Abstract
DNA transport is an essential life process. From chromosome separation during cell division or sporulation, to DNA virus ejection or encapsidation, to horizontal gene transfer, it is ubiquitous in all living organisms. Directed DNA translocation is often energetically unfavorable and requires an active process that uses energy, namely the action of molecular motors. In this review we present recent advances in the understanding of three molecular motors involved in DNA transport in prokaryotes, paying special attention to recent studies using single-molecule techniques. We first discuss DNA transport during cell division, then packaging of DNA in phage capsids, and then DNA import during bacterial transformation.
Introduction
Directed transport of macromolecules through nanometer-sized pores is essential for many cellular processes and will likely have applications in biotechnology. Microbes have evolved different molecular motors for transporting DNA during processes as diverse as cell division, horizontal gene transfer, and packaging of DNA into viral capsids. A combined effort of genetics, biochemistry, and structural biology has identified the essential components and structures of some of these motors. Recently, single molecule techniques have led to many advances in studying biophysical properties of these motors. The classic approach to studying mechano-chemistry of individual molecular motors is to purify the proteins and study their kinetics, directionality, and force generation [1–3]. Directional motor movement has then been quantitatively analyzed by recording relative length changes of DNA using laser tweezers or magnetic devices, allowing the individual consecutive chemical and mechanical steps of the motor enzymes to be dissected. At a higher level of complexity, packaging of DNA into bacteriophage capsids has been quantified, revealing such features as the forces resisting DNA confinement, a novel mechanism of coordination between motor subunits, and insights on structure-function relationships. DNA translocation systems across biological membranes have not been purified in a functional form so far. However, single-molecule techniques have been adapted to directly measuring import of DNA molecules by living bacteria, again allowing detailed study of a motor during its natural task. In this review, we concentrate on three different DNA transport motors representing each of the categories, namely the bacterial FtsK motor which has been studied in vitro, the bacteriophage packaging motor, and the motor driving DNA import into living bacteria during transformation.
DNA translocation during cell division: FtsK
Cell division in prokaryotes is not as well temporarily synchronized with replication as it is in eukaryotes [4]. Additionally due to the circular form of many bacterial chromosomes, homologous recombination produces chromosome dimers. These dimers have to be resolved before cell separation in order to avoid the lethal configuration where a dimer is trapped at the septum during its closure. FtsK is the essential protein that catalyzes this reaction. It pumps the trapped dimer until it encounters the recombinases XerC/D that bind specifically to the dif site [5], see figure 1C). XerC/D reaction is catalyzed by the mechanical contact of FtsK on XerD [5–7]. FtsK activity is increased when the motor is acting in the terminal region of the chromosome [8]. FtsK translocation is also related to cell division and replication by its ability, demonstrated in vitro [9], to interact with topoimoserase IV and stimulate decatenation and positive supercoils relaxation.
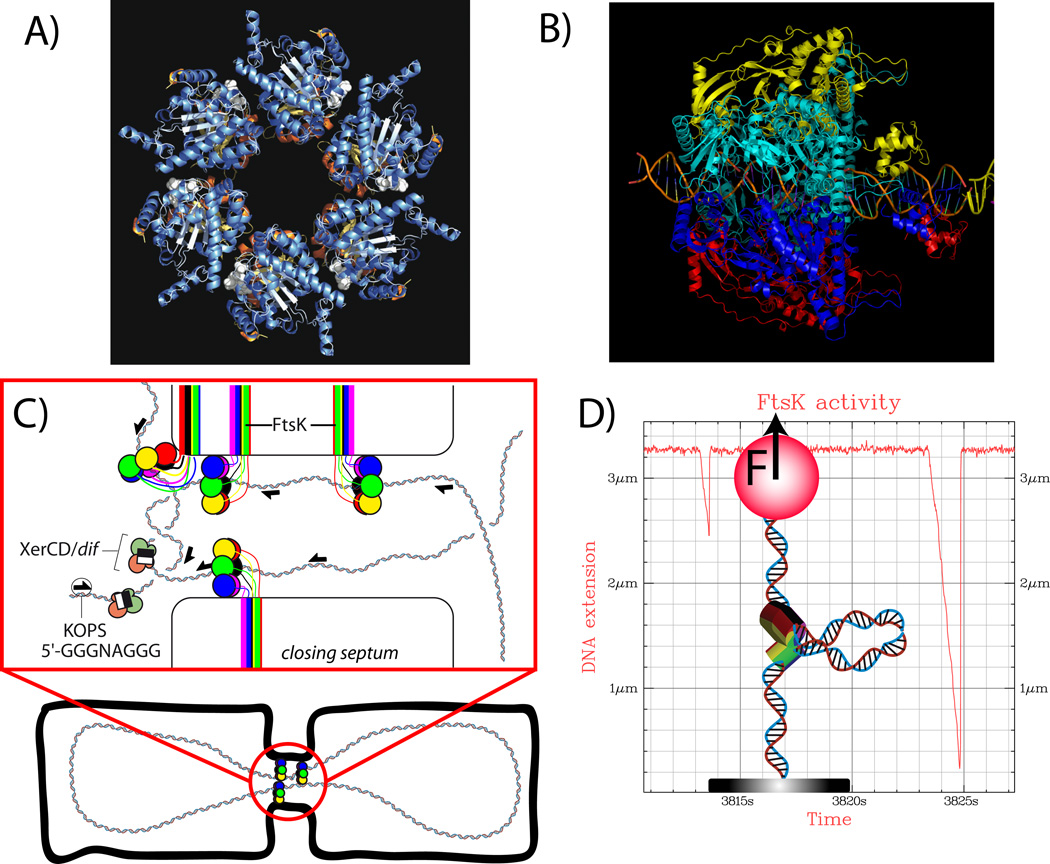
Figure 1.
