Abstract
The epidermal growth factor receptor (EGFR) is a central regulator of tumor progression in human cancers. Cetuximab is an anti-EGFR antibody that has been approved for use in oncology. Previously we investigated mechanisms of resistance to cetuximab using a model derived from the non-small cell lung cancer line NCI-H226. We demonstrated that cetuximab-resistant clones (CtxR) had increased nuclear localization of the EGFR. This process was mediated by Src family kinases (SFK), and nuclear EGFR played a role in resistance to cetuximab. To better understand SFK mediated nuclear translocation of EGFR, we investigated which SFK member(s) controlled this process as well as the EGFR tyrosine residues that are involved. Analyses of mRNA and protein expression indicated up-regulation of the SFK members Yes and Lyn in all CtxR clones. Further, immunoprecipitation analysis revealed that EGFR interacts with Yes and Lyn in CtxR clones, but not in cetuximab-sensitive (CtxS) parental cells. Using RNAi interference, we found that knockdown of either Yes or Lyn led to loss of EGFR translocation to the nucleus. Conversely, overexpression of Yes or Lyn in low nuclear EGFR expressing CtxS parental cells led to increased nuclear EGFR. Chromatin immunoprecipitation (ChIP) assays confirmed nuclear EGFR complexes associated with the promoter of the known EGFR target genes B-Myb and iNOS. Further, all CtxR clones exhibited up-regulation of B-Myb and iNOS at the mRNA and protein levels. siRNAs directed at Yes or Lyn led to decreased binding of EGFR complexes to the B-Myb and iNOS promoters based on ChIP analyses. SFKs have been shown to phosphorylate EGFR on tyrosines 845 and 1101 (Y845 and Y1101) and mutation of Y1101, but not Y845, impaired nuclear entry of the EGFR. Taken together, our findings demonstrate that Yes and Lyn phosphorylate EGFR at Y1101 which influences EGFR nuclear translocation in this model of cetuximab resistance.
Keywords: nuclear EGFR, SFK, Yes, Lyn
INTRODUCTION
Activation of the epidermal growth factor receptor (EGFR), a receptor tyrosine kinase (RTK), provides cells with potent growth and survival signals that enable tumors to manifest (1–3). Aberrant expression or activity of the EGFR is identified as a major etiological factor in many human epithelial cancers including colorectal cancer (CRC), head and neck squamous cell carcinoma (HNSCC), non-small cell lung cancer (NSCLC) and brain cancer (2, 4, 5). In the classical EGFR signaling pathway ligand binding to the EGFR allows for receptor homo- or hetero-dimerization at the plasma membrane. This interaction activates each receptor’s tyrosine kinase domain and induces autophosphorylation of each dimer’s cytoplasmic tail. The phosphorylated cytoplasmic tail of the EGFR serves as docking sites for numerous proteins that initiates key oncogenic pathways including the RAS/RAF/MEK/ERK and phosphatidylinositol 3-kinase (PI3K)-Akt pathways; however, the activation of src family tyrosine kinases (SFKs), Phospholipase C-gamma (PLC), Protein kinase C (PKC) and Signal Transducers and Activators of Transcription (STAT) proteins have also been documented (1, 6).
In addition to the classical signaling pathways initiated by the EGFR at the cell surface, there is now an emerging novel signaling pathway influenced by EGFR located in the nucleus. The full-length EGFR can be shuttled from the plasma membrane to the nucleus in a series of well-defined steps (7–9). These events include receptor internalization to the early endosome and interaction with importinβ1 via its tripartite nuclear localization sequence (NLS), followed by COPI-mediated retrograde trafficking to the Golgi apparatus and the endoplasmic reticulum (ER) (10, 11). Once in the ER the EGFR-importinβ1 complex moves to the outer nuclear membrane where importinβ1 interacts with nucleoporin 62 lining the nuclear pore channel to shuttle the EGFR-importinβ1 complex to the inner nuclear membrane. Here the complex interacts with the Sec61β translocon to be released from the membrane into the nucleus (12, 13).
Within the nucleus, EGFR serves as a transcriptional co-activator for a series of tumor promoting genes including cyclin D1, inducible nitric oxide synthase (iNOS), Aurora Kinase A, B-Myb, COX2, c-Myc, Breast Cancer Related Protein (BCRP) and GRP78 (14–21). Additionally, nuclear EGFR can phosphorylate and stabilize the proliferating cell nuclear antigen (PCNA) at the replication fork of the dividing cell (22), and activate DNA-PK to enhance DNA repair (23).
High levels of nuclear EGFR correlate with poor clinical outcome in breast cancer, oropharyngeal squamous cell cancer, ovarian cancer, and gallbladder cancer (24–28). Nuclear EGFR also contributes to cancer cells resistance to cetuximab (29), gefitinib (20), cisplatin and radiation therapy (30–33). Taken together these pieces of evidence suggest that nuclear EGFR plays a role in the promotion of cancer and provides a rationale for studying the mechanisms of EGFR nuclear translocation in order to target the nuclear functions of the EGFR.
It is well established that SFKs are necessary for full activation of the EGFR (34, 35). Src kinase is the prototype member of this family of non-RTKs that include Yes, Fyn, Lyn, Lck, Hck Fgr, Blk and Yrk (36, 37). These SFKs mediate mitogenic signals from a variety of RTKs (38, 39). It has been observed that SFKs can phosphorylate EGFR at both tyrosine 845 (Y845) and tyrosine 1101 (Y1101). EGFR Y845 is located in the activation loop of the kinase domain that is highly conserved among other RTKs. Phosphorylation of EGFR Y845 appears to be critical for EGFR-mediated mitogenesis, and is critical for the phosphorylation and activation of the STAT5b transcription factor (34, 40, 41). The second known Src-mediated phosphorylation site is Y1101, which lies within the carboxyl-terminal region of the EGFR; however the function of Y1101 has not been fully elucidated (34). Oncogenic cooperation between Src and EGFR has been well established in breast cancer (34, 42), glioblastoma (43), HNSCC and NSCLC (44–47).
We established six clonal CtxR variants of the NCI-H226 NSCLC line (29, 48, 49). In previous reports we found that CtxR clones had a dramatic increase in nuclear EGFR localization, in addition to having increased SFK activity (29, 44). Further, we reported that the SFK inhibitor dasatinib (BMS-354825, Sprycel™) could 1) block SFK activation, 2) decrease nuclear EGFR translocation, 3) increase plasma membrane levels of the EGFR, and 4) re-sensitize CtxR cell lines to cetuximab. Collectively these findings suggest that SFKs play a crucial role in nuclear translocation of the EGFR in this model of cetuximab-resistance. However, the specific SFKs involved in the mediation of EGFR nuclear translocation and how they mediate this process are unknown.
In the current study we demonstrate that CtxR clones had increased expression of the SFKs Yes and Lyn. Both Yes and Lyn were strongly associated with EGFR in CtxR clones as compared to the CtxS parental cell line. Depletion of either Yes or Lyn kinase decreased EGFR nuclear translocation, and reduced phosphorylation at Y845 and Y1101 of the EGFR. Reciprocally, overexpression of Yes or Lyn increased EGFR nuclear translocation in CtxS parental cell line. Furthermore, mutation of Y1101 of the EGFR impaired its nuclear translocation. Collectively these data suggest that Yes, Lyn and Y1101 of the EGFR are involved in EGFR nuclear translocation in this model of acquired resistance to cetuximab.
RESULTS
The SFK inhibitor Dasatinib blocks nuclear translocation of the EGFR
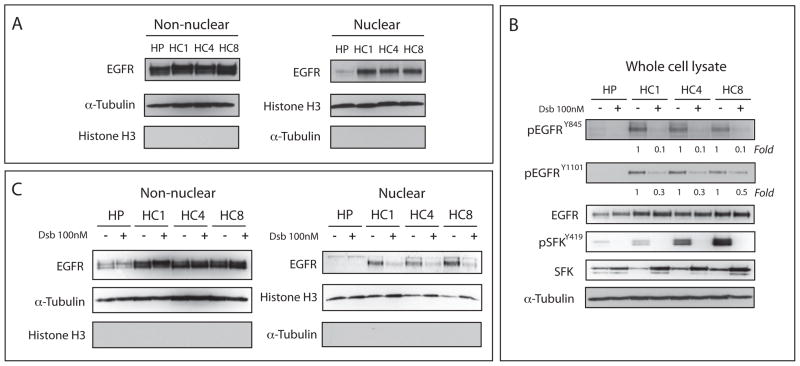
We have previously reported that CtxR clones have increased nuclear EGFR and activation of SFKs (Figure 1A, 1B) (29, 48). Using this model, we determined the effects of the SFK inhibitor dasatinib on the phosphorylation status of the EGFR in three CtxR clones (HC1, HC4 and HC8) and the CtxS parental clone (HP) after treatment with 100 nM of dasatinib for 24 hours. Dasatinib inhibited the full activation of SFKs as indicated by the loss of phospho-Y419 and decreased the phosphorylation of EGFR at the known SFK phosphorylation sites Y845 and Y1101 (Figure 1B). Also, treatment with dasatinib led to modest increases in steady state expression of total SFKs in all CtxR clones.
