The poly(ADP-ribose) glycohydrolase (PARG) catalyzes the removal of PAR chains from posttranslationally modified proteins by hydrolysis of α(122-22) O-glycosidic linkages, functioning as an endo-glycosidase to release oligo(ADP-ribose) and as an exo-glycosidase to release ADP-ribose1,2. Long PAR polymers are efficiently hydrolyzed (Km = ~ 1 μM) by a combination of endo- and exo-glycosidic activity3, whereas smaller digestion products are poor substrates for PARG (Km > 10 μM) allowing release of oligo(ADP-ribose) chains that are ligands for histones and DNA repair and damage checkpoint proteins such as XRCC1 and p534.
PARG comprises an N-terminal regulatory and targeting domain (A-domain; residues 1-456 in the rat protein), a central mitochondrial targeting sequence (MTS; residues 457-472), and a C-terminal catalytic domain (residues 473-972)5. The A-domain is required for recruitment of PARG to DNA damage sites6, and mice expressing a PARG isoform lacking the A-domain are hypersensitive to genotoxic stresses5. This isoform has constitutively high enzymatic activity7 and retains the MTS that is essential for PARG activity in vitro8, suggesting a regulatory and/or structural role of the A-domain and MTS.
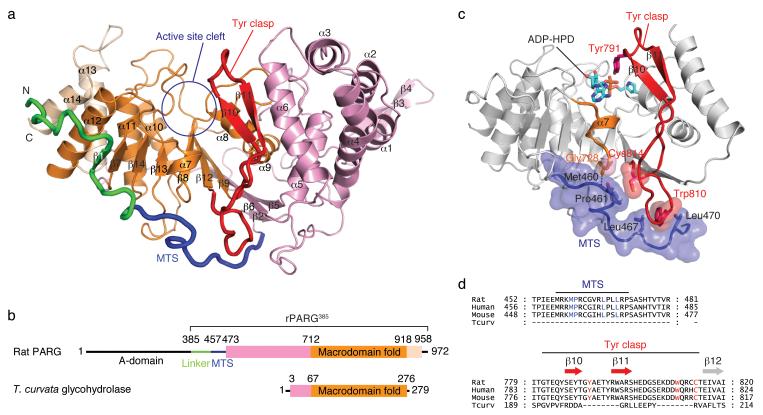
To better understand PARG enzymatic functions and their potential for regulation, we determined a 1.95 Å crystal structure of rat PARG (rPARG385; residues 385-972) lacking the poorly structured A-domain that does not contribute to PARG activity in vitro8 and its structure bound to the transition state analog ADP-HPD9 (Fig. 1 and Supplementary Figs.1, 2, and 3, and Supplementary Table 1). The rPARG385 catalytic domain adopts a bean-shaped structure with a deep central cleft containing the conserved PARG-signature motif (GGG-X6-8-QEE)10 and Tyr791 that contributes strongly to PARG catalytic efficiency and inhibitor binding11 (Fig. 1). The structure consists of a α-β-α fold with a nine-stranded, mixed β-sheet flanked by several layers of α-helices (Fig. 1a and Supplementary Fig. 4a). The active site cleft lies on one edge of the β-sheet and an extended N-terminal segment containing the MTS wraps around the other edge of the β-sheet, contributing to the PARG catalytic domain (Fig. 1). The core of rPARG385 resembles a canonical macrodomain found in ADP-ribose binding proteins, but the rPARG385 fold is more elaborate than other macrodomains (Fig. 1 and Supplementary Fig. 4)12-15 including a bacterial PAR glycohydrolase from Thermomonospora curvata14. The core structure of rPARG385 closely resembles the bacterial PARG homologue (DALI16 Z score = 16.5, rmsd. = 3.2 Å for 208 Cα atoms) with many of the catalytic residues superimposing well (Fig. 2a). However, the shape of the substrate-binding site differs substantially in mammalian and bacterialglycohydrolases, including a unique structural element we have named the “tyrosine clasp” (Tyr clasp) (Figs. 1 and 2).
Figure 1.