A) Structure of FtsK : α and β domains of FtsK form an hexameric complex [11]. B) The hexamer has a central hole that allows the pumping of DNA during bacterial cell division. Central DNA and γ domain have been modeled onto the structure of A). Images from A) and B) were provided by D. Sherratt. C) FtsK localizes at the septum and catalyzes XerCD recombination at dif sites to resolve dimers produced by homologous recombination[5]. The exact structure of FtsK at the septum is not yet known but specific pores, formed by the membrane bound N terminal part of the proteins, are not required [10]. D) FtsK activity can be studied in vitro using optical or magnetic tweezers[15,16]. When FtsK is active it forms a loop that transiently shortens DNA extension. Distribution of changes in extension gives the processivity of the motor. Distribution of the slopes gives the speed. Both can be studied as function of ATP concentration, force, sequence, etc.
FtsK is a member of the FtsK/HerA family related of the AAA+ proteins. It possesses a membrane bound N terminal domain that localizes the protein at the septum. Through a linker, that needs to be long enough for efficient chromosome segregation [10], this membrane bound part is related to the C terminal part made from 3 domains α, β, γ [11]. C terminal functional assembly is a hexamer presenting a hole in the center that allows double stranded DNA (dsDNA) to go through, see figure 1A) and 1B) [11]. This feature common to other DNA pumping systems like TrwB, involved in the E. coli R388 conjugative system [12], or TraB, involved in conjugation in Streptomyces [13].
This C-terminal part was demonstrated to use ATP and translocate on DNA [14]. Single molecule studies performed at room temperature measured translocation velocities up to 7 kbps/s [15,16] at saturating ATP concentration, see figure 1D). The motor could not be stalled even at forces higher than 50 pN, a value more than 10 times higher than myosin II stalling force. The putative role for this high force generation is to provide the motor an ability to displace strongly interacting proteins on DNA that would otherwise act as roadblocks impeding FtsK activity in dimer resolution [17]. The same capability applies to SpoIIIE, a homologous protein involved in sporulation in Bacillus subtilis [18]. It must be noted that translocation speed and ability to displace roadblocks are two different mechanical properties that are not related as was demonstrated with mutants in the Walker motifs in some monomers [17].
In a recent study, a kinetic analysis of ATP dependence of translocation rates demonstrated cooperative cycles of ATP hydrolysis with ~2 bp translocated per ATP hydrolyzed [19] implying an energy transduction efficiency of about 50% (estimated with a stall force of 60 pN and an energy of 20 kBT for ATP hydrolysis). Such a step size provides an origin for the property of FtsK hexamers to rotate around DNA during translocation[20]. A rotary inchworm model, involving α and β domains, was proposed to explain this behavior based on FtsK structure [11]. The current proposed mechanism [19] is a sequential hydrolysis around the hexamer with a step size of 2 bps hydrolyzing 1 ATP and with a slight rotation around DNA at each step. However, mutations in Walker motifs impeding ATP hydrolysis do not block the motor [17] implying the sequential hydrolysis is not crucial.
To fulfill its role in dimer resolution FtsK has to physically contact XerD that is bound at a specific location on the chromosome [6]. An essential element for FtsK translocation is thus its translocation directionality. Different explanations were initially considered with both bi-directional and partially directed movement [15,16]. It was later demonstrated by multiple means [21–24] that, through its γ domain, FtsK interacts with specific polar sequences, named KOPS (GGGNAGGG), that have statistically skewed orientations along the chromosome[25,26]. The resulting orientation of the movement is not 100% efficient: three consecutive KOPS sequences must be placed in a row to reach a ~40% efficiency, a value proposed to allow directional transport on average, while avoiding potential trapping in vivo between two anti oriented KOPS [26]. Similar mechanisms for recognition of polar sequences were also observed for SpoIIIE [27]. A recent study suggested that translocation orientation is the result of an oriented binding of the hexamer, while during translocation FtsK would be insensitive to KOPS orientation[19].
Viral encapsidation
Viruses are among the simplest life forms, typically consisting of just a nucleic acid molecule enclosed in a protein shell. A remarkable step in the assembly of many dsDNA viruses is the use of an ATP-powered DNA translocating motor that packages DNA into preassembled capsid shells (Fig. 2A)[28] [29–33]. This process has recently been studied via single DNA manipulation with optical tweezers. A capsid is attached to one trapped microsphere and the DNA terminus is attached to a second microsphere [34–37]. As packaging proceeds the microspheres are pulled together and translocation velocity and force are directly measured (Fig. 2B,C). Three different motors, from phages phi29, lambda, and T4, have been studied.