Figure 1. SFKs are essential for EGFR translocation to the nucleus.
(A) CtxR clones (HC1, HC4 and HC8) have increased nuclear EGFR localization as compared to Ctxs cell line (HP). Cells were harvested for non-nuclear and nuclear protein, and fractionated on SDS-PAGE followed by immunoblotting for indicated proteins. Histone H3 and α-tubulin were used as loading and purity controls for the nuclear and non-nuclear fractions, respectively. (B) Dasatinib decreased EGFR activity in CtxR cells. CtxR clones (HC1, HC4 and HC8) and CtxS HP cell line were treated with 100 nM dasatinib for 24 hr. Cells were harvested and protein lysates were fractionated on SDS-PAGE followed by immunoblotting for the indicated proteins. α-tubulin was used as a loading control. Expression was quantitated using ImageJ software. (C) Dasatinib can inhibit EGFR nuclear translocation in CtxR clones. CtxR clones (HC1, HC4, and HC8) and CtxS cell line were treated with 100 nM dasatinib for 24 hr. Cells were harvested for non-nuclear and nuclear protein, and fractionated on SDS-PAGE followed by immunoblotting for indicated proteins. Histone H3 and α-tubulin were used as loading and purity controls for the nuclear and non-nuclear fractions, respectively.
To determine the effects of dasatinib treatment on nuclear translocation of EGFR, we treated the CtxR clones and the CtxS parental clone with 100 nM of dasatinib for 24 hours followed by nuclear fractionation. As illustrated in Figure 1C, dasatinib treatment reduced EGFR nuclear translocation in CtxR clones. The CtxS parental clone has very low levels of nuclear EGFR and dasatinib treatment had no effect. Thus, the inhibition of SFK activity decreased the phosphorylation of EGFR at Y845 and Y1101 as well as impaired nuclear entry of the EGFR. These results suggested that SFK phosphorylation of EGFR may play a role in inducing its nuclear translocation.
Yes and Lyn are overexpressed and associate with the EGFR in CtxR clones
Based on our previous findings with clonal CtxR variants of the NCI-H226 NSCLC line we hypothesized that SFK member(s) may regulate EGFR nuclear translocation. To identify the specific SFKs that are necessary for EGFR nuclear translocation, we performed microarray analysis comparing CtxS HP parental cells to the three CtxR clones (HC1, HC4 and HC8). Microarray analysis demonstrated a ~4-fold up-regulation of Yes and Lyn kinases and ~3-fold down-regulation of Src kinase in all of three CtxR cells (data not shown). Other SFK family members did not exhibit significant expression level changes in the three CtxR clones (HC1, HC4 and HC8) compared to sensitive parental line (HP).
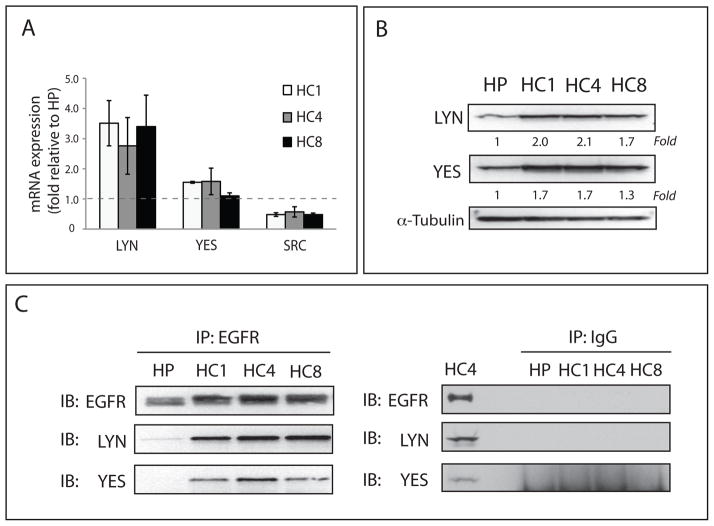
To validate the microarray findings we performed qPCR analysis. These results confirmed the microarray data indicating up-regulation of Lyn (~ 3-fold) and Yes (~1.5 fold), and down-regulation of Src kinase (~2-fold) in all three CtxR clones (Figure 2A). Next, we examined if these increased mRNA levels reflected total protein levels in CtxR when compared to CtxS cells. We found that total protein levels of Yes and Lyn were increased 1.3- to 2.1-fold in CtxR clones compared to parental cells (Figure 2B). Finally, we investigated whether EGFR associated with Yes and Lyn. Immunoprecipitation analysis of EGFR binding partners indicated that EGFR displayed increased association to Yes and Lyn in all three CtxR clones as compared to the CtxS HP cell line (Figure 2C). Collectively, these results indicate that Yes and Lyn are up-regulated in CtxR clones and have increased association with the EGFR.
Figure 2. Yes and Lyn SFK family members are overexpressed in CtxR cells.
(A) Yes and Lyn mRNA is up-regulated in CtxR clones (HC1, HC4, and HC8). Lyn (3.6~4.5-fold), Yes (~1.5-fold) and Src (−2.3~−3.5-fold) mRNA expression levels were compared to that of the CtxS cell line (HP) by qPCR. (B) Total protein levels of Yes and Lyn were increased (1.3~2.1-fold) in CtxR clones (HC1, HC4, and HC8) as compared to the CtxS cell line (HP). Cells were harvested and protein lysates were fractionated on SDS-PAGE followed by immunoblotting for the indicated proteins. α-tubulin was used as a loading control. Protein expression was quantitated using ImageJ software. (C) Analysis of EGFR binding partners in CtxR cells using immunoprecipitation assay indicated that EGFR displayed increased binding with Yes and Lyn as compared to the CtxS parental cell line. Cells were harvested and EGFR or IgG were immunoprecipitated with anti-mouse EGFR antibody or normal mouse IgG. The immunoprecipitate complexes were fractionated on SDS-PAGE followed by immunoblotting for indicated proteins.
Yes and Lyn are necessary for nuclear translocation of EGFR in CtxR cells
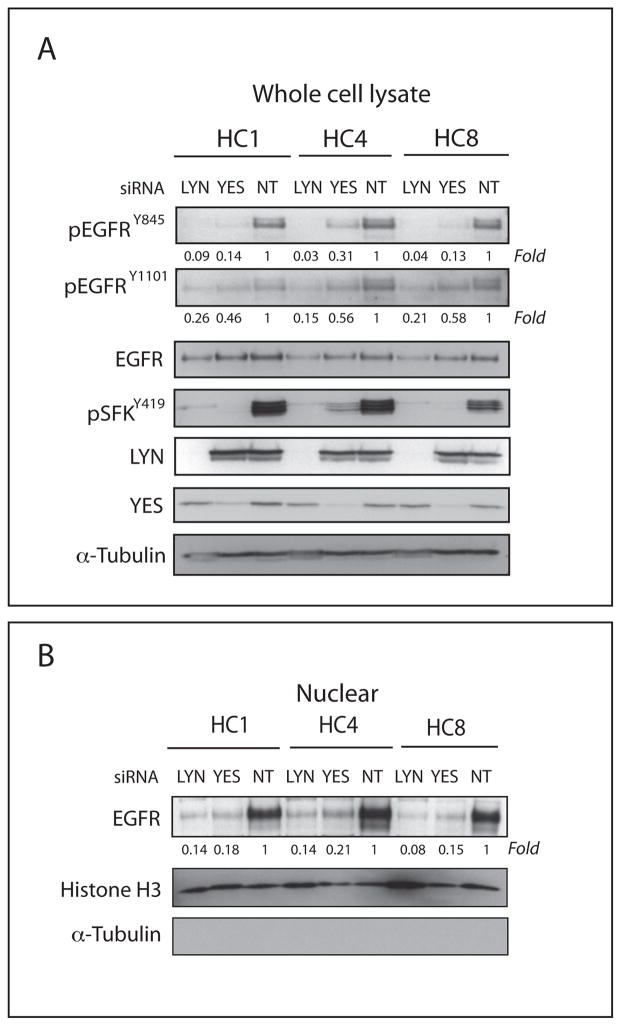
To further investigate if Yes and/or Lyn expression altered EGFR activation and nuclear translocation we performed gene-silencing experiments using siRNA directed against Yes or Lyn in CtxR clones (HC1, HC4 and HC8). CtxS cells were not included in these siRNA studies because the cells have negligible levels of nuclear EGFR. After treatment with siYES or siLYN in CtxR clones for 72 hours we observed decreased phosphorylation of EGFR Y845 (70–99%) and EGFR Y1101 (40–85%) relative to control non-targeting siRNA (NT) (Figure 3A). Moreover, siYES and siLYN decreased the nuclear localization of EGFR in CtxR clones (Figure 3B).
Figure 3. siYES and siLYN can reduce the nuclear localization of the EGFR.