Mammalian poly(ADP-ribose) glycohydrolase (PARG) structure. (a) The catalytic domain of rat PARG (rPARG385; residues 385-972) consists of a core macrodomain fold (orange) sandwiched between flanking N-terminal (pink) and C-terminal (beige) helical bundles. The mitochondrial targeting sequence (MTS; blue) wraps around the catalytic domain and stabilizes the tyrosine clasp (Tyr clasp; red), a unique substrate-binding element of mammalian PARG. (b) Domain organization of rat PARG and Thermomonospora curvata glycohydrolase. (c) The MTS buttresses the Tyr clasp, orienting Tyr791 towards the active site cleft, explaining why the MTS is required for PARG activity. (d) Sequence alignment reveals conserved residues in mammalian PARG that participate in the interaction between the MTS and the Tyr clasp.
Figure 2.
PARG-inhibitor complex structure reveals the Tyr clasp as a unique substrate-binding element of mammalian PARG. (a) The superposition of rPARG385 (beige) and T. curvata glycohydrolase (cyan) active sites shows that the substrate analogue ADP-HPD binds in the same orientation with respect to conserved active site residues. The mammalian Tyr clasp, which is not found in the bacterial enzyme, positions Tyr791 at its apex to coordinate with the O5′ of the diphosphate of ADP-HPD and edge-stack with its adenine ring. (b) Stereo diagram of the active site of rPARG385. A matrix of van der Waals contacts and polar interactions secures ADP-HPD for catalysis by Glu752. Two regions of the unliganded rPARG385 structure (purple) change conformation slightly in the ligand-bound structure (beige). Several hydrogen bonds from main chain atoms to the ligand were omitted for clarity.
The conserved macrodomain topology centers on a seven-stranded sheet (strand order 1-2-7-6-3-5-4; Supplementary Figs. 2 and 4a)12,14 with five conserved α-helices of the macrodomain fold (H1-H5) inserted in the connections between strands where they pack against either side of the β-sheet. These helices and connecting loops include the majority of residues participating in the ADP-ribose binding12, deacetylase15, and glycohydrolase14 activities of different macrodomains. The rPARG385 macrodomain lacks strand S1, but has three additional β-strands (β2, β5, and β6) adjacent to strand S4 that expand the width of the sheet and provide additional surface for helices α1-α6 to pack against (Fig. 1a and Supplementary Fig. 4a). These additional structural elements, which are contributed by residues located between the MTS and the macrodomain core of rPARG385, pack against one face of the central β-sheet, giving a comma-shaped appearance and accounting for nearly one-third of the residues in the catalytic domain. The helical bundle comprising α1-α6 has no structural homologs in the Protein Data Bank. Helices α10-α14 located in the C-terminal half of the catalytic domain pack against the opposite face of the β-sheet to complete the fold. The N- and C-terminal helical bundles of PARG form the boundaries of a broad cleft that contains the active site (Fig. 1a) and are suggestive of specialized functions of the mammalian PARG including its substrate preference for long PAR polymers3.
The ADP-HPD inhibitor is a tight-binding mimic of ADP-ribose9 that was crystallized in the PARG active site (Fig. 2 and Supplementary Fig. 3). The pyrrolidine ring of ADP-HPD mimics the positively charged oxocarbenium ion of the transition state for glycosidase reactions17 and is similarly positioned in the rat PARG and T. curvata glycohydrolase structures (Fig. 2). The catalytic residue Glu75210,14 lies proximal to the anomeric C1’ position (Fig. 2b) where Glu752 could function as a general acid or base to protonate the 2′-OH of the ribose’ of the leaving group then activate water for nucleophilic attack. A water molecule close to Glu752 in the unliganded rPARG385 structure is in a position compatible with a nucleophilic attack of the ribose22 C12 of a PAR substrate (Supplementary Fig. 5a,b), as proposed for a similarly positioned water in the T. curvata glycohydrolase structure (Supplementary Fig. 5c)14. The PARG-signature motif (GGG-X6-8-GEE)10 extends from the glycine-rich loop and precisely orients the catalytic Glu752 towards the scissile O-glycosidic bond of the ribose22 moiety (Supplementary Fig. 4b); Two neighboring main chain nitrogen atoms from Gly742 and Val749 form hydrogen bonds to the side chain of Glu752 (Fig. 2b). This microenvironment may shift the effective pKa of Glu752 to enable protonation of the leaving group as the first step of the proposed mechanism (Supplementary Fig. 5d). The orientation of Glu752 relative to the substrate analogue in the crystal structure could support a water attack from either side of the ribose ring, generating either the ADP-α-ribose22 (retaining mechanism) or the ADP-β-ribose22 (inverting mechanism) (Supplementary Fig. 5d).