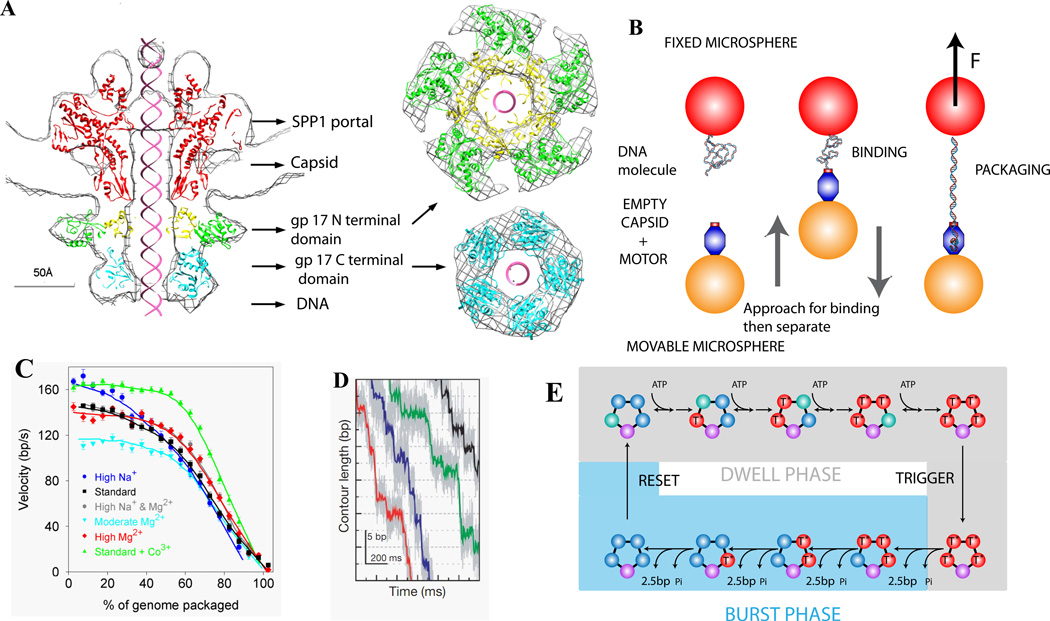
Figure 2.
(A) Structure of the Phage T4 DNA packaging motor [28]. (B) Measuring single phi29 viral DNA packaging using optical tweezers [34,35]. (C) Average motor velocity vs. % of genome packaged at 5 pN load. (D) High-resolution measurements of DNA translocation, showing phi29 packaging occurs in bursts of four ~2.5 bp steps [43]. (E) Schematic model of motor subunit coordination [43].
All three motors exhibited high force generation (>50–80 pN), high processivity (translocating >10–50 kbp without slipping), and decreasing velocity with increasing load and increasing capsid filling. The average motor velocity at low load ranged from 145 bp/s for phi29 (19.3 kbp genome) to 590 bp/s for lambda (48.5 kbp) to 770 bp/s for T4 (171 kbp).
The measured decrease in velocity with capsid filling indicate that a large force of ~50–100 pN resists DNA confinement [36–39], consistent with theoretically predicted forces arising from DNA electrostatic self-repulsion, bending rigidity, and entropy loss [40–42]. Studies of the effect of ionic screening of DNA charge confirm that electrostatic repulsion is the main factor [39]. Two complexities were observed in lambda packaging [36]. A measured dip in motor velocity at ~30% filling suggests that capsid expansion temporarily relieves internal force buildup and is triggered by ~4 pN of force on average. Second, a sudden increase in velocity above 90% filling is consistent with capsid rupture in the absence of a putative stabilizing protein, gpD.
High resolution optical tweezers measurements revealed that the motor translocates in bursts of four 2.5 bp steps, suggesting that the motor subunits are highly coordinated and four subunits bind ATP before a burst is triggered (Fig. 2E,F) [43]. Studies of the dependence of motor velocity on [ATP], [ADP], and [Pi] suggest that power strokes follow Pi release [44]. The motor was also shown to traverse single-stranded gaps, unpaired bulges, and synthetic polymer inserts, suggesting that translocation is mainly driven by non-specific steric interactions [45].
Effects on motor dynamics of residue changes in the lambda gpA motor subunit have provided information on mechanism [46,47]. Changes in velocity, processivity, and/or velocity-force dependence support the assignment of a Walker A-like phosphate-binding motif, an adenine binding Q motif analogous to that recently identified in RNA helicases, and a C motif that couples ATP hydrolysis to DNA translocation. Interestingly, a loop-helix-loop region predicted to position key residues of Walker B and C catalytic motifs also appears to regulate motor velocity in both the lambda virus motor and the related SpoIIIE/FtsK motor discussed above [48].
DNA import during horizontal gene transfer
Horizontal gene transfer enables bacteria to speed up adaptive evolution. Three different mechanisms have been identified for exchanging DNA between bacteria, namely transformation, conjugation, and transduction. Individual events of DNA transfer have been recently observed during transformation and conjugation [48][49].