(A) siYES and siLYN decreased phosphorylation of EGFR tyrosine 845 (Y845), tyrosine 1101 (Y1101) and SFK tyrosine 419. Cells were harvested 72 hr after treatment with either siYES or siLYN, and protein lysates were fractionated on SDS-PAGE followed by immunoblotting for the indicated proteins. The non-targeting siRNA (NT) was used as a control. α-tubulin was used as a loading control. Protein expression was quantitated using ImageJ software. (B) siYES and siLYN can reduce the nuclear localization of the EGFR. Cells were harvested for non-nuclear and nuclear protein, and fractionated on SDS-PAGE followed by immunoblotting for indicated proteins after 72hr treatment with either siYES or siLYN. The non-targeting siRNA (NT) was used as a control. Histone H3 and α-tubulin were used as loading and purity controls for the nuclear and non-nuclear fractions, respectively. Protein expression was quantitated using ImageJ software.
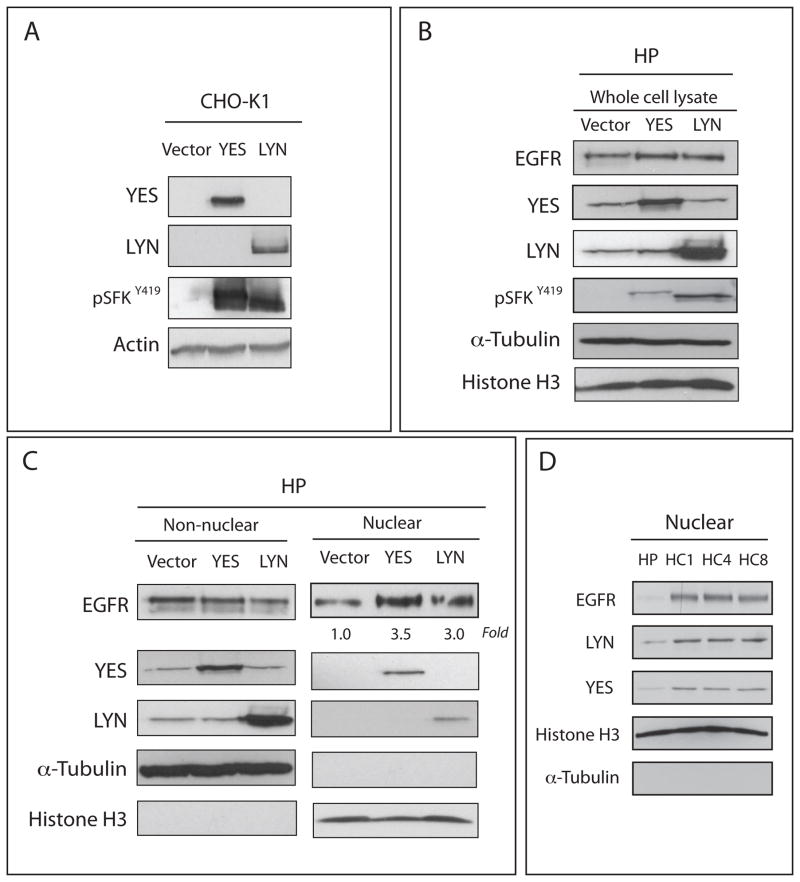
Knockdown studies of Yes or Lyn in cells with high nuclear EGFR expression led to decreased nuclear EGFR levels. Therefore, we hypothesized that the overexpression of Yes or Lyn could increase the level of nuclear EGFR in a cell line with low levels of EGFR in the nucleus. To test this hypothesis we overexpressed Yes or Lyn in CtxS cells, which express low-levels of nuclear EGFR. First, Yes and Lyn were cloned into mammalian expression vectors, expressed in CHO-K1 cells and characterized for increased total Yes and Lyn protein expression and activity (Figure 4A). Comparable increases in Yes and Lyn expression and activation were observed in CtxS cells after transfection compared to vector only (Figure 4B). Consistent with siRNA observations, transient transfection of Yes or Lyn into CtxS cells significantly increased (3~3.5 fold) nuclear EGFR translocation (Figure 4C). Interestingly, Yes and Lyn were also detected in nucleus of HP cells after transfection. To determine if CtxR cells with increased of nuclear EGFR also express more Yes and Lyn in the nucleus compared to CtxS cells, we determined nuclear Yes and Lyn levels in CtxR clones and CtxS cells. As seen with EGFR, increased levels of both Yes and Lyn were found in the nucleus of CtxR clones compared to CtxS cells (Figure 4D). These siRNA and overexpression results suggest that Yes and Lyn play a role in EGFR nuclear translocation.
Figure 4. YES and LYN can induce the nuclear localization of the EGFR.
(A) Increased levels of total protein expression of Yes and Lyn were detected in Chinese hamster ovary (CHO-K1) cells post 48hr transfection with either a Yes or Lyn mammalian expression vector. (B) Increased Yes and Lyn expression as well as increased levels of pSFK Y419 were observed in CtxS cells after 48hr transfection compared to vector only. (C) Nuclear EGFR translocation was increased by overexpression of Yes and Lyn. Cells were transfected with a Yes or Lyn mammalian expression vector and harvested for non-nuclear and nuclear protein. Each protein was fractionated on SDS-PAGE followed by immunoblotting. Histone H3 and α-tubulin were used as loading and purity controls for the nuclear and non-nuclear fractions, respectively. (D) CtxR clones (HC1, HC4 and HC8) have increased nuclear Yes and Lyn localization as compared to Ctxs cell line (HP). Cells were harvested for non-nuclear and nuclear protein, and fractionated on SDS-PAGE followed by immunoblotting for indicated proteins. Histone H3 and α-tubulin were used as loading and purity controls for the nuclear and non-nuclear fractions, respectively.
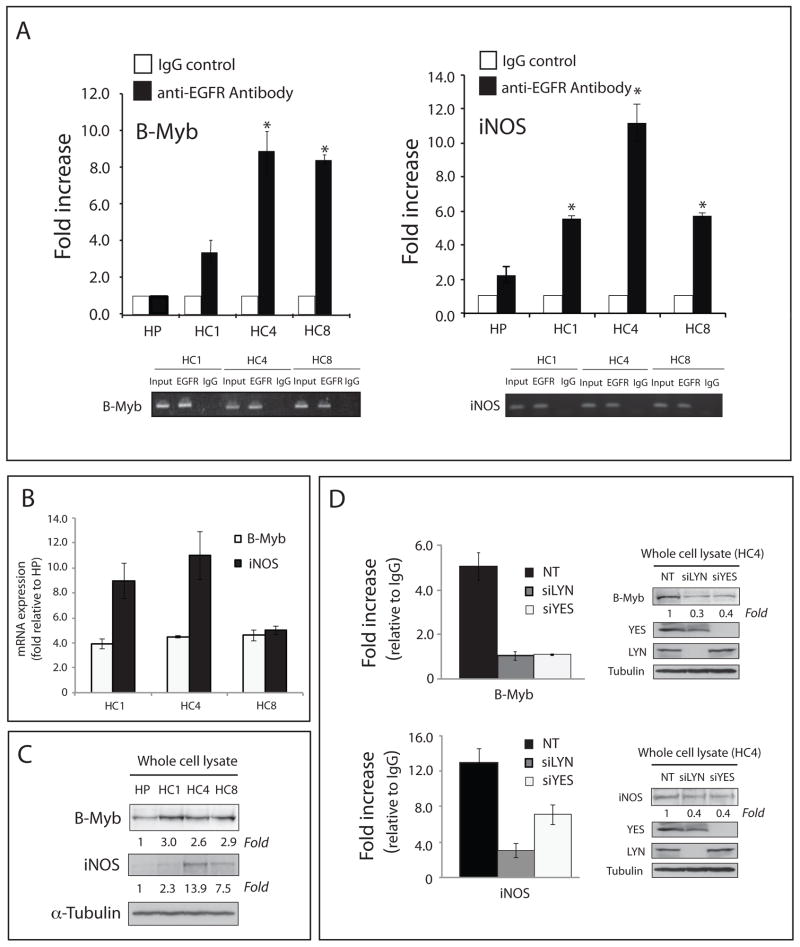
Depletion of Yes or Lyn decreases binding of nuclear EGFR complexes to the B-Myb and iNOS promoter regions
Nuclear EGFR and various transcription factor complexes have been shown to bind promoter regions and regulate the transcription of multiple genes including cyclin D1, iNOS, B-Myb, Aurora Kinase A, COX2, c-Myc, BCRP and GRP78 (14–21). To confirm that nuclear EGFR complexes in CtxR clones bound to known EGFR target gene promoters we performed Chromatin immunoprecipitation-quantitative PCR (ChIP-qPCR) analysis of the B-Myb and iNOS promoters. We demonstrated that CtxR clones have increased EGFR association with the B-Myb (3–9-fold increase in EGFR binding) and iNOS (6–12-fold increase in EGFR binding) promoter regions as compared to CtxS parental cell line (Figure 5A). These results indicate that nuclear EGFR complexes bind known EGFR target promoters in CtxR clones more strongly than in CtxS HP cells.
Figure 5. Yes and Lyn influence the binding of EGFR complexes to the B-Myb and iNOS promoter regions.