The orientation of ADP-HPD is similar in rPARG385 and T. curvata glycohydrolases (Fig. 2a), but the adenine ring and pyrrolidine substituents are more solvent accessible in rPARG385, enabling this enzyme to access internal sites of irregularly branching PAR chains. Mammalian PARG makes additional contacts to the adenylate moiety using the Tyr clasp and residues in the adenine-binding pocket (Fig. 2a and Supplementary Fig. 2), which may compensate for the comparatively exposed and unencumbered binding of the proximal ribose’. The Tyr clasp forms a β-hairpin (β10 and β11) with an apical Tyr791 pointing into the substrate binding cleft where it can crosslink to a photoaffinity conjugate of ADP-HPD11. Besides stacking with the adenine ring, the protruding Tyr791 side chain also forms a hydrogen bond with O5′ of the diphosphate of ADP-HPD (Fig. 2b). These interactions provide a structural rationale for the observed ~20-fold decrease in affinity for ADP-HPD in the bovine mutant equivalent to Y791A in the rat enzyme11. Importantly, the intricate interactions between rPARG385 and the substrate analogue impart the correct binding register with the enzyme and re-enforce proper alignment for catalysis.
The MTS proposed to function in mitochondrial import of PARG18 is also required for enzymatic activity in vitro, despite its distant location in the primary structure from the active site residues8. Thirty-five residues preceding and including the MTS traverse a more than 80 Å, taking an L-shaped path along the outer surface of the rPARG385 catalytic domain (Fig. 1). In this extended conformation, the MTS buttresses helix α7 and the base of the Tyr clasp (Fig. 1c). These interactions include a hydrogen bond between the main chain nitrogen atom of Met460 and the Gly728 main chain carbonyl on a loop following α7. Pro461 is within van der Waals contact distance of the Cys814 side chain of the Tyr clasp. Two conserved leucine residues (Leu467 and Leu470) pack against Trp810 side chain of the Tyr clasp (Fig. 1c). Deletion of the MTS or mutation of the conserved leucine residues would remove an anchor point and potentially destabilize helix α7 and the Tyr clasp, unraveling the adenine-binding pocket and explaining the loss of enzymatic activity caused by this truncation8. The adjacent A-domain is proposed to regulate PARG activity7,10 and is connected to the catalytic domain by an extended, solvent-exposed linker incorporating the MTS motif. Protein interactions or post-translational modifications of the A-domain might alter this connection to the catalytic domain of PARG and change enzymatic activity.
At the opposite end of the substrate-binding channel, rPARG385 makes extensive contacts with the adenine ring. Recognition of the adenine base is accomplished by α7 and the Tyr clasp (Fig. 2b). A total of eight residues (Tyr788 and Tyr791 from the Tyr clasp; Thr721, Ile722 and Glu723 from α7; Gln750 from α8; and Phe898) interact with the adenine ring, explaining the binding selectivity for adenine nucleosides1,19. Both side chain dihedral angles of Phe898 rotate approximately 180° relative to the unliganded enzyme position, opening the pocket to allow the adenine ring to bind and positioning the phenylalanine side chain for an edge-stacking π-π interaction with the ligand. Similar perpendicular π-π stacking is observed between the side chains of Tyr791 and Phe734 and the adenine moiety. Substitution of Tyr791 with tryptophan does not markedly affect catalytic activity11, revealing the importance of stacking interactions with the Tyr clasp namesake residue. The chemical identity of the adenine base is read out by hydrogen bonds between the side chain of Glu723 and N6, and the backbone nitrogen of Ile722 and N1 (Fig. 2b).