Naturally competent bacteria generate a multi-protein machine that allows them to bind extracellular DNA, transport it through their cell envelope, and finally integrate it into their chromosome. Most of the characterized DNA import systems rely on the type IV pilus / type II secretion system. Through a series of studies with deletion mutants in different gram-positive and gram negative species, the components of the DNA import system have been identified [50]. The individual steps of DNA import and the mechanistic involvement of the molecular components is best understood in the gram-positive Bacillus subtilis [51] (Fig. 3a), although the molecular mechanism driving DNA import is still elusive [52]. The first steps are binding of DNA to the cell surface and recruitment into a DNase resistant state [53]. One single strand is imported through the membrane channel ComEC into the cytoplasm, where the DNA is rapidly covered by single-strand binding proteins for protection and further DNA processing [54]. The only known naturally competent bacterial species which does not employ type IV pilus proteins for DNA import during transformation is the gram-negative Helicobacter pylori. The latter has adapted a type IV secretion system for DNA import (Fig. 3b) [55,56]. It is therefore interesting to compare the physical characteristics of these two DNA import machines.
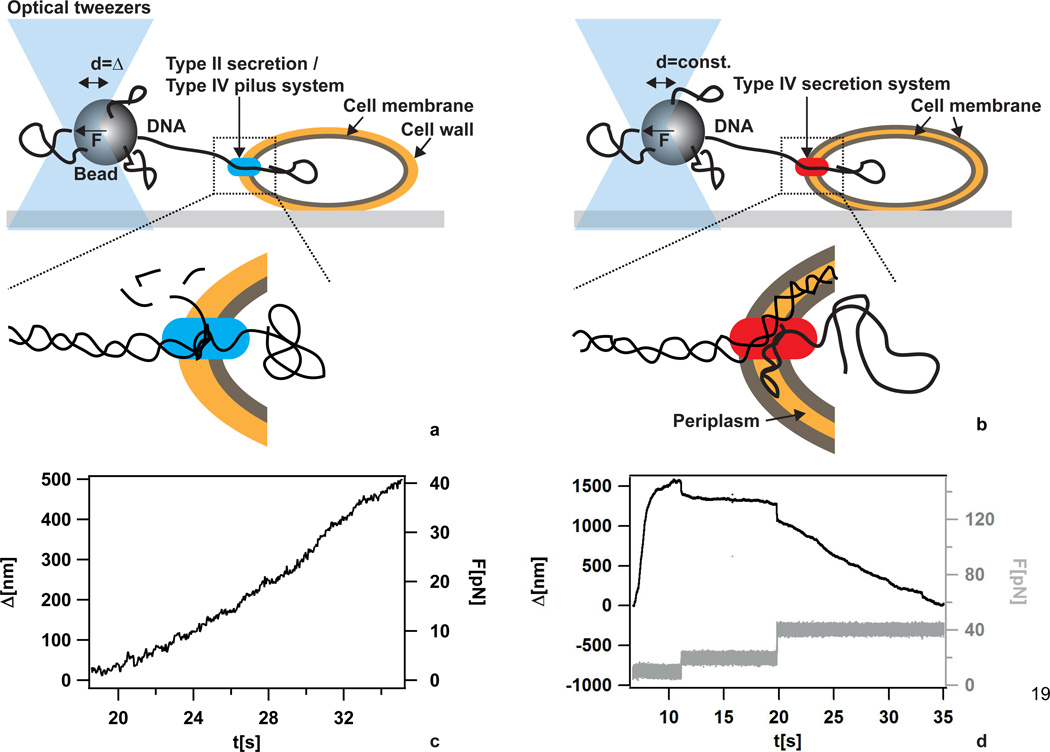
Figure 3.
DNA import during transformation. a) Scheme of the setup for measuring DNA import kinetics through the single membrane of B. subtilis. No force clamp was applied. b) Scheme of the setup for measuring DNA import kinetics through the two membranes of H. pylori. A force clamp was applied. c) Typical DNA import event in B. subtilis. Δ denotes the length of imported DNA after monitoring was started. With increasing Δ, the deflection of the bead from the center of the laser trap increased leading to increasing force F. d) Typical DNA import event in H. pylori. Δ denotes the length of imported DNA after monitoring was started. The force F was clamped at different values.
Using laser tweezers, DNA import was characterized at the single molecule level [57–59]. The experimental approach was to bind DNA to a micrometer-sized bead at one extremity and to offer the other extremity to bacteria that were firmly attached to a glass cover slide (Fig. 3a, b). As bacteria bound single DNA molecules and imported them, the shortening of external DNA as a function of time was measured at different forces (Fig. 3c, d). Both B. subtilis and H. pylori used highly processive machines for DNA import. The speed of import of B. subtilis was ≈ 80 bp/s and largely independent of external force up to forces of 40 pN [57]. With H. pylori, the speed of import was larger by an order of magnitude (≈ 1.3 kbp/s at F = 10 pN) but was strongly dependent on the external force [59]. In contrast to B. subtilis, DNA import was reversible by application of a force exceeding ≈ 23 pN in H. pylori.