(A) EGFR regulated gene promoter regions are more strongly associated with EGFR in CtxR clones (HC1, HC4, and HC8) as compared to CtxS cells (HP). EGFR-ChIP and subsequent qPCR from the ChIP sample for the presence of B-Myb and iNOS promoter sequences. Data points are represented as mean ± SEM (n = 3). *p< 0.05. qPCR specificity for the B-Myb and iNOS promoter regions was also confirmed by agarose gel electrophoresis of semi-quantitative PCR products. (B) B-Myb and iNOS mRNA levels were significantly up-regulated in CtxR cells (HC1, HC4, and HC8) as compared to the CtxS cell line (HP) by qPCR. The mRNA expression of B-Myb and iNOS in HP, HC1, HC4 and HC8 were determined by qPCR. Data points are represented as mean ± SEM (n = 3). (C) B-Myb and iNOS protein levels were increased in CtxR cells (HC1, HC4, and HC8) as compared to the CtxS cell line (HP) by immunoblot analysis. Cells were harvested and protein lysates were fractionated on SDS-PAGE followed by immunoblotting for the indicated proteins. α-tubulin was used as a loading control. (D) Loss of Yes or Lyn prevents EGFR association with B-Myb and iNOS promoters, and corresponds with a decrease in protein expression. EGFR-ChIP and subsequent qPCR from the ChIP sample for the presence of B-Myb and iNOS promoter sequences. The non-targeting siRNA (NT) was used as a control. B-Myb and iNOS protein levels were decreased in HC4 after siYES or siLYN treatment by immunoblot analysis. Cells were harvested after treatment with siLYN or siYES for 72 hr and protein lysates were fractionated on SDS-PAGE followed by immunoblotting for the indicated proteins. α-tubulin was used as a loading control.
Given the high binding of nuclear EGFR complexes to the B-Myb and iNOS promoters we performed qPCR to determine whether this binding resulted in increased expression of B-Myb and iNOS genes as previously reported (15, 16). Results in Figure 5B indicated that B-Myb mRNA expression was increased approximately 4-fold in all CtxR clones as compared to CtxS parental cells. Whereas, iNOS mRNA expression was increased 4–11-fold in CtxR clones when compared to CtxS parental cells. Furthermore, B-Myb protein expression was up-regulated approximately 3-fold and iNOS protein expression was up-regulated 2–14-fold in CtxR clones (Figure 5C). Using the CtxR HC4 clone we demonstrate that silencing of Yes or Lyn using siRNA reduced nuclear EGFR complex formation with the B-Myb and iNOS promoters as detected by ChIP-qPCR (Figure 5D). Additionally, the protein expression of both B-Myb and iNOS were decreased (60–70 %) after siYES or siLYN transfection compared to control non-targeting siRNA in HC4 (Figure 5D). Collectively, these data demonstrate that CtxR clones with high levels of nuclear EGFR associate more strongly with known EGFR regulated promoter regions, and that these association (demonstrated with B-Myb and iNOS) can be prevented upon depletion of Yes or Lyn.
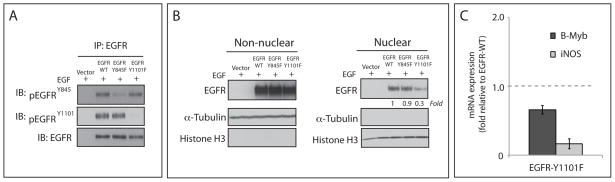
EGFR Y1101 is involved in nuclear translocation
Analysis of known SFK phosphorylation sites on the EGFR showed increased phosphorylation of Y845 and Y1101 in CtxR clones (Figure 1B). However, the relevance of these two tyrosine sites for EGFR nuclear translocation is unknown. To determine if phosphorylation of one or both of these tyrosine residues is involved in EGFR nuclear translocation, MCF-7 breast cancer cells (which express very low EGFR levels) were transiently transfected with cDNAs encoding wild-type EGFR (WT) or the following EGFR mutants: EGFR-Y845F or EGFR-Y1101F. Immunoprecipitation of WT and mutant EGFR followed by immunoblotting with antibodies directed against pEGFRY845 or pEGFRY1101 showed reduced phosphorylation of Y845 and Y1101. (Figure 6A). To induce nuclear translocation of the EGFR we treated the transfected cells with EGF for 45 minutes prior to cell lysis and nuclear fractionation. The results of this experiment indicated that EGF was able to induce nuclear translocation of EGFR in both EGFR-WT and EGFR-Y845F mutant cells (Figure 6B). However, EGF-induced EGFR nuclear translocation was diminished ~70% in EGFR-Y1101F mutant cells as compared to EGFR-WT expressing cells (Figure 6B). Furthermore qPCR analysis revealed that B-Myb and iNOS mRNA expression were downregulated in cells transfected with EGFR-Y1101F mutant compared to EGFR-WT transfected cells (Figure 6C). Collectively, these data suggest that the phosphorylation of Y1101 is important for the nuclear translocation of EGFR while the phosphorylation of Y845 does not appear to be essential for this process.
Figure 6. Phosphorylation of EGFR at Y1101 is involved in the nuclear localization of EGFR.
(A) Immunoprecipitation of total EGFR followed by immunoblotting with anti-pEGFR Y845, anti-pEGFR Y1101 or pan-EGFR antibodies. MCF7 Cells overexpressing EGFR-WT, EGFR-Y845F or EGFR-Y1101F were harvested after stimulation with 100 ng/mL of EGF for 45 min. A total of 500 ug of cell lysate was immunoprecipitated with pan-EGFR antibody. The immunoprecipitates were fractionated on SDS-PAGE followed by immunoblotting for the indicated proteins. (B) Mutation of Y1101, but not Y845, reduces the nuclear localization of EGFR in MCF-7 breast cancer cells. Cells were transiently transfected with plasmids encoding the EGFR wild-type (WT), EGFR-Y845F, EGFR-Y1101F or vector only. 48hr post-transfection the cells were incubated with EGF (100 ng/ml) for 45min, harvested for whole cell lysate, non-nuclear, and nuclear protein, and fractionated on SDS-PAGE followed by immunoblotting for indicated proteins. Histone H3 and α-tubulin were used as loading and purity controls for the nuclear and non-nuclear fractions, respectively. (C) B-Myb and iNOS mRNA levels were down-regulated in HC4 cells transfected with EGFR-Y1101F mutant compared to EGFR-WT transfected cells by qPCR. Cells were transiently transfected with plasmids encoding the EGFR wild-type (WT) or EGFR-Y1101F. 24hr post-transfection the cells were treated with EGF (100 ng/ml) for 45min, and harvested for RNA. The mRNA expression of B-Myb and iNOS was determined by qPCR.
Discussion
The nuclear localization of receptor tyrosine kinases (RTKs) have been observed for over 20 years, however only in the last 10 years has research begun to focus on how they translocate from the cell surface to the nucleus and what functions they perform there. All four HER family members have been identified in the nucleus of various types of human cancer cells and tumor specimens (9, 50–53). Currently, eight target genes of nuclear EGFR have been identified (14–21), and nuclear EGFR has been correlated with resistance to cetuximab, radiation, cisplatin and gefitinib therapies (20, 23, 29–33). Collectively, these results suggest an emerging role of the nuclear EGFR signaling network in cancer progression and response to therapeutic modalities.
Several studies have examined how EGFR moves from the plasma membrane to the nucleus of the cell. It has been shown that the full-length EGFR can be shuttled from the plasma membrane to the nucleus through associations with importinβ1, the nuclear pore complex, and the Sec61β translocon (7, 10, 13). Despite this mechanism of EGFR nuclear translocation, the early events at the plasma membrane that may serve as critical initiating signals for the movement of the EGFR to the nucleus have yet to be defined and form the basis of the current study.
To further elucidate the molecular requirements for EGFR nuclear transport we utilized a previously established model of acquired resistance to cetuximab in the NCI-H226 NSCLC cell line (48). In this model, cetuximab-resistant cells were observed to have increased levels of nuclear EGFR as compared to their cetuximab-sensitive parental cells, making it an ideal model for investigating events involved in nuclear translocation of the EGFR (29). Additionally, cetuximab-resistant cell lines were shown to have increased expression and activity of SFKs (44). Further investigation using dasatinib, an inhibitor of SFKs, demonstrated that SFK activity was necessary for the nuclear transport of EGFR in this model of cetuximab-resistance (29). In the current study, we identified Yes and Lyn to have increased expression and association with the EGFR (Figure 2). This result is consistent with other reports identifying Yes and Lyn interaction and activation of the EGFR (54–57). In addition, loss of Yes and Lyn expression using siRNA technology, led to reduced phosphorylation of Y845 and Y1101 of the EGFR and more importantly impaired nuclear EGFR accumulation (Figure 3). Consistent with this observation, overexpression of Yes and Lyn in CtxS cells that express low-levels of nuclear EGFR significantly increased (3~3.5 fold) nuclear EGFR translocation (Figure 4B). ChIP assays demonstrated that nuclear EGFR complexes bind to B-Myb and iNOS promoter regions and siYES and siLYN decreased binding to these promoters (Figure 5). Mutagenesis studies of Y845 and Y1101 indicated that Y1101, not Y845, might be necessary for nuclear translocation of the EGFR from the membrane to the nucleus (Figure 6). Recently, Jaganathan et al. reported that Src and EGFR associate in the nucleus with the transcription factor STAT3 to regulate the expression of the c-Myc gene in pancreatic cancer (19). Consistent with this report, we found that Yes or Lyn not only increased the levels of nuclear EGFR but also had increased nuclear localization themselves, suggesting that they may have been imported into the nucleus with the EGFR. Collectively, these studies provide evidence for the role of SFKs in mediating nuclear translocation of the EGFR. However, it remains to be investigated whether Yes or Lyn are solely responsible for this nuclear translocation, or if SFKs exhibit a functional redundancy where the overexpression of one or more SFK may result in the induction of nuclear EGFR in various cancers.
