Abstract
Cellular fusion of macrophages into multinucleated giant cells is a distinguishing feature of the granulomatous response to inflammation, infection and foreign bodies (1). We observed a marked increase in fusion of macrophages genetically deficient in Dicer, an enzyme required for canonical miRNA biogenesis. Gene expression profiling of miRNA deficient macrophages revealed an up-regulation of the IL4 responsive fusion protein Tm7sf4, analyses identify miR-7a-1 as a negative regulator of macrophage fusion, functioning by directly targeting Tm7sf4 mRNA. miR-7a-1 is itself an IL4 responsive gene in macrophages, suggesting feedback control of cellular fusion. Collectively these data indicate that miR-7a-1 functions to regulate IL4 directed multinucleated giant cell formation.
Introduction
Circulating monocytes and tissue resident macrophages serve as sentinels of the immune system by sensing the presence of pathogens using a diverse array of membrane anchored and cytosolic detectors. Upon pathogen exposure, macrophages coordinate both intrinsic and extrinsic host defense mechanisms to eliminate the intruding organism and establish protective immunity against reinfection (1, 2). Macrophages are remarkably adaptable in their response to infection and activate distinct effector mechanisms commensurate with the pathogen detected. For example, detection of viruses through cytosolic sensors triggers a rapid type 1 interferon dominated response, whereas pro-inflammatory cytokine expression is induced following TLR2 recognition of peptidoglycan, a component of Gram-positive bacteria (1, 3).
Macrophage responses are further regulated by leukocyte derived soluble and cell surface molecules. Exposure of macrophages to TLR ligands in the presence of IFNγ induces a classically activated state characterized by expression of TNFα and IL12, which together promote TH1 or TH17 dominated responses. In contrast, macrophages activated by the TH2 cytokines IL4 and IL13 participate in wound repair, immune suppression and defense against parasites (2, 4, 5). Additionally, IL4 and IL13 can induce macrophage fusion into multinucleated giant cells that serve to isolate and/or eliminate foreign bodies and parasites and are a hallmark of the granulomatous response (6, 7). Studies of molecular mechanisms that regulate mammalian cell fusion have identified several key fusogenic molecules (i.e. CD9, CD47, CD44, Tm7sf4/DC-STAMP, and SIRPα) (8) (9). Whereas macrophage activation is generally considered a plastic and reversible response, the formation of multinucleated cells represents a terminally differentiated state and is thus likely subject to unique regulatory mechanisms.
miRNAs are 21–23 nucleotide non-coding RNAs generated by sequential nuclear and cytosolic processing of precursor transcripts by the RNAseIII enzymes Drosha and Dicer, respectively (10). They negatively regulate gene expression by binding to conserved, partially complementary sequences most often found in the 3’ non coding regions of target mRNAs. Several miRNAs are responsive to TLR agonists and function to dampen or reshape innate immune responses within macrophages (11). Here, we demonstrate a role for miR-7a-1 in regulating multinucleated giant cell formation by targeting the fusogenic cell surface protein Tm7sf4.
Materials and Methods
Mice
Mice in which myeloid cells were selectively deficient in Dicer were generated by crossing C57BL/6.129 Dicerfl/fl (12) with the C57BL/6.129 Lyz2tm1(cre) deletor strain (13) to generate C57BL/6 Dicerfl/flLyz2tm1(cre)/+ mice. Throughout experiments, age and sex matched C57BL/6 Dicerfl/+Lyz2tm1(cre)/+ mice were used as controls. Mice obtained from The Jackson Laboratory (Bar Harbor, ME) were housed under SPF conditions with approval of IACUC at the Institute for Systems Biology.
Tissue culture
Bone marrow-derived macrophages (BMM) were cultured in complete DMEM (cDMEM; plus 10% FBS, 2 mM l-glutamine, penicillin and streptomycin) with recombinant human CSF-1 (50 ng/ml) for 6 d. To generate giant cells, bone-marrow cells were cultured as above for 4 d then mouse-IL-4 (50 ng/mL; Peprotech) was added for a further 6 d. Human embryonic kidney cells (HEK 293) for luciferase assays were grown in cDMEM. Macrophages derived from conditionally immortalized Hoxb8 progenitors were generated from C57BL/6 mice essentially as described (14) and maintained in their myeloid progenitor stage in RPMI, 10% FBS, 2 mM l-glutamine, 10ng/mL GM-CSF-1, 1μM β-estradiol. To generate Hoxb8 macrophages from wild-type and miRNA transduced lines, progenitors were harvested, washed 2 X in 1X PBS and resuspended in fresh BMM culture medium containing CSF-1 and cultured for 6 d as per the BMM protocol.