Although the proteins essential for transformation have been identified, it is still unclear, which of them is the dominant molecular motor driving DNA translocation. Potential mechanisms include a translocation ratchet, type IV pilus retraction, and cyclic ATP consuming motors [52]. Chemical treatment indicated that DNA transport was inhibited by depletion proton motive force in B. subtilis. In H. pylori both uncouplers and inhibitors of the ATP synthase inhibited DNA transport, showing that DNA import is an energy consuming process. Several lines of evidence indicate that different competent species share a common mechanism for inner membrane transport of DNA, whereas the outer membrane transport has evolved differently for different species [60]. The inner membrane channel of H. pylori shares homology with ComEC, the channel involved in DNA import in B. subtilis [61]. Concomitantly, in both species structurally modified DNA was not imported into the cytoplasm [59]. Furthermore, there is good evidence that dsDNA (both modified and unmodified) can accumulate in the periplasm of H. pylori prior to transport into the cytoplasm, indicating a two-step import. Future experiments need to elucidate the role of the type of secretion system for the physical properties of DNA import by characterizing DNA import in a gram-negative bacterium based on the type IV pilus system.
Conclusion
Molecular motors involved in DNA translocation share a high processivity and a high stall force and a complex structure. Nevertheless many molecular details on which such properties are based are still to be discovered as well as the way those multimeric motors assemble during the cell cycle. Single-molecule techniques have proved to be a powerful approach to complement traditional biochemistry and cell biology studies of these systems.
Highlights.
Single molecule techniques enable mechanistic insight into DNA translocation motors.
DNA transport in bacteria and phages is driven by powerful molecular motors.
During chromosome segregation or dimer resolution SpoIIIE/FtsK translocation directionality is set by polar DNA sequences.
Translocation velocity decreases with capsid filling during viral encapsidation.
Force-dependent DNA import rate during transformation depends on bacterial species.
Acknowledgements
J-F A. wishes to thanks O. A. Saleh, F.X. Barre, F. Cornet, D. Sherratt, S. Bigot, E. Crozat, A. Meglio, L. Bonne for their collaboration and ANR, IUF for their financial support. D.E.S. thanks D. Fuller, J. Rickgauer, D. Raymer, J. Tsay, D. del Toro and members of the Bustamante, Anderson, Grimes, Jardine, Catalano, Feiss, and Rao labs for collaboration, and NSF and NIH for support. B.M. is grateful for collaborations with D. Dubnau and M. Koomey, and for support by the Deutsche Forschungsgemienschaft through grant MA3898.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Greenleaf WJ, Woodside MT, Block SM. High-resolution, single-molecule measurements of biomolecular motion. Annu Rev Biophys Biomol Struct. 2007;36:171–190. doi: 10.1146/annurev.biophys.36.101106.101451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Manosas M, Meglio A, Spiering MM, Ding F, Benkovic SJ, Barre FX, Saleh OA, Allemand JF, Bensimon D, Croquette V. Magnetic tweezers for the study of DNA tracking motors. Methods Enzymol. 2010;475:297–320. doi: 10.1016/S0076-6879(10)75013-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bustamante C, Cheng W, Mejia YX. Revisiting the central dogma one molecule at a time. Cell. 2011;144:480–497. doi: 10.1016/j.cell.2011.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sherratt DJ. Bacterial chromosome dynamics. Science. 2003;301:780–785. doi: 10.1126/science.1084780. [DOI] [PubMed] [Google Scholar]
- 5.Grainge I, Lesterlin C, Sherratt DJ. Activation of XerCD-dif recombination by the FtsK DNA translocase. Nucleic Acids Res. 2011;39:5140–5148. doi: 10.1093/nar/gkr078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Graham JE, Sivanathan V, Sherratt DJ, Arciszewska LK. FtsK translocation on DNA stops at XerCD-dif. Nucleic Acids Res. 2010;38:72–81. doi: 10.1093/nar/gkp843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bonne L, Bigot S, Chevalier F, Allemand JF, Barre FX. Asymmetric DNA requirements in Xer recombination activation by FtsK. Nucleic Acids Res. 2009;37:2371–2380. doi: 10.1093/nar/gkp104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deghorain M, Pages C, Meile JC, Stouf M, Capiaux H, Mercier R, Lesterlin C, Hallet B, Cornet F. A defined terminal region of the E. coli chromosome shows late segregation and high FtsK activity. PLoS One. 2011;6:e22164. doi: 10.1371/journal.pone.0022164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bigot S, Marians KJ. DNA chirality-dependent stimulation of topoisomerase IV activity by the C-terminal AAA+ domain of FtsK. Nucleic Acids Res. 2010;38:3031–3040. doi: 10.1093/nar/gkp1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dubarry N, Barre FX. Fully efficient chromosome dimer resolution in Escherichia coli cells lacking the integral membrane domain of FtsK. EMBO J. 2010;29:597–605. doi: 10.1038/emboj.2009.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Massey TH, Mercogliano CP, Yates J, Sherratt DJ, Lowe J. Double-stranded DNA translocation: structure and mechanism of hexameric FtsK. Mol Cell. 2006;23:457–469. doi: 10.1016/j.molcel.2006.06.019. [DOI] [PubMed] [Google Scholar]