Y845 and Y1101 of the EGFR are phosphorylated by SFKs (34). Biscardi et al. utilized a GST bound SH2 domain of the c-Src protein to demonstrate its specific binding to the EGFR via affinity chromatography (34). Subsequently, these investigators identified and validated that EGFR was indeed phosphorylated by c-Src at Y845 and Y1101. Breast cancer cell lines with high levels of Src activity also had increased levels of phospho Y845 and Y1101 of the EGFR. Researchers further showed that phospho-Y845 was necessary for full EGFR activation and EGF-induced DNA synthesis (34). This study represented a landmark finding by identifying novel Src phosphorylation sites on the EGFR and the role of tyrosine 845 in the complete activation of the EGFR. Further studies looking at the function of EGFR Y845 demonstrated that Y845 mediated EGFR binding to the mitochondrial protein cytochrome c oxidase subunit II at the mitochondria; however, EGFR Y845 was not necessary for its movement to the mitochondria (58). These findings support our data that EGFR Y845 may not be required for the intracellular trafficking of the EGFR.
In the current study, we corroborate findings of Biscardi et al. by showing that Y845 and Y1101 are Src specific phosphorylation sites through the use of the SFK inhibitor dasatinib (Figure 1B). In addition, siRNA directed towards Yes and Lyn decreased the phosphorylation of EGFR Y845 and Y1101 (Figure 3A). Our data further suggests that Y1101, not Y845, may be a critical molecular determinant in the localization of nuclear EGFR as indicated by site-directed mutagenesis (Figure 6B). It should be noted, however, that mutation of Y1101 did not completely block translocation of the EGFR to the nucleus, suggesting that other post-translational modifications of the EGFR may be necessary. Recent evidence has identified another key phosphorylation site on the EGFR, serine 229 (S229), as being necessary for EGFR translocation to the nucleus (20). It was reported that the serine/threonine kinase AKT can influence the nuclear translocation of the EGFR by phosphorylating S229 on the EGFR in a model of gefitinib resistance. In this model, gefitinib-resistant A431 cells have both increased AKT activity and increased nuclear EGFR as compared to gefitinib-sensitive A431 cells. Using an antibody that recognizes the phosphorylated consensus motif of AKT substrates and subsequent mass spectrometry, Huang et al. revealed that EGFR was phosphorylated by AKT at S229. Inhibition of AKT kinase activity prevented this phosphorylation event, and decreased the nuclear transport of EGFR providing evidence for the role of alternative kinases and post-translational modifications of the EGFR that indeed affect its nuclear translocation. Collectively, these findings suggest that the phosphorylation of the EGFR on Y1101 by Yes and Lyn together with AKT phosphorylation of S229 may be critical molecular determinants that influence the nuclear localization of the EGFR.
EGFR is tightly linked to the etiology of HNSCC, NSCLC, CRC, breast and brain cancers. Accordingly, five EGFR inhibitors, three tyrosine kinase inhibitors, and two monoclonal antibodies have been developed for clinical use to inhibit EGFR activation and downstream signaling. Despite the successes of these agents, many tumors do not respond to EGFR inhibition, or eventually become resistant to this therapeutic strategy. Accumulating evidence suggests that nuclear EGFR plays a role in resistance to radiation, cetuximab, cisplatin and gefitinib therapies (20, 23, 29–33). The mechanisms for how nuclear EGFR leads to this resistance are not clear. However work from our laboratory suggests that nuclear translocation can protect EGFR from the inhibitory effects of cetuximab causing resistance to this therapy (29). The results presented in this study provide a potential mechanism for the key molecules involved in nuclear localization of the EGFR providing rational targets to prevent nuclear translocation and thus nuclear function of the EGFR.
In summary, the data presented in the current study has identified the SFKs Yes and Lyn to play a crucial role in nuclear translocation of the EGFR in a model of cetuximab-resistance. In addition, the SFK phosphorylation site Y1101 of the EGFR appears to be involved in translocation of the EGFR from the plasma membrane to the nucleus. These findings are of instrumental value in understanding the molecular requirements for nuclear EGFR transport, and for potentially targeting nuclear EGFR in the future.
MATERIALS AND METHODS
Cell lines
The human NSCLC line NCI-H226, the human breast cancer line MCF-7 and Chinese hamster ovary K1 (CHO-K1) cells were purchased from ATCC (Manassas, VA, USA). The cells were maintained in 10% fetal bovine serum (FBS) in RPMI-1640 for H226, DMEM/F12K for MCF-7 and F12K for CHO-K1 (Mediatech Inc., Manassas, VA, USA) with 1% penicillin and streptomycin. The development of cells with acquired resistance to cetuximab has been previously described (48).
Plasmid constructs and transfection
EGFR wild-type (WT), Y845F and Y1101F mutants, were kindly provided from Dr. Julie Boerner (Wayne State University School of Medicine, Karmanos Cancer Institute, MI). The presence of 845F and 1101F mutations were confirmed by DNA sequencing. For transient transfections, MCF-7 cells were transfected with plasmid DNA for each construct or pcDNA3.1 vector using Lipofectamine LTX and Opti-MEM I (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s recommendations. Either 24 hr (for RNA) or 48 hr (for protein) after transfection, EGF (100 ng/ml, Millipore, Billerica, MA, USA) was added to the plates for 45 min. Cells were collected, isolated RNA or fractionated and screened for their EGFR expression levels by quantitative PCR (qPCR) or immunoblotting as described below. For siRNAs, CtxR cells (HC1, HC4 and HC8) were transiently transfected with siYES (ON-TARGETplus SMARTpool YES1: L-003184-00, Dharmacon, Lafayette, CO, USA) or siLYN (ON-TARGETplus SMARTpool LYN: L-003153-00) using Lipofectamine RNAiMAX according to the manufacture’s instructions (Invitrogen). The non-targeting siRNA (ON-TARGETplus Non-targeting Pool, D-001810-10) was obtained from Dharmacon as a control. Cells were then lysed for analysis of protein knockdown by immunoblotting 72hr after siRNA transfection. Wild-type human YES (source ID: 5260751) and LYN (source ID: 8992174) cDNAs were purchased from Open Biosystems (Lafayette, CO, USA) and cloned into the NOTI/PACI restriction sites of the pQCXIP expression vector (Clontech, Mountain View, CA, USA). YES-PQCXIP, LYN-PQCXIP, or PQXCIP vector were transiently transfected into CHO-K1 cells with using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s recommendations. Forty-eight hours after transfection, cells were collected and lysed. HP parental cells were transiently transfected with the same constructs using Lipofectamine LTX (Invitrogen) according to the manufacturer’s recommendations. Forty-eight hours after transfection, cells were collected and fractionated for nuclear protein. Nuclear EGFR expression levels were then detected via immunoblot analysis.
Compounds
Dasatinib (BMS-354825, Sprycel™) was generously provided by Bristol-Myers Squibb (New York, NY, USA).
Antibodies
All antibodies were purchased from commercial sources as indicated below: EGFR, B-Myb, Actin, Histone H3, HRP-conjugated goat-anti-rabbit IgG, goat-anti-mouse IgG and donkey-anti-goat IgG were obtained from Santa Cruz Biotechnology Inc., (Santa Cruz, CA, USA). SFK, YES, LYN, pSFK (Y419) and normal mouse IgG were obtained from Cell Signaling Technology (Beverly, MA, USA). pEGFR (Y1101) was purchased by Abcam (Cambridge, MA, USA). Anti-mouse EGFR and pEGFR (Y845) were purchased from Invitrogen. Polyclonal iNOS was obtained from BD Biosciences (San Jose, CA, USA). α-tubulin was purchased from Calbiochem (San Diego, CA, USA).