Gene expression arrays
Total RNA was isolated using TRIzol (Invitrogen) and mRNA was labeled and hybridized to an Affymetrix GeneChip Mouse Exon ST 1.0 array. Expression data were acquired from six microarray hybridization experiments comprising three Dicerfl/flLyz2tm1(cre)/+ mutant-derived macrophages, and three Dicerfl/+Lyz2tm1(cre)/+ controls. Data were background-subtracted and normalized, then averaged across biological replicates using the log2 intensities (15, 16). Significance testing was performed using log2 intensities and expression cut-off value of 6 with a student’s t-test; p-value < 0.01 regarded as significant. Gene array data are deposited in NCBI-GEO, accession number: GSE36585 (http://www.ncbi.nlm.nih.gov/projects/geo/query/acc.cgi?acc=GSE36585)
Retroviral transduction of miR-7a-1
Genomic sequences encoding pri-miR-7a-1 were cloned corresponding to genetic coordinates Chr13:58493908 and Chr13:58494429. The product was cloned into a MSCV-PIG vector (17) and a control construct in the same backbone was generated encoding a pri-miR-30 formatted shRNA against Tlr5, a gene not expressed by mouse BMM. Retroviral constructs were packaged using the Phoenix ecotropic 293 line. Macrophages were transduced and cells selected in puromycin for 21 days prior to analysis.
Tm7sf4 3’UTR cloning, mutagenesis and luciferase reporter assays
The 3’UTR of Tm7sf4 was cloned from C57BL/6 mouse genomic DNA by PCR corresponding to Ensembl coordinates 15071 and 15621. The product was cloned 3’ of the firefly luciferase ORF in a pVitro-blasti (Invivogen) backbone carrying a control Renilla luciferase gene. Point mutations in the Tm7sf4 miR-7a target sequence were introduced by mutagenesis PCR. HEK 293 cells were co-transfected with luciferase and miRNA expressing vectors and assayed using a luminometer.
DNA staining to determine giant cell ploidy
Ploidy staining was performed as described (18). Flow cytometry was performed using a Becton Dickinson FACSAria II and ploidy classes for giant cells determined.
Gene expression analysis
To analyze recombination of Dicer1 alleles, expression of genes and microRNAs, total RNA was isolated from macrophages using TRIzol (Invitrogen), treated with DNAase (Ambion), and used as template for reverse transcription (Superscript II, Invitrogen). Quantitative PCR (qPCR) reactions were performed using an ABI 2900 HT thermocycler and expression levels were calculated using the ΔΔCT method relative to control Ef1a gene. MicroRNA-specific reverse transcription was performed for each miRNA using gene-specific primers (Applied Biosystems). MicroRNA levels were normalized to small-nucleolar-RNA sno202. The levels of Sno202 were unchanged by CSF-1, IL-4, or by a variety of TLR agonists in the primary bone-marrow derived macrophage or Hox macrophage systems.
Western blotting analyses
Were performed using standard techniques, membranes were probed with relevant primary antibodies: rabbit anti-Stat6 (Cell signaling technologies, MA), rabbit anti-phospho-Stat6 (Abcam, MA), rabbit anti-mouse-Tm7sf4 (Santa Cruz, CA), rabbit anti-mouse beta-actin1-HRP (Jackson Immunoresearch labs).
Statistical analysis
Significance was determined using an unpaired 2-tailed Student’s t test.