Mammalian PARG functions as both an endo- and exo-glycohydrolase, initially cleaving PAR polymers internally into shorter chains that are subsequently hydrolyzed by an inefficient exo-glycohydrolase activity, whereas the T. curvata glycohydrolase is proposed to only cleave the terminal PAR linkage14. The substantially expanded structure of the mammalian PARG catalytic domain in comparison to other macrodomain structures is probably linked to PARG’s substrate preference for large polymers of ADP-ribose3. The grooves extending from the ADP-HPD binding site are attractive candidates for additional interactions (Fig. 3) that support endo-glycohydrolase activity and the production of oligo(ADP-ribose) chains functioning in damage signaling20 and the regulation of chromatin structure4. Structural comparison of the ADP-HPD complexes of rPARG385 and the T. curvata glycohydrolase reveals that Arg268 of the bacterial enzyme caps the 2′-OH group of adenosine ribose, effectively blocking access to internal glycosidic linkages of the PAR polymer14 (Fig. 3a,b). The position of Arg268 is reinforced by an ion pair with Asp261 in the loop connecting conserved S7 and H5 (S7-H5 loop), a hydrogen bond to the main chain carbonyl of Cys224, and multiple van der Waals contacts14. This matrix of interactions readily explains why the bacterial enzyme is an obligate exo-glycohydrolase.
Figure 3.
The mammalian PARG structure supports endo-glycosidic cleavage of poly(ADP-ribose). (a) Sequence alignments of rat PARG, human PARG8, macrodomain Af152112, macrodomain D115, macrodomain H2A1.113, and T. curvata glycohydrolase14 reveal unique C-terminal helices (α13 and α14) in mammalian PARG that pack against α12. Interacting residues of α12, α13, and α14 of rat PARG are labeled with black dots (•. (b) The 2′-OH of ADP-HPD is exposed in the rPARG385 structure (orange) to enable binding and internal cleavage of PAR polymers by endo-glycohydrolase activity. One of three representative (n+1) ADP-ribose conformers from panel (c) is shown as a stick model (black). Residues from the S7-H5 loop ofthe T. curvata glycohydrolase (white) cap the ribose ring, limiting binding activity to the terminal ADP-ribose of a PAR substrate for exo-glycohydrolase activity. (c) Comparison of solvent accessible surfaces reveals an open platform in rat PARG (left) that can accommodate the (n+1) ADP-ribose, whereas the 2′-OH of the adenosine ribose is completely blocked by the ribose cap in the T. curvata glycohydrolase (right). Three representative (n+1) ADP-ribose conformers from computational simulations are shown in rat PARG surface.
By contrast, α12 of rPARG385, corresponding to the C-terminal helix H5 of the conserved macrodomain fold, is rotated ~20° and translated ~5 Å with respect to the corresponding helix of the bacterial glycohydrolase (Fig. 3b). These distinctive packing arrangements resemble the movement of the analogous C-terminal helix in the macroH2A1.1 protein, which is triggered by binding to ADP-ribose and completely masks the 2′-OH group of adenosine ribose13.
Consequently, this alternative conformation of α12 exposes the 2′-OH group of adenosine ribose, giving rise to an open platform that accommodates binding of an additional ADP-ribose molecule [(n+1) ADP-ribose] and is consistent with this enzyme’s ability to bind and cleave internal sites of PAR chains (Fig. 3b,c). Computational simulations of di-ADP-ribose binding reveal an unencumbered substrate-binding site, suggesting that additional electrostatic interactions with the phosphate backbone of PAR chains help to secure substrates for cleavage (Fig. 3c).
Collectively structures and computational analyses of rPARG385 reveal unique features of mammalian PARG, including the Tyr clasp buttressed by the MTS and endo-glycosidic PAR cleavage. The shared macrodomain fold and similar constellations of catalytic residues of mammalian PARG and the T. curvata glycohydrolase14 predict similar catalytic mechanisms of PARG across the organisms. However, the unique Tyr clasp that orients Tyr791 for interactions with ADP-HPD11 (Fig. 3) could be exploited for the development of small molecule inhibitors of mammalian PARG. PARG inhibitors have potential therapeutic applications in cancer, diabetes, and other inflammatory diseases as well as the potential to advance our understanding of the role of PAR turnover in normal physiology and disease states.