Cellular fractionation and Immunoblotting analysis
Cells were swelled in cytoplasmic lysis buffer (20 mM HEPES, pH 7.0, 10 mM KCl, 2 mM MgCl2, 0.5% NP40, 1 mM Na3VO4, 1 mM PMSF, 1mM BGP, 10 μg/ml of leupeptin and aprotinin) for 10 min on ice and homogenized by 20–30 strokes in a tightly fitting Dounce homogenizer. The homogenate was centrifuged at 1,500 g for 5 min at 4°C to sediment the nuclei. The supernatant was then centrifuged at 15,000 g for 10 min at 4°C, and the resulting supernatant formed the non-nuclear fraction. The nuclear pellet was washed three times in cytoplasmic lysis buffer and re-suspended in the same buffer containing 0.5 M NaCl to extract nuclear proteins. After sonication and vortex, the extracted sample was centrifuged at 15,000 g for 10 min at 4°C. Whole cell protein lysate was obtained by tween-20 lysis buffer (50 mM HEPES, pH 7.4, 150 mM NaCl, 0.1% Tween-20, 10% glycerol, 2.5 mM EGTA, 1 mM EDTA, 1 mM DTT, 1mM Na3VO4, 1 mM PMSF, 1mM BGP and 10 μg/ml of leupeptin and aprotinin). Samples were sonicated and then centrifuged at 15,000 g for 10 min at 4°C. Protein concentrations were determined by Bradford assay (Bio-Rad Laboratories, Hercules, CA). Equal amounts of protein were fractionated by SDS-PAGE, transferred to a PVDF membrane (Millipore), and analyzed by incubation with the appropriate primary antibody. Proteins were detected via incubation with HRP-conjugated secondary antibodies and ECL chemiluminescence detection system (GE Healthcare Life Sciences, Piscataway, NJ, USA), ECL Western Blotting Substrate (Promega Cooperation, Madison, WI, USA), SuperSignal* West Dura Extended Duration Chemiluminescent Substrate or SuperSignal* West Femto Maximum Sensitivity Chemiluminescent Substrate (Thermo Fisher Scientific, Waltham, MA, USA).
Immunoprecipitation
Cells were lysed with NP-40 lysis buffer (50 mM HEPES, pH 7.4, 150 mM NaCl, 1% NP-40, 0.5% Deoxycholic acid, 10% glycerol, 2.5 mM EGTA, 1 mM EDTA, 1 mM DTT, 1 mM PMSF, 1mM BGP and 10 μg/ml of leupeptin and aprotinin). Cell lysates containing 0.5 mg of protein were incubated overnight at 4°C with 1 μg of anti-mouse EGFR antibody (Invitrogen) or normal mouse IgG (Cell Signaling Technology). After adding 25 μl of protein A/G agarose beads (Santa Cruz), cell lysates were incubated for another 2 hours at 4°C. The immunoprecipitates were pelleted by centrifugation and washed several times with NP-40 lysis buffer. The captured immunocomplexes were then eluted by boiling the beads in 2x SDS sample buffer for 5 minutes and subjected to immunoblot analysis as described above.
Microarray analysis
Total RNAs extracted from HP, HC1, HC4 and HC8 using an RNeasy kit (Qiagen Inc., Valencia, CA, USA). Gene expression profiling using the HT-HG-U133 Human Genome Array (Affymetrix, Santa Clara, CA, USA) containing over 22,000 well characterized genes. After RMA normalization, data were analyzed using Partek Discovery Suite (St. Louis, MO, USA) and signature genes were genes increased or decreased > 2-fold expression levels in three CtxR clones (HC1, HC4 and HC8) compared to sensitive parental line (HP) with p value <0.05.
cDNA synthesis and quantitative PCR
cDNA from total RNA of HP, HC1, HC4 and HC8 were synthesized using SuperScript III First-Strand Synthesis System (Invitrogen). Quantitative PCR (qPCR) analysis was performed using a Bio-Rad iQ5 Real-Time PCR Detection System (Bio-Rad Laboratories) using the iQ Supermix as recommended by manufacturer. All reactions were performed in triplicate. The sequences of primer sets used for this analysis are as follows: Lyn-F: 5′-GGCTCCAGA AGCAATCAACT-3′, Lyn-R: 5′-TCACGTCGGCATTAGTTCTC-3′; Yes-F: 5′-CTAGTAACA AAGGGCCGAGTG-3′, Yes-R: 5′-ATCCTGTATCCTCGCTCCAC-3′; Src-F: 5 ′-GAGGAG CCCATTTACATCGT-3′, Src-R: 5′-TGAGAAAGTCCAGCAAACTCC-3′, B-Myb-F: 5′-ATG TCCAGTGCCTGGAAGAC-3′, B-Myb-R: 5′-AGATGAGGGTCCGAGATGTG-3′. iNOS–F: 5′-CCATAAGGCCAAAGGGATTT-3′, iNOS-R: 5 ′-ATCTGGAGGGGTAGGCTTGT-3′. Fold increases or decreases in gene expression were determined by quantitation of cDNA from target samples (HC1, HC4 and HC8) relative to a calibrator sample (HP). Human β-actin gene (F: 5′-CAGCCATGTACGTTGCTATCCAGG-3′, R: 5′-AGGTCCAGA CGCAGGATGGCATG-3′) was used as the endogenous control for normalization of initial RNA levels. To determine this normalized value, 2−ΔΔCT values were compared between target and calibrator samples, where the change in crossing threshold (ΔCt)=Cttarget gene − Ctb-actin and ΔΔCt=ΔCtHC1, HC4 or HC8 − ΔCtHP.
Chromatin immunoprecipitation (ChIP) assay
Cells were fixed with formaldehyde at a final concentration of 1 % for 15 min at room temperature, stopped fixation by 1.25 M glycine for 5 min. Subsequently, cells were washed with ice-cold PBS and collected in the tube and centrifuge at 4°C for 5 min. The cell pellets was lysed in cell lysis buffer (5 mM HEPES, pH 8.0, 85 mM KCl, 0.5% NP-40 and 10 mM sodium pyrophosphate) by a Dounce homogenizer. After centrifuge, supernatant was removed, and the nuclei pellets were lysed in nuclei lysis buffer (Tris-HCl 50 mM, pH 8.1, 10 mM EDTA, 1% SDS and 10 mM sodium pyrophosphate). The lysate was sonicated on ice to shear DNA, and the supernatant was pre-cleared with protein A/G agarose beads (Santa Cruz) in dilution buffer (16.7 mM Tris-HCl, pH 8.1, 1.2 mM EDTA, 167 mM NaCl, 1.1% Triton X-100, 0.01% SDS and 10 mM sodium pyrophosphate) for 1h at 4°C. The pre-cleared lysates were immunoprecipitated by incubating with protein A/G beads containing 1 ug of anti-EGFR antibody or IgG and rotated at 4°C for overnight. The beads were washed with wash buffer I (25 mM Tris-HCl, pH 8.0, 2 mM EDTA, 150 mM NaCl, 1% Triton X-100, 0.1% SDS and 10 mM sodium pyrophosphate), wash buffer II (25 mM Tris-HCl, pH 8.0, 2 mM EDTA, 500 mM NaCl, 1% Triton X-100, 0.1% SDS and 10 mM sodium pyrophosphate), wash buffer III (10 mM Tris-HCl, pH 8.0, 1 mM EDTA, 250 mM LiCl, 1% NP-40, 1% deoxycholic acid and 10 mM sodium pyrophosphate) and TE buffer (10 mM Tris-HCl, pH8.0, 1 mM EDTA, 10 mM sodium pyrophosphate). The bound protein was eluted twice with elute buffer (100 mM NaHCO3 and 1% SDS). Then, 5 M NaCl was added to the pooled eluent and incubated at 68°C overnight. The DNA was recovered and purified using DNA purification kit (Qiagen). The purified chromatin immunoprecipitated DNA was used as a template for the qPCR of the promoter regions using the following primer pairs: B-Myb-F: 5′-CTGGTCTTAGCTACCCGTGAG TTGA–3′ and B-Myb-R: 5′-CAGGAGTATCCCACATAGCGAACAC-3′ (15), iNOS-F: 5′-TGATGAACTGCCACCTTGGAC–3′ and iNOS-R: 5 ′-TTCACCCAACCC ACCTCTTTC-3′ (16). The qPCR program was: 95°C for 3min, followed by 40 cycles of 95°C for 15 s and 60°C for 30 s for B-Myb or 55°C for 30 s for iNOS. The qPCR was performed using the iQ5 Real-time PCR Detection system (Bio-Rad).
Acknowledgments
This project was supported, in part, by grant P30CA014520 from the National Cancer Institute, grant 1UL1RR025011 from the Clinical and Translational Science Award program of the National Center for Research Resources and the National Institutes of Health (D.L.W.) by grant RSG-10-193-01-TBG from the American Cancer Society (D.L.W.), and by NIH grant 1732 GM08.1061-01A2 from Graduate Training in Cellular and Molecular Pathogenesis of Human Diseases (T.M.B.).
The abbreviations used are
- BCRP
breast cancer related protein
- CRC
colorectal cancer
- CtxR
cetuximab-resistant
- CtxS
cetuximab-sensitive
- DMSO
dimethyl sulfoxide
- EGF
epidermal growth factor
- EGFR
epidermal growth factor receptor
- ER
endoplasmic reticulum
- FBS
fetal bovine serum
- FGFR
fibroblast growth factor receptor
- HNSCC
head and neck squamous cell carcinoma
- iNOS
inducible nitric oxide synthase
- Lyn
v-yes-1 Yamaguchi sarcoma viral related oncogene homolog
- mAb
monoclonal antibody
- NSCLC
non-small cell lung cancer
- PI3K
phosphatidylinositol 3-kinase
- PKC
protein kinase C
- PLCγ
phospholipase C-gamma
- qPCR
quantitative PCR
- PGFR
platelet derived growth factor
- RTK
receptor tyrosine kinase
- SFK
Src-family kinases
- STAT
signal transducers and activators of transcription
- Yes
v-Yes-1 yamaguchi sarcoma viral oncogene
Footnotes
Conflict of interest
The authors declare no conflict of interest.