Results and Discussion
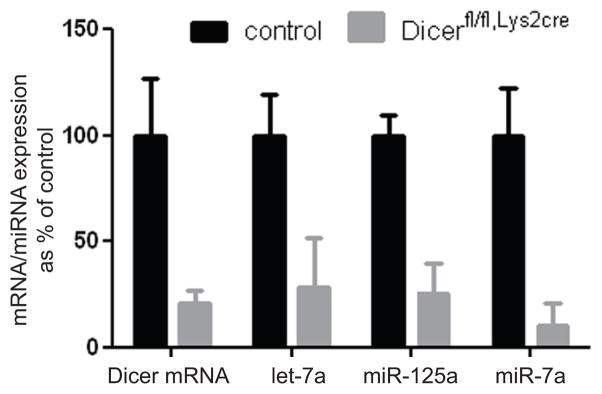
miRNAs have established roles in regulating classic innate immune responses. We took a genetic approach to globally assess a role for miRNAs in regulating macrophage fusion into multinucleated giant cells, a specialized TH2 driven response of macrophages to intrusion by parasites or synthetic foreign material. Mice carrying a floxed allele of Dicer were crossed to a myeloid restricted cre expressing strain (Lys2Cre). The resulting mice, Dicerfl/fl Lys2Cre, were born at the expected Mendelian frequencies from the appropriate crosses and were overtly indistinguishable from littermate controls heterozygous for the floxed Dicer allele, Dicerfl/+ Lys2Cre. Dicer mRNA levels were reduced approximately 75 % in Dicerfl/fl, Lys2Cre bone marrow macrophages (BMM) relative to controls (Fig. 1). The extent of Dicer deletion could not be further increased by using mice homozygous for Lyz2cre, prolonged culture or LPS stimulation (unpublished observations) and are in agreement with previous reports using the Lyz2-cre deletor strain (19). A corresponding decrease in the expression of miRNAs abundantly expressed in BMMs was observed (Fig. 1A).
Figure 1. Deletion of Dicer mRNA in bone-marrow derived macrophages (BMM) reduces miRNA levels.
Quantification of Dicer mRNA levels in control (Dicerfl/+,Lys2cre) and Dicer deficient (Dicerfl/fl,Lys2cre) macrophages, and analysis of microRNAs let-7a, miR-125a-5p and miR-7a expression in control and Dicerfl/fl,Lys2cre macrophages by qPCR.
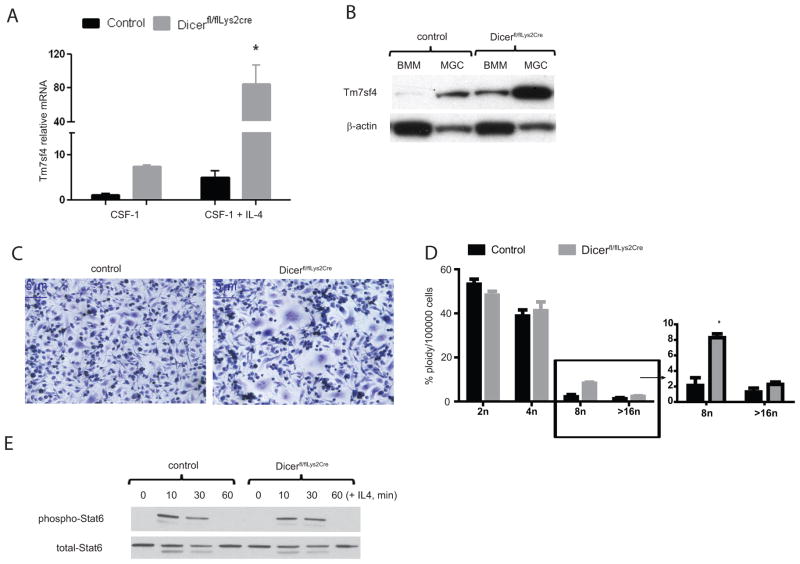
We used genome wide expression profiling to analyze the consequences of Dicer deficiency on macrophage responses to IL4. IL4 programs macrophages to dampen immune responses, repair damaged tissue and participate in defense against parasites (2, 4). The expression of classic markers of IL4 macrophage activation including Arginase-1 (Arg1), Chitinase-3 like-3 (Chi3l3/Ym1) and Resistin-like molecule-α (Retnl1a/Fizz), is unaffected by Dicer depletion (Supplement 1B). IL4 additionally induces the expression of genes involved in macrophage fusion. Amongst this panel of genes, only Tm7sf4, a molecule required for macrophage fusion (20–22), was significantly altered in Dicer deficient cells (Supplement 1A) These data were confirmed by qRT-PCR and western blot analyses of resting and IL4 stimulated macrophages (Fig. 2A and 2B) and suggests a unique role for Dicer, and hence miRNAs, in the control of Tm7sf4 expression. Extended culture of BMMs in IL4 containing media leads to the generation of multinucleated giant cells. Under these conditions, we noticed a marked increase in the number of multinucleated cells in Dicerfl/fl, Lys2Cre relative to control cells (Fig. 2C and 2D). This suggests a direct link between the expression of the fusogenic molecule, Tm7sf4, and the fusion phenotype. Considering that IL-4 highly induces Tm7sf4 in Dicerfl/fl, Lys2Cre cultures relative to controls, we examined whether Dicerfl/fl, Lys2Cre macrophages were globally hyper-responsive to IL-4. This was not