- 12.Gomis-Ruth FX, Moncalian G, Perez-Luque R, Gonzalez A, Cabezon E, de la Cruz F, Coll M. The bacterial conjugation protein TrwB resembles ring helicases and F1-ATPase. Nature. 2001;409:637–641. doi: 10.1038/35054586. [DOI] [PubMed] [Google Scholar]
- 13.Vogelmann J, Ammelburg M, Finger C, Guezguez J, Linke D, Flotenmeyer M, Stierhof YD, Wohlleben W, Muth G. Conjugal plasmid transfer in Streptomyces resembles bacterial chromosome segregation by FtsK/SpoIIIE. EMBO J. 2011;30:2246–2254. doi: 10.1038/emboj.2011.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aussel L, Barre FX, Aroyo M, Stasiak A, Stasiak AZ, Sherratt D. FtsK Is a DNA motor protein that activates chromosome dimer resolution by switching the catalytic state of the XerC and XerD recombinases. Cell. 2002;108:195–205. doi: 10.1016/s0092-8674(02)00624-4. [DOI] [PubMed] [Google Scholar]
- 15.Saleh OA, Perals C, Barre FX, Allemand JF. Fast, DNA-sequence independent translocation by FtsK in a single-molecule experiment. EMBO J. 2004;23:2430–2439. doi: 10.1038/sj.emboj.7600242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pease PJ, Levy O, Cost GJ, Gore J, Ptacin JL, Sherratt D, Bustamante C, Cozzarelli NR. Sequence-directed DNA translocation by purified FtsK. Science. 2005;307:586–590. doi: 10.1126/science.1104885. [DOI] [PubMed] [Google Scholar]
- 17. Crozat E, Meglio A, Allemand JF, Chivers CE, Howarth M, Venien-Bryan C, Grainge I, Sherratt DJ. Separating speed and ability to displace roadblocks during DNA translocation by FtsK. EMBO J. 2010;29:1423–1433. doi: 10.1038/emboj.2010.29. *This work demonstrate how some mutations can affect the force produced by the motor without affecting its speed. This rules out some translocation models.
- 18.Marquis KA, Burton BM, Nollmann M, Ptacin JL, Bustamante C, Ben-Yehuda S, Rudner DZ. SpoIIIE strips proteins off the DNA during chromosome translocation. Genes Dev. 2008;22:1786–1795. doi: 10.1101/gad.1684008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Graham JE, Sherratt DJ, Szczelkun MD. Sequence-specific assembly of FtsK hexamers establishes directional translocation on DNA. Proc Natl Acad Sci U S A. 2010;107:20263–20268. doi: 10.1073/pnas.1007518107. * Proposes a possible model for assembly, initiation and translocation of FtSK based on millisecond stop flow analysis.
- 20.Saleh OA, Bigot S, Barre FX, Allemand JF. Analysis of DNA supercoil induction by FtsK indicates translocation without groove-tracking. Nat Struct Mol Biol. 2005;12:436–440. doi: 10.1038/nsmb926. [DOI] [PubMed] [Google Scholar]
- 21.Sivanathan V, Allen MD, de Bekker C, Baker R, Arciszewska LK, Freund SM, Bycroft M, Lowe J, Sherratt DJ. The FtsK gamma domain directs oriented DNA translocation by interacting with KOPS. Nat Struct Mol Biol. 2006;13:965–972. doi: 10.1038/nsmb1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bigot S, Saleh OA, Cornet F, Allemand JF, Barre FX. Oriented loading of FtsK on KOPS. Nat Struct Mol Biol. 2006;13:1026–1028. doi: 10.1038/nsmb1159. [DOI] [PubMed] [Google Scholar]
- 23.Ptacin JL, Nollmann M, Bustamante C, Cozzarelli NR. Identification of the FtsK sequence-recognition domain. Nat Struct Mol Biol. 2006;13:1023–1025. doi: 10.1038/nsmb1157. [DOI] [PubMed] [Google Scholar]
- 24.Lowe J, Ellonen A, Allen MD, Atkinson C, Sherratt DJ, Grainge I. Molecular mechanism of sequence-directed DNA loading and translocation by FtsK. Mol Cell. 2008;31:498–509. doi: 10.1016/j.molcel.2008.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bigot S, Saleh OA, Lesterlin C, Pages C, El Karoui M, Dennis C, Grigoriev M, Allemand JF, Barre FX, Cornet F. KOPS: DNA motifs that control E. coli chromosome segregation by orienting the FtsK translocase. EMBO J. 2005;24:3770–3780. doi: 10.1038/sj.emboj.7600835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Levy O, Ptacin JL, Pease PJ, Gore J, Eisen MB, Bustamante C, Cozzarelli NR. Identification of oligonucleotide sequences that direct the movement of the Escherichia coli FtsK translocase. Proc Natl Acad Sci U S A. 2005;102:17618–17623. doi: 10.1073/pnas.0508932102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ptacin JL, Nollmann M, Becker EC, Cozzarelli NR, Pogliano K, Bustamante C. Sequence-directed DNA export guides chromosome translocation during sporulation in Bacillus subtilis. Nat Struct Mol Biol. 2008;15:485–493. doi: 10.1038/nsmb.1412. * Combination of single molecule studies with in vivo studies to compare DNA transport by SpoIIIE and FtsK. Specie sequence specificity was nicely switched by exchange of the γ domain of SpoIIIE and FtsK.
