP, Prudhomme J, Le Roch K, Fairchild CR, Franzblau SG, Aalbersberg W, Hay ME, Kubanek J. Bioorg Med Chem Lett. 2010;20:5662–5665. doi: 10.1016/j.bmcl.2010.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dorn A, Stoffel R, Matile H, Bubendorf A, Ridley RG. Nature. 1995;374:269–271. doi: 10.1038/374269a0. [DOI] [PubMed] [Google Scholar]
- 28.Srivastava IK, Morrisey JM, Darrouzet E, Daldal F, Vaidya AB. Mol Microbiol. 1999;33:704–711. doi: 10.1046/j.1365-2958.1999.01515.x. [DOI] [PubMed] [Google Scholar]
- 29.Kumar S, Guha M, Choubey V, Maity P, Bandyopadhyay U. Life Sci. 2007;80:813–828. doi: 10.1016/j.lfs.2006.11.008. [DOI] [PubMed] [Google Scholar]
- 30.Hawley SR, Bray PG, Mungthin M, Atkinson JD, O’Neill PM, Ward SA. Antimicrob Agents Chemother. 1998;42:682–686. doi: 10.1128/aac.42.3.682. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.