the case. For example, IL4-induced Stat6 phosphorylation and dephosphorylation were identical in Dicerfl/fl, Lys2Cre and control macrophages (Fig. 2E) Furthermore, another IL4-responsive fusogenic molecule, E-cadherin (Cdh1), was unaffected in Dicerfl/fl, Lys2Cre macrophages (Fig. 1B), demonstrating the specificity of regulation of Tm7sf4 by miRNA. Finally, the expression of a number of macrophage fusogenic molecules was unaffected in Dicerfl/fl, Lys2Cre macrophages (Supplement 1A).
Figure 2. IL-4 induces Tm7sf4 expression and excess giant cell formation in Dicerfl/fl,Lys2cre macrophages.
A) qPCR of resting and IL4-induced (24 h) Tm7sf4 mRNA in control relative to Dicerfl/fl,Lys2cre macrophages. Error bars denote SEM, asterisk represents significance; p value of < 0.05, representative of five independent experiments. B) Immunoblotting of Tm7sf4 protein expression in macrophages (BMM; CSF-1 derived) or IL-4 derived multinucleated giant cells (MGC; CSF-1/IL-4 derived) in control or Dicerfl/fl,Lys2cre cells, representative of n = 3 experiments. C) Dicer deficiency increases cellular fusion and multinucleated giant cell formation. Hematoxylin staining of control or Dicerfl/fl,Lys2cre macrophages stimulated with IL-4 for 144h; representative of five experiments. Images are 100X original magnification. D) Flow cytometric enumeration of the frequency of multinucleated cells in control or Dicerfl/fl,Lys2cre multinucleated giant cells. Cells were stained with propidium iodide (PI). Bars indicate percent of cells against chromosomal copy number. E) Immunoblotting analysis of Dicerfl/fl,Lys2cre cultures for total Stat6 protein and phospho-Stat6 response to IL-4, representative of n = 5 experiments.
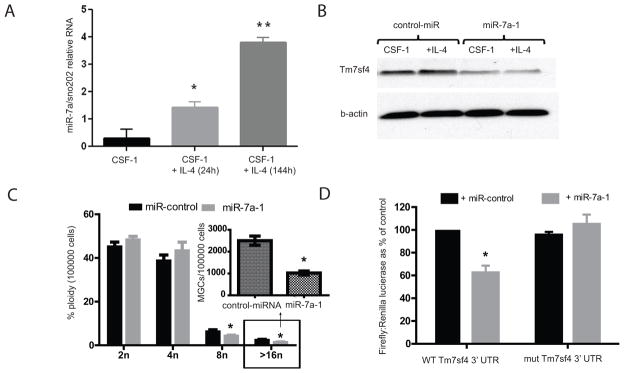
Examination of the Tm7sf4 3’UTR using various target prediction algorithms (23) demonstrated a number of miRNA target sites. However, only the predicted miR-7a target sequence was conserved amongst mammalian species. There are three isoforms of miR-7, miR-7a-1, miR-7a-2 and miR-7b; miR-7a and 7b can be distinguished by PCR based on their mature form, but the miR-7a-1 and miR-7a-2 isoforms cannot. miR-7b is not expressed in macrophages whereas miR-7a-(1/2) are (data not shown). Moreover, miR-7a-(1/2) are strongly induced by IL-4 (Fig. 3A), suggesting a potential role in regulating Tm7sf4 (Fig. 3A). In order to distinguish between miR-7a-1 and miR-7a-2 we turned to epigenetic mapping (Supplementary Figure 2). These two miRNA’s are found on chromosomes 13 and 7, respectively. Histone acetylation results in an open chromosome structure that facilitates transcription. ChIP-seq experiments in IL4-treated macrophages demonstrated substantial histone acetylation in the vicinity of miR-7a-1 on chromosome 13 whereas there was no detectable histone acetylation in the vicinity of miR-7a-2 on chromosome 7. This suggests that miR-7a-1 is the IL-4 responsive species (Supplement 2). We therefore considered miR-7a-1 to be a candidate miRNA regulating both Tm7sf4 expression and IL4 induced multinucleated giant cell formation.