- 28.Sun S, Kondabagil K, Draper B, Alam TI, Bowman VD, Zhang Z, Hegde S, Fokine A, Rossmann MG, Rao VB. The Structure of the Phage T4 DNA Packaging Motor Suggests a Mechanism Dependent on Electrostatic Forces. Cell. 2008;135:1251–1262. doi: 10.1016/j.cell.2008.11.015. [DOI] [PubMed] [Google Scholar]
- 29.Rao VB, Feiss M. The Bacteriophage DNA Packaging Motor. Annu Rev Genet. 2008;42:647–681. doi: 10.1146/annurev.genet.42.110807.091545. [DOI] [PubMed] [Google Scholar]
- 30.Catalano CE, editor. Viral Genome Packaging Machines: Genetics, Structure, and Mechanism. 2005 [Google Scholar]
- 31.Jardine PJ, Anderson DL. DNA packaging in double-stranded DNA bacteriophages. In: Calendar R, editor. The Bacteriophages. Oxford Press; 2006. p. 49. [Google Scholar]
- 32.Rao VB, Black LW. Bacteriophage T4 DNA Packaging. In: Catalano CE, editor. Viral Genome Packaging Machines: Genetics, Structure and Mechanism. Landes Biosciences; 2005. [Google Scholar]
- 33.Smith DE. Single-molecule studies of viral DNA packaging. Current Opinion in Virology. 2011;1:134. doi: 10.1016/j.coviro.2011.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smith DE, Tans SJ, Smith SB, Grimes S, Anderson DL, Bustamante C. The bacteriophage phi29 portal motor can package DNA against a large internal force. Nature. 2001;413:748–752. doi: 10.1038/35099581. [DOI] [PubMed] [Google Scholar]
- 35.Rickgauer JP, Fuller DN, Grimes S, Jardine PJ, Anderson DL, Smith DE. Portal motor velocity and internal force resisting viral DNA packaging in bacteriophage phi29. Biophys J. 2008;94:159–167. doi: 10.1529/biophysj.107.104612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fuller DN, Raymer DM, Rickgauer JP, Robertson RM, Catalano CE, Anderson DL, Grimes S, Smith DE. Measurements of single DNA molecule packaging dynamics in bacteriophage lambda reveal high forces, high motor processivity, and capsid transformations. J Mol Biol. 2007;373:1113–1122. doi: 10.1016/j.jmb.2007.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fuller DN, Raymer DM, Kottadiel VI, Rao VB, Smith DE. Single phage T4 DNA packaging motors exhibit barge force generation, high velocity, and dynamic variability. Proc Natl Acad Sci U S A. 2007;104:16868–16873. doi: 10.1073/pnas.0704008104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rickgauer JP, Smith DE. Single-Molecule Studies of DNA Visualization and Manipulation of Individual DNA Molecules with Fluorescence Microscopy and Optical Tweezers. In: Borsali R, Pecora R, editors. Soft Matter: Scattering, Imaging And Manipulation. Springer; 2008. [Google Scholar]
- 39.Fuller DN, Rickgauer JP, Jardine PJ, Grimes S, Anderson DL, Smith DE. Ionic effects on viral DNA packaging and portal motor function in bacteriophage phi 29. Proc Natl Acad Sci U S A. 2007;104:11245–11250. doi: 10.1073/pnas.0701323104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tzlil S, Kindt JT, Gelbart WM, Ben-Shaul A. Forces and pressures in DNA packaging and release from viral capsids. Biophys J. 2003;84:1616–1627. doi: 10.1016/S0006-3495(03)74971-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Purohit PK, Inamdar MM, Grayson PD, Squires TM, Kondev J, Phillips R. Forces during bacteriophage DNA packaging and ejection. Biophys J. 2005;88:851–866. doi: 10.1529/biophysj.104.047134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Petrov AS, Harvey SC. Packaging double-helical DNA into viral capsids: Structures, forces, and energetics. Biophys J. 2008;95:497–502. doi: 10.1529/biophysj.108.131797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Moffitt JR, Chemla YR, Aathavan K, Grimes S, Jardine PJ, Anderson DL, Bustamante C. Intersubunit coordination in a homomeric ring ATPase. Nature. 2009;457:446–450. doi: 10.1038/nature07637. ** Direct high-resolution measurement of bursts of 2.5 bp steps of the phi29 packaging motor, leading to a model for motor kinetics and subunit coordination.