ONLINE METHODS
Protein purification, crystallization, and data Collection
The rat PARG (residues 385-972) was expressed from pET28a (Novagen) with N-terminal his-tag in E. coli BL21 expressing GroESL chaperone in the presence of 0.5 M urea. The protein was purified by affinity capture on a Ni-NTA (Qiagen) column. After elution with imidazole, the protein was loaded onto a heparin column and was eluted by a salt gradient (0 – 1M NaCl). Finally, the protein was applied to a Superdex 200 (GE healthcare) size exclusion column. Selenomethionine labeled rPARG385 protein was purified by the same protocol as the native protein. Native and selenium-containing proteins were concentrated to 15 mg ml−1 in 25 mM HEPES pH 7.5, 150 mM NaCl, 5% glycerol and 2 mM DTT and was stored at −80°C. Crystals were grown by hanging drop vapor diffusion. The protein solution was mixed with an equal volume of well solution (16-20% (w/v) PEG2000 monomethylether, 0.1 M Tris-HCl pH 7.5, 0.1 M NaCl, and 0.2 M potassium thiocyanate) and incubated at 22°C. Crystals were briefly equilibrated in well solution then transferred to a cryoprotectant solution containing 20% PEG2000 monomethylether, 0.1 M Tris-HCl pH 7.5, 0.1 M NaCl, 0.2 M potassium thiocyanate, and 20% glycerol, then were flash-cooled in liquid nitrogen. To form the PARG-inhibitor complex, crystals of unliganded rPARG385 were soaked in 20% PEG2000 monomethylether, 0.1 M Tris-HCl pH 7.5, 0.1 M NaCl, 0.2 M potassium thiocyanate, and 200 μM ADP-HPD (EMD bioscience) for 24 hours. PARG–ADP-HPD complex crystals were then transferred to a cryoprotectant solution containing 16-20% PEG2000 monomethylether, 0.1 M Tris-HCl pH 7.5, 0.1 M NaCl, 0.2 M potassium thiocyanate, 200 μM ADP-HPD, and 20% glycerol, then were flash-cooled in liquid nitrogen. X-ray diffraction data were collected at the Advanced Light Source (ALS) SIBYLS beamline 12.3.1 LBNL Berkeley, California. Native crystals (C2221, a =126.5 Å, b =199.5 Å, c =50.5 Å, α=β=γ=90°; one PARG molecule per asymmetric unit) diffracted to 1.95Å resolution, and PARG-inhibitor complex crystals (C2221, a = 130.8 Å, b =196.0 Å, c =163.5 Å, α=β=γ=90°; three PARG molecules per asymmetric unit) diffracted to 3.0 Å resolution. X-ray diffraction data statistics are shown in Supplementary Table 1 of the online supplementary information.
Structure determination
The apo rPARG385 structure was determined by MAD (multiple-wavelength anomalous diffraction) phasing of a selenomethionine protein derivative. X-ray data were processed using HKL200021 and SCALEPACK21,22. Nine selenium sites were located by the automated Patterson searches implemented in SOLVE23. Experimentally phased maps had a well-defined solvent boundary and readily interpretable electron density for protein. The crystallographic model was constructed using COOT24 and refined with TLS constraints in REFMAC25. The apo PARG385 structure was refined to a crystallographic Rfactor=18.0% and Rfree=21.6%. The N-terminal 53 residues (residues 385-437) and C-terminal 14 residues (959–972) were disordered. The PARG-ADP-HPD complex structure was solved by molecular replacement using CNS26 with the apo PARG385 structure as a search model. The asymmetric unit contains three PARG molecules, and they all show a strong electron density for ADP-HPD. The model was built using COOT with refinement in REFMAC. The structure was refined to an Rfactor = 24.5% and an Rfree = 27.5%. Crystallographic data statistics are shown in Supplementary Table 1. 100% (apo rPARG385) and 99.4% (rPARG385-ADP-HPD complex) of all residues are in favored and allowed region of the Ramachandran plot. All structural figures were prepared using PyMOL (www.pymol.org).