Figure 3. miR-7a is regulated by IL-4 and ectopic expression of miR-7a regulates Tm7sf4 mRNA and protein levels, leading to restricted giant cell formation.
A) qPCR analysis of miR-7a response to IL-4 in macrophages. Representative of three independent experiments. Asterisk denotes significance; p < 0.05. B) Immunoblotting of Tm7sf4 in macrophages expressing a control miRNA or miR-7a, cells were stimulated with IL-4 for 24 h and examined for Tm7sf4 expression relative to beta-actin. C) Flow cytometric analysis of multinucleated cells from control miRNA or miR-7a expressing cells. Asterisk denotes significance; p < 0.05. D). HEK 293 cells co-transfected with WT or mutated Tm7sf4 3′UTR and miR-7a, then assessed for luciferase activity. Asterisk denotes significance; p < 0.01.
To directly examine a role for miR-7a-1 in regulating Tm7sf4 expression, we transduced immortalized BMMs with retroviruses encoding either pri-miR-7a-1 or a control pri-miR-30. Decreased Tm7sf4 expression was specifically observed in miR-7a-1 over-expressing BMMs (Fig 3B). Moreover, IL4 induced multinucleated giant cell formation was attenuated in miR-7a-1 over-expressing BMMs (Fig. 3C). To determine if the ability of miR-7a-1 to regulate Tm7sf4 expression is through direct targeting of the 3’UTR, we fused these sequences downstream of a luciferase reporter. The resulting construct was tested in HEK 293 cells co-transfected with either miR-7a-1 or a control miRNA. As shown in Fig 3D, the Tm7sf4 3’UTR imparted miR-7a-1 specific suppression of luciferase expression. Transfection of the Tm7sf4 3’UTR containing a point mutation in the miR-7a-1 target sequence had no effect on luciferase activity (Fig. 3D). These experiments demonstrate that miR-7a-1 interacts directly and specifically with the Tm7sf4 3’UTR to regulate its expression.
Our study identifies miR-7a-1, an IL4 responsive miRNA, as critical negative regulator of IL4-mediated gene transcription of Tm7sf4. This novel mechanism allows for precise control of macrophage differentiation into multinucleated giant cells. In support of a role for miRNA in regulation of cellular differentiation, Dicer dependent pathways are required for the differentiation of Langerhans cells and T-cells (24–26). More specifically, miR-181 is required for human hematopoietic cell differentiation (27), while miR-150 controls B-cell differentiation (28), and miR-223 regulates neutrophils (29). By generating a myeloid specific deletion of Dicer, we have identified for the first time that miRNA can fine-tune the macrophage differentiation response to IL4. More specifically, miR-7a-1 acts as a negative regulator of Tm7sf4 and as a macrophage fusion rheostat.
An environment of chronic TH2 inflammation programs macrophages to differentiate to multinucleated giant cells, a hallmark of granulomatous disease. However, the agents driving giant cell formation in granulomatous diseases such as sarcoidosis remain elusive. It is possible that IL-4 regulated Tm7sf4 via miR-7a-1 may play a role in the etiology of chronic inflammatory diseases.
Supplementary Material
Acknowledgments
Grant support:. This work was funded by NIH grants #5RO1AI032971-20, #5RO1AI025032- 23, HHSN272200700038C.