References
- 1.Yarden Y, Sliwkowski MX. Untangling the ErbB signalling network. Nat Rev Mol Cell Biol. 2001 Feb;2(2):127–37. doi: 10.1038/35052073. [DOI] [PubMed] [Google Scholar]
- 2.Wheeler DL, Dunn EF, Harari PM. Understanding resistance to EGFR inhibitors-impact on future treatment strategies. Nat Rev Clin Oncol. 2010 Sep;7(9):493–507. doi: 10.1038/nrclinonc.2010.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brand TM, Iida M, Wheeler DL. Molecular mechanisms of resistance to the EGFR monoclonal antibody cetuximab. Cancer Biol Ther. 2011 May 1;11(9):777–92. doi: 10.4161/cbt.11.9.15050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pao W, Chmielecki J. Rational, biologically based treatment of EGFR-mutant non–small-cell lung cancer. Nat Rev Cancer. 2010 Nov;10(11):760–74. doi: 10.1038/nrc2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gschwind A, Fischer OM, Ullrich A. The discovery of receptor tyrosine kinases: targets for cancer therapy. Nat Rev Cancer. 2004 May;4(5):361–70. doi: 10.1038/nrc1360. [DOI] [PubMed] [Google Scholar]
- 6.Marmor MD, Skaria KB, Yarden Y. Signal transduction and oncogenesis by ErbB/HER receptors. Int J Radiat Oncol Biol Phys. 2004 Mar 1;58(3):903–13. doi: 10.1016/j.ijrobp.2003.06.002. [DOI] [PubMed] [Google Scholar]
- 7.Wang YN, Yamaguchi H, Hsu JM, Hung MC. Nuclear trafficking of the epidermal growth factor receptor family membrane proteins. Oncogene. 2010 Jul 15;29(28):3997–4006. doi: 10.1038/onc.2010.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brand TM, Iida M, Li C, Wheeler DL. The nuclear epidermal growth factor receptor signaling network and its role in cancer. Discov Med. 2011 Nov;12(66):419–32. [PMC free article] [PubMed] [Google Scholar]
- 9.Carpenter G, Liao HJ. Trafficking of receptor tyrosine kinases to the nucleus. Exp Cell Res. 2009 May 15;315(9):1556–66. doi: 10.1016/j.yexcr.2008.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lo HW, Ali-Seyed M, Wu Y, Bartholomeusz G, Hsu SC, Hung MC. Nuclear-cytoplasmic transport of EGFR involves receptor endocytosis, importin beta1 and CRM1. J Cell Biochem. 2006 Aug 15;98(6):1570–83. doi: 10.1002/jcb.20876. [DOI] [PubMed] [Google Scholar]
- 11.Hsu SC, Hung MC. Characterization of a novel tripartite nuclear localization sequence in the EGFR family. J Biol Chem. 2007 Apr 6;282(14):10432–40. doi: 10.1074/jbc.M610014200. [DOI] [PubMed] [Google Scholar]
- 12.Wang YN, Yamaguchi H, Huo L, Du Y, Lee HJ, Lee HH, et al. The translocon Sec61beta localized in the inner nuclear membrane transports membrane-embedded EGF receptor to the nucleus. J Biol Chem. 2010 Dec 3;285(49):38720–9. doi: 10.1074/jbc.M110.158659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liao HJ, Carpenter G. Role of the Sec61 translocon in EGF receptor trafficking to the nucleus and gene expression. Mol Biol Cell. 2007 Mar;18(3):1064–72. doi: 10.1091/mbc.E06-09-0802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin SY, Makino K, Xia W, Matin A, Wen Y, Kwong KY, et al. Nuclear localization of EGF receptor and its potential new role as a transcription factor. Nat Cell Biol. 2001 Sep;3(9):802–8. doi: 10.1038/ncb0901-802. [DOI] [PubMed] [Google Scholar]
- 15.Hanada N, Lo HW, Day CP, Pan Y, Nakajima Y, Hung MC. Co-regulation of B-Myb expression by E2F1 and EGF receptor. Mol Carcinog. 2006 Jan;45(1):10–7. doi: 10.1002/mc.20147. [DOI] [PubMed] [Google Scholar]
- 16.Lo HW, Hsu SC, Ali-Seyed M, Gunduz M, Xia W, Wei Y, et al. Nuclear interaction of EGFR and STAT3 in the activation of the iNOS/NO pathway. Cancer Cell. 2005 Jun;7(6):575–89. doi: 10.1016/j.ccr.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 17.Hung LY, Tseng JT, Lee YC, Xia W, Wang YN, Wu ML, et al. Nuclear epidermal growth factor receptor (EGFR) interacts with signal transducer and activator of transcription 5 (STAT5) in activating Aurora-A gene expression. Nucleic Acids Res. 2008 Aug;36(13):4337–51. doi: 10.1093/nar/gkn417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lo HW, Cao X, Zhu H, Ali-Osman F. Cyclooxygenase-2 is a novel transcriptional target of the nuclear EGFR-STAT3 and EGFRvIII-STAT3 signaling axes. Mol Cancer Res. 2010 Feb;8(2):232–45. doi: 10.1158/1541-7786.MCR-09-0391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jaganathan S, Yue P, Paladino DC, Bogdanovic J, Huo Q, Turkson J. A functional nuclear epidermal growth factor receptor, SRC and Stat3 heteromeric complex in pancreatic cancer cells. PLoS One. 2011;6(5):e19605. doi: 10.1371/journal.pone.0019605. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 20.Huang WC, Chen YJ, Li LY, Wei YL, Hsu SC, Tsai SL, et al. Nuclear Translocation of Epidermal Growth Factor Receptor by Akt-dependent Phosphorylation Enhances Breast Cancer-resistant Protein Expression in Gefitinib-resistant Cells. J Biol Chem. 2011 Jun 10;286(23):20558–68. doi: 10.1074/jbc.M111.240796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Piccione EC, Lieu TJ, Gentile CF, Williams TR, Connolly AJ, Godwin AK, et al. A novel epidermal growth factor receptor variant lacking multiple domains directly activates transcription and is overexpressed in tumors. Oncogene. 2011 Oct 10; doi: 10.1038/onc.2011.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang SC, Nakajima Y, Yu YL, Xia W, Chen CT, Yang CC, et al. Tyrosine phosphorylation controls PCNA function through protein stability. Nat Cell Biol. 2006 Dec;8(12):1359–68. doi: 10.1038/ncb1501. [DOI] [PubMed] [Google Scholar]
- 23.Dittmann K, Mayer C, Fehrenbacher B, Schaller M, Raju U, Milas L, et al. Radiation-induced epidermal growth factor receptor nuclear import is linked to activation of DNA-dependent protein kinase. J Biol Chem. 2005 Sep 2;280(35):31182–9. doi: 10.1074/jbc.M506591200. [DOI] [PubMed] [Google Scholar]
- 24.Lo HW, Xia W, Wei Y, Ali-Seyed M, Huang SF, Hung MC. Novel prognostic value of nuclear epidermal growth factor receptor in breast cancer. Cancer Res. 2005 Jan 1;65(1):338–48. [PubMed] [Google Scholar]
- 25.Hadzisejdic I, Mustac E, Jonjic N, Petkovic M, Grahovac B. Nuclear EGFR in ductal invasive breast cancer: correlation with cyclin-D1 and prognosis. Mod Pathol. 2010 Mar;23(3):392–403. doi: 10.1038/modpathol.2009.166. [DOI] [PubMed] [Google Scholar]
- 26.Xia W, Wei Y, Du Y, Liu J, Chang B, Yu YL, et al. Nuclear expression of epidermal growth factor receptor is a novel prognostic value in patients with ovarian cancer. Mol Carcinog. 2009 Jul;48(7):610–7. doi: 10.1002/mc.20504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Psyrri A, Yu Z, Weinberger PM, Sasaki C, Haffty B, Camp R, et al. Quantitative determination of nuclear and cytoplasmic epidermal growth factor receptor expression in oropharyngeal squamous cell cancer by using automated quantitative analysis. Clin Cancer Res. 2005 Aug 15;11(16):5856–62. doi: 10.1158/1078-0432.CCR-05-0420. [DOI] [PubMed] [Google Scholar]
- 28.Li CF, Fang FM, Wang JM, Tzeng CC, Tai HC, Wei YC, et al. EGFR Nuclear Import in Gallbladder Carcinoma: Nuclear Phosphorylated EGFR Upregulates iNOS Expression and Confers Independent Prognostic Impact. Ann Surg Oncol. 2012 Feb;19(2):443–54. doi: 10.1245/s10434-011-1942-6. [DOI] [PubMed] [Google Scholar]
- 29.Li C, Iida M, Dunn EF, Ghia AJ, Wheeler DL. Nuclear EGFR contributes to acquired resistance to cetuximab. Oncogene. 2009 Oct 29;28(43):3801–13. doi: 10.1038/onc.2009.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li C, Iida M, Dunn EF, Wheeler DL. Dasatinib blocks cetuximab- and radiation-induced nuclear translocation of the epidermal growth factor receptor in head and neck squamous cell carcinoma. Radiother Oncol. 2010 Nov;97(2):330–7. doi: 10.1016/j.radonc.2010.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hsu SC, Miller SA, Wang Y, Hung MC. Nuclear EGFR is required for cisplatin resistance and DNA repair. Am J Transl Res. 2009;1(3):249–58. [PMC free article] [PubMed] [Google Scholar]