PARG activity assay
PARG activity was measured against PARylated PARP1. PARylation of PARP1 was performed at 30°C in 50 mM Tris-HCl pH 7.5, 100 mM NaCl, 10 mM MgCl2, and 2 mM DTT. To PARylate PARP1, PARP1375-1014 (2 μM), DNA-binding domain (residues 1-374) (1 μM) and a nicked DNA (2 μM) were pre-incubated for 10 min on ice. 200 μM of NAD+ was then added to the reaction, and the mixture was incubated at 30°C for 30 min. The reaction was quenched by adding 80 mM nicotinamide. Different concentrations of purified PARG proteins were then treated to PARylated PARP1 and incubated for 30 min at 30°C. The level of modification of PARP1 was visualized by SDS-PAGE (Supplementary Fig. 1).
Computational simulations of poly(ADP-ribose)
Conformational sampling of an ADP-ribose dimer model was used to predict binding sites for the (n+1) ADP-ribose moiety while fixing the adjacent (n) ADP-ribose group at the binding site for the ADP-HPD ligand. The ADP-ribose dimer was generated by splicing an ADP-ribose molecule onto the 2′-OH group of the adenosine ribosé moiety of the ADP-HPD inhibitor. The MindRocket modeling package (Drug Design Methodologies, LLC.) was then used to perform an exhaustive search of chemically reasonable conformations of the ADP-ribose tail to identify favorable binding sites on the rPARG385 surface nearby. The search consisted of eleven rotatable bonds and was constrained within 8 Å of the protein surface to prevent unnecessary sampling out in solvent. Docking poses were scored using a modified potential function based upon the VALIDATE scoring function27. The search was performed after scaling the van der Waals radii (scale factor = 0.85) to allow more complete sampling, especially near the junction between the ribose moieties. The side chains of rPARG385 were fixed during the search, which generated approximately two million conformers of the ADP-ribose moiety. The top 5000 structures were minimized using the Tripos Force Field28 and visually clustered into three conformational families where the ADPR tail could reside. Three representative low-energy conformers of the (n+1) ADP-ribose shown in Fig. 4c exhibit diverse binding modes for the diphosphate and adenosine moieties, and no specific binding pocket was identified for (n+1) ADP-ribose. These results suggest that internal cleavage of PAR polymers is accommodated by conformationally unrestrictive, electrostatic interactions with PAR’s phosphoribose backbone at sites flanking the enzyme active site.
Supplementary Material
Reversible posttranslational modification by poly(ADP-ribose) (PAR) regulates chromatin structure, DNA repair, and cell fate in response to genotoxic stresses. PAR glycohydrolase (PARG) removes PAR chains from poly(ADP-ribose)ylated proteins to restore protein function and release oligo(ADP-ribose) chains to signal damage. Here we report crystal structures of mammalian PARG and its complex with a substrate mimic revealing an open substrate-binding site and a unique “tyrosine clasp” that enables endo-glycosidic cleavage of branched PAR chains.
ACKNOWLEDGEMENTS
We thank Dr. W. Lee Kraus at University of Texas Southwestern Medical Center for providing a pET28a-ratPARG(1-972) plasmid. This work was supported in part by grants from the National Institutes of Health (NIH) including (5R01 GM052504; TE), and The Structural Cell Biology of DNA Repair Program (P01 CA92584; TE, JAT). X-ray diffraction and scattering technologies and their applications to the determination of macromolecular structures and conformations at the SIBYLS beamline at the Advanced Light Source, Lawrence Berkeley National Laboratory, are supported in part by the DOE program Integrated Diffraction Analysis Technologies (IDAT) under Contract Number DE-AC02-05CH11231 with the U.S. Department of Energy.
Footnotes
Accession codes. Coordinates for the apo rPARG385 (accession code 3UEK) and rPARG385-ADP-HPD complex (accession code 3UEL) structure have been deposited in the Protein Data Bank.
Note: Supplementary information is available on the Nature Structural & Molecular Biology website.