Bruz Marzolf and Pamela Troisch for array hybridization; Hien Duong for technical assistance; David Rodriguez and vivarium staff.
Non-standard condensed words
- miRNA
microRNA
- miR-7a
miRNA-7a
References
- 1.Kawai T, Akira S. Toll-like receptors and their crosstalk with other innate receptors in infection and immunity. Immunity. 2011;34:637–650. doi: 10.1016/j.immuni.2011.05.006. [DOI] [PubMed] [Google Scholar]
- 2.Gordon SMF. Alternative activation of macrophages: mechanism and functions. Immunity. 2010;32:593–604. doi: 10.1016/j.immuni.2010.05.007. [DOI] [PubMed] [Google Scholar]
- 3.Underhill DM, Ozinsky A, Hajjar AM, Stevens A, Wilson CB, Bassetti M, Aderem A. The Toll-like receptor 2 is recruited to macrophage phagosomes and discriminates between pathogens. Nature. 1999;401:811–815. doi: 10.1038/44605. [DOI] [PubMed] [Google Scholar]
- 4.Martinez FO, Sica A, Mantovani A, Locati M. Macrophage activation and polarization. Front Biosci. 2008;13:453–461. doi: 10.2741/2692. [DOI] [PubMed] [Google Scholar]
- 5.McNally AK, Anderson JM. Foreign body-type multinucleated giant cells induced by interleukin-4 express select lymphocyte co-stimulatory molecules and are phenotypically distinct from osteoclasts and dendritic cells. Exp Mol Pathol. 2011 doi: 10.1016/j.yexmp.2011.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chensue SW, Terebuh PD, Warmington KS, Hershey SD, Evanoff HL, Kunkel SL, Higashi GI. Role of IL-4 and IFN-gamma in Schistosoma mansoni egg-induced hypersensitivity granuloma formation. Orchestration, relative contribution, and relationship to macrophage function. J Immunol. 1992;148:900–906. [PubMed] [Google Scholar]
- 7.Kao WJ, McNally AK, Hiltner A, Anderson JM. Role for interleukin-4 in foreign-body giant cell formation on a poly(etherurethane urea) in vivo. J Biomed Mater Res. 1995;29:1267–1275. doi: 10.1002/jbm.820291014. [DOI] [PubMed] [Google Scholar]
- 8.Vignery A. Macrophage fusion: molecular mechanisms. Methods Mol Biol. 2008;475:149–161. doi: 10.1007/978-1-59745-250-2_9. [DOI] [PubMed] [Google Scholar]
- 9.Lundberg P, Koskinen C, Baldock PA, Lothgren H, Stenberg A, Lerner UH, Oldenborg PA. Osteoclast formation is strongly reduced both in vivo and in vitro in the absence of CD47/SIRPalpha-interaction. Biochem Biophys Res Commun. 2007;352:444–448. doi: 10.1016/j.bbrc.2006.11.057. [DOI] [PubMed] [Google Scholar]
- 10.Lodish HF, Zhou B, Liu G, Chen CZ. Micromanagement of the immune system by microRNAs. Nat Rev Immunol. 2008;8:120–130. doi: 10.1038/nri2252. [DOI] [PubMed] [Google Scholar]
- 11.Baltimore D, Boldin MP, O'Connell RM, Rao DS, Taganov KD. MicroRNAs: new regulators of immune cell development and function. Nat Immunol. 2008;9:839–845. doi: 10.1038/ni.f.209. [DOI] [PubMed] [Google Scholar]
- 12.Harfe BD, McManus MT, Mansfield JH, Hornstein E, Tabin CJ. The RNaseIII enzyme Dicer is required for morphogenesis but not patterning of the vertebrate limb. Proc Natl Acad Sci U S A. 2005;102:10898–10903. doi: 10.1073/pnas.0504834102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clausen BE, Burkhardt C, Reith W, Renkawitz R, Forster I. Conditional gene targeting in macrophages and granulocytes using LysMcre mice. Transgenic Res. 1999;8:265–277. doi: 10.1023/a:1008942828960. [DOI] [PubMed] [Google Scholar]
- 14.Wang GG, Calvo KR, Pasillas MP, Sykes DB, Hacker H, Kamps MP. Quantitative production of macrophages or neutrophils ex vivo using conditional Hoxb8. Nat Methods. 2006;3:287–293. doi: 10.1038/nmeth865. [DOI] [PubMed] [Google Scholar]