- 32.Liccardi G, Hartley JA, Hochhauser D. EGFR nuclear translocation modulates DNA repair following cisplatin and ionizing radiation treatment. Cancer Res. 2011 Feb 1;71(3):1103–14. doi: 10.1158/0008-5472.CAN-10-2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dittmann K, Mayer C, Kehlbach R, Rodemann HP. Radiation-induced caveolin-1 associated EGFR internalization is linked with nuclear EGFR transport and activation of DNA-PK. Mol Cancer. 2008;7:69. doi: 10.1186/1476-4598-7-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Biscardi JS, Maa MC, Tice DA, Cox ME, Leu TH, Parsons SJ. c-Src-mediated phosphorylation of the epidermal growth factor receptor on Tyr845 and Tyr1101 is associated with modulation of receptor function. J Biol Chem. 1999 Mar 19;274(12):8335–43. doi: 10.1074/jbc.274.12.8335. [DOI] [PubMed] [Google Scholar]
- 35.Maa MC, Leu TH, McCarley DJ, Schatzman RC, Parsons SJ. Potentiation of epidermal growth factor receptor-mediated oncogenesis by c-Src: implications for the etiology of multiple human cancers. Proc Natl Acad Sci U S A. 1995 Jul 18;92(15):6981–5. doi: 10.1073/pnas.92.15.6981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wheeler DL, Iida M, Dunn EF. The role of Src in solid tumors. Oncologist. 2009 Jul;14(7):667–78. doi: 10.1634/theoncologist.2009-0009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim LC, Song L, Haura EB. Src kinases as therapeutic targets for cancer. Nat Rev Clin Oncol. 2009 Oct;6(10):587–95. doi: 10.1038/nrclinonc.2009.129. [DOI] [PubMed] [Google Scholar]
- 38.Thomas SM, Brugge JS. Cellular functions regulated by Src family kinases. Annu Rev Cell Dev Biol. 1997;13:513–609. doi: 10.1146/annurev.cellbio.13.1.513. [DOI] [PubMed] [Google Scholar]
- 39.Bromann PA, Korkaya H, Courtneidge SA. The interplay between Src family kinases and receptor tyrosine kinases. Oncogene. 2004 Oct 18;23(48):7957–68. doi: 10.1038/sj.onc.1208079. [DOI] [PubMed] [Google Scholar]
- 40.Tice DA, Biscardi JS, Nickles AL, Parsons SJ. Mechanism of biological synergy between cellular Src and epidermal growth factor receptor. Proc Natl Acad Sci U S A. 1999 Feb 16;96(4):1415–20. doi: 10.1073/pnas.96.4.1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kloth MT, Laughlin KK, Biscardi JS, Boerner JL, Parsons SJ, Silva CM. STAT5b, a Mediator of Synergism between c-Src and the Epidermal Growth Factor Receptor. J Biol Chem. 2003 Jan 17;278(3):1671–9. doi: 10.1074/jbc.M207289200. [DOI] [PubMed] [Google Scholar]
- 42.Ottenhoff-Kalff AE, Rijksen G, van Beurden EA, Hennipman A, Michels AA, Staal GE. Characterization of protein tyrosine kinases from human breast cancer: involvement of the c-src oncogene product. Cancer Res. 1992 Sep 1;52(17):4773–8. [PubMed] [Google Scholar]
- 43.Lu KV, Zhu S, Cvrljevic A, Huang TT, Sarkaria S, Ahkavan D, et al. Fyn and SRC are effectors of oncogenic epidermal growth factor receptor signaling in glioblastoma patients. Cancer Res. 2009 Sep 1;69(17):6889–98. doi: 10.1158/0008-5472.CAN-09-0347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wheeler DL, Iida M, Kruser TJ, Nechrebecki MM, Dunn EF, Armstrong EA, et al. Epidermal growth factor receptor cooperates with Src family kinases in acquired resistance to cetuximab. Cancer Biol Ther. 2009 Apr;8(8):696–703. doi: 10.4161/cbt.8.8.7903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang J, Kalyankrishna S, Wislez M, Thilaganathan N, Saigal B, Wei W, et al. SRC-family kinases are activated in non-small cell lung cancer and promote the survival of epidermal growth factor receptor-dependent cell lines. Am J Pathol. 2007 Jan;170(1):366–76. doi: 10.2353/ajpath.2007.060706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fu YN, Yeh CL, Cheng HH, Yang CH, Tsai SF, Huang SF, et al. EGFR mutants found in non-small cell lung cancer show different levels of sensitivity to suppression of Src: implications in targeting therapy. Oncogene. 2008 Feb 7;27(7):957–65. doi: 10.1038/sj.onc.1210684. [DOI] [PubMed] [Google Scholar]
- 47.Koppikar P, Choi SH, Egloff AM, Cai Q, Suzuki S, Freilino M, et al. Combined inhibition of c-Src and epidermal growth factor receptor abrogates growth and invasion of head and neck squamous cell carcinoma. Clin Cancer Res. 2008 Jul 1;14(13):4284–91. doi: 10.1158/1078-0432.CCR-07-5226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wheeler DL, Huang S, Kruser TJ, Nechrebecki MM, Armstrong EA, Benavente S, et al. Mechanisms of acquired resistance to cetuximab: role of HER (ErbB) family members. Oncogene. 2008 Jun 26;27(28):3944–56. doi: 10.1038/onc.2008.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brand TM, Dunn EF, Iida M, Myers RA, Kostopoulos KT, Li C, et al. Erlotinib is a viable treatment for tumors with acquired resistance to cetuximab. Cancer Biol Ther. 2011 Sep 1;12(5):436–46. doi: 10.4161/cbt.12.5.16394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Massie C, Mills IG. The developing role of receptors and adaptors. Nat Rev Cancer. 2006 May;6(5):403–9. doi: 10.1038/nrc1882. [DOI] [PubMed] [Google Scholar]
- 51.Carpenter G. Nuclear localization and possible functions of receptor tyrosine kinases. Curr Opin Cell Biol. 2003 Apr;15(2):143–8. doi: 10.1016/s0955-0674(03)00015-2. [DOI] [PubMed] [Google Scholar]
- 52.Wang SC, Hung MC. Nuclear translocation of the epidermal growth factor receptor family membrane tyrosine kinase receptors. Clin Cancer Res. 2009 Nov 1;15(21):6484–9. doi: 10.1158/1078-0432.CCR-08-2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lo HW, Hung MC. Nuclear EGFR signalling network in cancers: linking EGFR pathway to cell cycle progression, nitric oxide pathway and patient survival. Br J Cancer. 2006 Jan 30;94(2):184–8. doi: 10.1038/sj.bjc.6602941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhao Y, He D, Saatian B, Watkins T, Spannhake EW, Pyne NJ, et al. Regulation of lysophosphatidic acid-induced epidermal growth factor receptor transactivation and interleukin-8 secretion in human bronchial epithelial cells by protein kinase Cdelta, Lyn kinase, and matrix metalloproteinases. J Biol Chem. 2006 Jul 14;281(28):19501–11. doi: 10.1074/jbc.M511224200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xi S, Zhang Q, Dyer KF, Lerner EC, Smithgall TE, Gooding WE, et al. Src kinases mediate STAT growth pathways in squamous cell carcinoma of the head and neck. J Biol Chem. 2003 Aug 22;278(34):31574–83. doi: 10.1074/jbc.M303499200. [DOI] [PubMed] [Google Scholar]
- 56.Kasai A, Shima T, Okada M. Role of Src family tyrosine kinases in the down-regulation of epidermal growth factor signaling in PC12 cells. Genes Cells. 2005 Dec;10(12):1175–87. doi: 10.1111/j.1365-2443.2005.00909.x. [DOI] [PubMed] [Google Scholar]
- 57.Su T, Bryant DM, Luton F, Verges M, Ulrich SM, Hansen KC, et al. A kinase cascade leading to Rab11-FIP5 controls transcytosis of the polymeric immunoglobulin receptor. Nat Cell Biol. 2010 Dec;12(12):1143–53. doi: 10.1038/ncb2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Boerner JL, Demory ML, Silva C, Parsons SJ. Phosphorylation of Y845 on the epidermal growth factor receptor mediates binding to the mitochondrial protein cytochrome c oxidase subunit II. Mol Cell Biol. 2004 Aug;24(16):7059–71. doi: 10.1128/MCB.24.16.7059-7071.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]