AUTHOR CONTRIBUTIONS
I.K.K. designed experiments, determined the crystal structures, analyzed results and wrote the manuscript. J.R.K. designed experiments, analyzed results and wrote the manuscript. C.M.W.H. performed the computational simulations. R.A.S. purified and crystallized the proteins. S.C. and J.A.T. helped with the manuscript and provided synchrotron facilities. T.E. analyzed results, wrote the manuscript, and provided financial support.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Miwa M, Tanaka M, Matsushima T, Sugimura T. J Biol Chem. 1974;249:3475–82. [PubMed] [Google Scholar]
- 2.Davidovic L, Vodenicharov M, Affar EB, Poirier GG. Exp Cell Res. 2001;268:7–13. doi: 10.1006/excr.2001.5263. [DOI] [PubMed] [Google Scholar]
- 3.Hatakeyama K, Nemoto Y, Ueda K, Hayaishi O. J Biol Chem. 1986;261:14902–11. [PubMed] [Google Scholar]
- 4.Malanga M, Althaus FR. Biochem Cell Biol. 2005;83:354–64. doi: 10.1139/o05-038. [DOI] [PubMed] [Google Scholar]
- 5.Cortes U, et al. Mol Cell Biol. 2004;24:7163–78. doi: 10.1128/MCB.24.16.7163-7178.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mortusewicz O, Fouquerel E, Amé JC, Leonhardt H, Schreiber V. Nucleic acids research. 2011;39:5045–5056. doi: 10.1093/nar/gkr099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gao H, et al. Exp Cell Res. 2007;313:984–96. doi: 10.1016/j.yexcr.2006.12.025. [DOI] [PubMed] [Google Scholar]
- 8.Botta D, Jacobson MK. Biochemistry. 2010;49:7674–7682. doi: 10.1021/bi100973m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Slama JT, et al. J Med Chem. 1995;38:389–93. doi: 10.1021/jm00002a021. [DOI] [PubMed] [Google Scholar]
- 10.Patel CN, Koh DW, Jacobson MK, Oliveira MA. Biochem J. 2005;388:493–500. doi: 10.1042/BJ20040942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koh DW, et al. Biochemistry. 2003;42:4855–63. doi: 10.1021/bi0272048. [DOI] [PubMed] [Google Scholar]
- 12.Karras GI, et al. EMBO J. 2005;24:1911–20. doi: 10.1038/sj.emboj.7600664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Timinszky G, et al. Nat Struct Mol Biol. 2009;16:923–929. doi: 10.1038/nsmb.1664. [DOI] [PubMed] [Google Scholar]
- 14.Slade D, et al. Nature. 2011;477:616–20. doi: 10.1038/nature10404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen D, et al. J Biol Chem. 2011;286:13261–71. doi: 10.1074/jbc.M110.206771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holm L, Sander C. Trends Biochem Sci. 1995;20:478–80. doi: 10.1016/s0968-0004(00)89105-7. [DOI] [PubMed] [Google Scholar]
- 17.Lau AY, Scharer OD, Samson L, Verdine GL, Ellenberger T. Cell. 1998;95:249–58. doi: 10.1016/s0092-8674(00)81755-9. [DOI] [PubMed] [Google Scholar]
- 18.Meyer RG, Meyer-Ficca ML, Whatcott CJ, Jacobson EL, Jacobson MK. Exp Cell Res. 2007;313:2920–36. doi: 10.1016/j.yexcr.2007.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tavassoli M, Tavassoli MH, Shall S. Eur J Biochem. 1983;135:449–55. doi: 10.1111/j.1432-1033.1983.tb07672.x. [DOI] [PubMed] [Google Scholar]
- 20.Andrabi SA, et al. Proc Natl Acad Sci U S A. 2006;103:18308–13. doi: 10.1073/pnas.0606526103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Otwinowski Z, Minor W. Methods in enzymology. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 22.Pflugrath JW. Acta Crystallogr D Biol Crystallogr. 1999;55:1718–25. doi: 10.1107/s090744499900935x. [DOI] [PubMed] [Google Scholar]
- 23.Terwilliger TC, Berendzen J. Acta Crystallogr D Biol Crystallogr. 1999;55:849–61. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Emsley P, Cowtan K. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–32. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 25.Murshudov GN, Vagin AA, Dodson EJ. Acta Crystallogr D Biol Crystallogr. 1997;53:240–55. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 26.Brunger AT, et al. Acta Crystallogr D Biol Crystallogr. 1998;54:905–21. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 27.Head RD, et al. J Am Chem Soc. 1996;118:3959–3969. [Google Scholar]
- 28.Clark M, Cramer RD, Opdenbosch NV. J Comp Chem. 1989;10:982–1012. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.