- 15.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 16.Gentleman RC, V, Carey J, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini AJ, Sawitzki G, Smith C, Smyth G, Tierney L, Yang JY, Zhang J. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dickins RA, Hemann MT, Zilfou JT, Simpson DR, Ibarra I, Hannon GJ, Lowe SW. Probing tumor phenotypes using stable and regulated synthetic microRNA precursors. Nat Genet. 2005;37:1289–1295. doi: 10.1038/ng1651. [DOI] [PubMed] [Google Scholar]
- 18.Drachman JG, Jarvik GP, Mehaffey MG. Autosomal dominant thrombocytopenia: incomplete megakaryocyte differentiation and linkage to human chromosome 10. Blood. 2000;96:118–125. [PubMed] [Google Scholar]
- 19.Yang Y, Liu B, Dai J, Srivastava PK, Zammit DJ, Lefrancois L, Li Z. Heat shock protein gp96 is a master chaperone for toll-like receptors and is important in the innate function of macrophages. Immunity. 2007;26:215–226. doi: 10.1016/j.immuni.2006.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yagi M, Miyamoto T, Sawatani Y, Iwamoto K, Hosogane N, Fujita N, Morita K, Ninomiya K, Suzuki T, Miyamoto K, Oike Y, Takeya M, Toyama Y, Suda T. DC-STAMP is essential for cell-cell fusion in osteoclasts and foreign body giant cells. J Exp Med. 2005;202:345–351. doi: 10.1084/jem.20050645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yagi M, Miyamoto T, Toyama Y, Suda T. Role of DC-STAMP in cellular fusion of osteoclasts and macrophage giant cells. J Bone Miner Metab. 2006;24:355–358. doi: 10.1007/s00774-006-0697-9. [DOI] [PubMed] [Google Scholar]
- 22.Yagi M, Ninomiya K, Fujita N, Suzuki T, Iwasaki R, Morita K, Hosogane N, Matsuo K, Toyama Y, Suda T, Miyamoto T. Induction of DC-STAMP by alternative activation and downstream signaling mechanisms. J Bone Miner Res. 2007;22:992–1001. doi: 10.1359/jbmr.070401. [DOI] [PubMed] [Google Scholar]
- 23.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 24.Fedeli M, Napolitano A, Wong MP, Marcais A, de Lalla C, Colucci F, Merkenschlager M, Dellabona P, Casorati G. Dicer-dependent microRNA pathway controls invariant NKT cell development. J Immunol. 2009;183:2506–2512. doi: 10.4049/jimmunol.0901361. [DOI] [PubMed] [Google Scholar]
- 25.Kuipers H, Schnorfeil FM, Fehling HJ, Bartels H, Brocker T. Dicer-dependent microRNAs control maturation, function, and maintenance of Langerhans cells in vivo. J Immunol. 185:400–409. doi: 10.4049/jimmunol.0903912. [DOI] [PubMed] [Google Scholar]
- 26.Cobb BS, Nesterova TB, Thompson E, Hertweck A, O'Connor E, Godwin J, Wilson CB, Brockdorff N, Fisher AG, Smale ST, Merkenschlager M. T cell lineage choice and differentiation in the absence of the RNase III enzyme Dicer. J Exp Med. 2005;201:1367–1373. doi: 10.1084/jem.20050572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- 28.Zhou B, Wang S, Mayr C, Bartel DP, Lodish HF. miR-150, a microRNA expressed in mature B and T cells, blocks early B cell development when expressed prematurely. Proc Natl Acad Sci U S A. 2007;104:7080–7085. doi: 10.1073/pnas.0702409104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johnnidis JB, Harris MH, Wheeler RT, Stehling-Sun S, Lam MH, Kirak O, Brummelkamp TR, Fleming MD, Camargo FD. Regulation of progenitor cell proliferation and granulocyte function by microRNA-223. Nature. 2008;451:1125–1129. doi: 10.1038/nature06607. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.