- 44.Chemla YR, Aathavan K, Michaelis J, Grimes S, Jardine PJ, Anderson DL, Bustamante C. Mechanism of force generation of a viral DNA packaging motor. Cell. 2005;122:683–692. doi: 10.1016/j.cell.2005.06.024. [DOI] [PubMed] [Google Scholar]
- 45. Aathavan K, Politzer AT, Kaplan A, Moffitt JR, Chemla YR, Grimes S, Jardine PJ, Anderson DL, Bustamante C. Substrate interactions and promiscuity in a viral DNA packaging motor. Nature. 2009;461:669–673. doi: 10.1038/nature08443. * Insights of the nature of motor-DNA interactions from studies of the dynamics of packaging of DNA templates with inserts containing structural modifications.
- 46. Tsay JM, Sippy J, Feiss M, Smith DE. The Q motif of a viral packaging motor governs its force generation and communicates ATP recognition to DNA interaction. Proc Natl Acad Sci U S A. 2009;106:14355–14360. doi: 10.1073/pnas.0904364106. * Lambda motor mutant studies provided evidence for a Walker A-like phosphate-binding motif and an adenine binding motif that regulates substrate affinity and motor power, analogous to the Q motif recently identified in RNA helicases.
- 47. Tsay JM, Sippy J, DelToro D, Andrews BT, Draper B, Rao V, Catalano CE, Feiss M, Smith DE. Mutations altering a structurally conserved loop-helix-loop region of a viral packaging motor change DNA translocation velocity and processivity. J Biol Chem. 2010;285:24282–24289. doi: 10.1074/jbc.M110.129395. ** Lambda motor mutant studies provided evidence for a C (coupling) motif regulating motor processivity and a loop-helix-loop region regulating motor velocity.
- 48.Babic A, Lindner AB, Vulic M, Stewart EJ, Radman M. Direct visualization of horizontal gene transfer. Science. 2008;319:1533–1536. doi: 10.1126/science.1153498. [DOI] [PubMed] [Google Scholar]
- 49.Clarke M, Maddera L, Harris RL, Silverman PM. F-pili dynamics by live-cell imaging. Proc Natl Acad Sci U S A. 2008;105:17978–17981. doi: 10.1073/pnas.0806786105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Burton B, Dubnau D. Membrane-associated DNA transport machines. Cold Spring Harb Perspect Biol. 2010;2:a000406. doi: 10.1101/cshperspect.a000406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Maier B. In: Competence and Transformation. Graumann P, editor. Caister University Press; 2011. [Google Scholar]
- 52.Allemand JF, Maier B. Bacterial translocation motors investigated by single molecule techniques. FEMS Microbiol Rev. 2009;33:593–610. doi: 10.1111/j.1574-6976.2009.00166.x. [DOI] [PubMed] [Google Scholar]
- 53.Briley K, Jr, Dorsey-Oresto A, Prepiak P, Dias MJ, Mann JM, Dubnau D. The secretion ATPase ComGA is required for the binding and transport of transforming DNA. Mol Microbiol. 2011;81:818–830. doi: 10.1111/j.1365-2958.2011.07730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Attaiech L, Olivier A, Mortier-Barriere I, Soulet AL, Granadel C, Martin B, Polard P, Claverys JP. Role of the single-stranded DNA-binding protein SsbB in pneumococcal transformation: maintenance of a reservoir for genetic plasticity. PLoS Genet. 2011;7:e1002156. doi: 10.1371/journal.pgen.1002156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hofreuter D, Odenbreit S, Haas R. Natural transformation competence in Helicobacter pylori is mediated by the basic components of a type IV secretion system. Mol Microbiol. 2001;41:379–391. doi: 10.1046/j.1365-2958.2001.02502.x. [DOI] [PubMed] [Google Scholar]
- 56.Karnholz A, Hoefler C, Odenbreit S, Fischer W, Hofreuter D, Haas R. Functional and topological characterization of novel components of the comB DNA transformation competence system in Helicobacter pylori. J Bacteriol. 2006;188:882–893. doi: 10.1128/JB.188.3.882-893.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Maier B, Chen I, Dubnau D, Sheetz MP. DNA transport into Bacillus subtilis requires proton motive force to generate large molecular forces. Nat Struct Mol Biol. 2004;11:643–649. doi: 10.1038/nsmb783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hahn J, Maier B, Haijema BJ, Sheetz M, Dubnau D. Transformation proteins and DNA uptake localize to the cell poles in Bacillus subtilis. Cell. 2005;122:59–71. doi: 10.1016/j.cell.2005.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Stingl K, Muller S, Scheidgen-Kleyboldt G, Clausen M, Maier B. Composite system mediates two-step DNA uptake into Helicobacter pylori. Proc Natl Acad Sci U S A. 2010;107:1184–1189. doi: 10.1073/pnas.0909955107. ** Combination of single molecule manipulation, fluorescence, and genetics showing that H. pylori imports DNA fast, reversibly, and in a two-step process.
- 60.Kruger NJ, Stingl K. Two steps away from novelty--principles of bacterial DNA uptake. Mol Microbiol. 2011;80:860–867. doi: 10.1111/j.1365-2958.2011.07647.x. [DOI] [PubMed] [Google Scholar]
- 61.Yeh YC, Lin TL, Chang KC, Wang JT. Characterization of a ComE3 homologue essential for DNA transformation in Helicobacter pylori. Infect Immun. 2003;71:5427–5431. doi: 10.1128/IAI.71.9.5427-5431.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]