Abstract
All three translation termination codons, or nonsense codons, contain a uridine residue at the first position of the codon1–3. Here, we demonstrate that pseudouridylation [conversion of uridine into pseudouridine (Ψ) 4] of nonsense codons suppresses translation termination both in vitro and in vivo. In vivo targeting of nonsense codons is accomplished by the expression of an H/ACA RNA capable of directing the isomerization of uridine to Ψ within the nonsense codon. Thus, targeted pseudouridylation represents a novel approach for promoting nonsense suppression in vivo. Remarkably, we also show that pseudouridylated nonsense codons code for amino acids with similar properties. Specifically, ΨAA and ΨAG code for serine and threonine, whereas ΨGA codes for tyrosine and phenylalanine, thus suggesting a novel mode of decoding. Our results suggest that RNA modification, as a naturally occurring mechanism, may offer a new way to expand the genetic code.
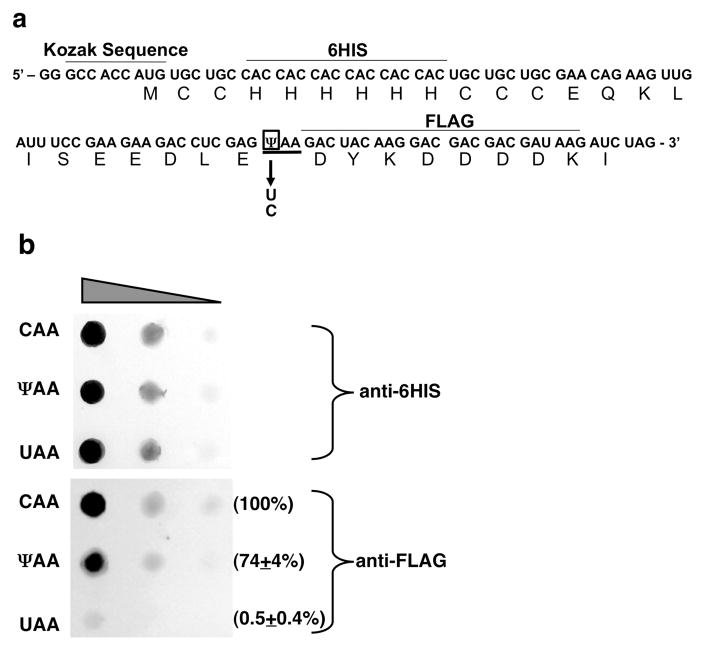
Ψ, the C5-glycoside isomer of uridine, exhibits many structural and biochemical differences from uridine5 (Supplementary Fig. 1). Thus, it is possible that replacement of the uridine within a nonsense codon with Ψ may affect translation termination. To investigate the possible effect of Ψ on translation termination, we developed an in vitro nonsense suppression assay (Fig. 1a). Briefly, we synthesized an artificial mRNA that encoded a 6X histidine (6HIS) tag at the N-terminus and a FLAG tag at the C-terminus. In between the 6HIS tag and FLAG tag, a pseudouridylated nonsense codon (ΨAA) was inserted (Fig. 1a). In addition, two control transcripts were created with the same sequence except that the Ψ of the nonsense codon was either changed to uridine (U), thus forming an authentic nonsense codon (UAA), or substituted with a cytidine (C), thus eliminating the nonsense codon (CAA) (Fig. 1a). Anti-6HIS immunoblot analysis indicated that all three RNAs were equally translated in rabbit reticulocyte lysate (Fig. 1b, top panel). Remarkably, however, according to the anti-FLAG blot, the presence of a Ψ within the termination codon resulted in robust nonsense suppression (Fig. 1b, lower panel). Specifically, the ΨAA-containing transcript produced a strong FLAG signal which is almost comparable to that produced by the CAA-containing transcript (Fig. 1b). In contrast, only a background level of FLAG was produced when the UAA-containing transcript was used (Fig. 1b). Our results thus indicate that presence of Ψ in a termination codon effectively suppresses translation termination in vitro.
Fig. 1.
Pseudouridylation of a termination codon promotes nonsense suppression in vitro. (a) Nucleotide sequence of the in vitro transcription product and its translated sequence are shown. Positions of the Kozak sequence, as well as epitopes (6HIS and FLAG) within the nucleotide and protein sequences are labeled. The pseudouridylated nonsense codon is indicated. Changes of Ψ to U and Ψ to C are also indicated. (b) Anti-6HIS and anti-FLAG immunoblot analysis of the in vitro translation lysate following translation of an RNA lacking a termination codon (CAA), an RNA containing a pseudouridylated termination codon (ΨAA), or an RNA containing an authentic termination codon (UAA). Relative efficiency of read-through (anti-FLAG/anti-6HIS) was calculated and indicated in parentheses (the control, CAA, is set to 100%). Error is given as the standard deviation of three independent experiments.
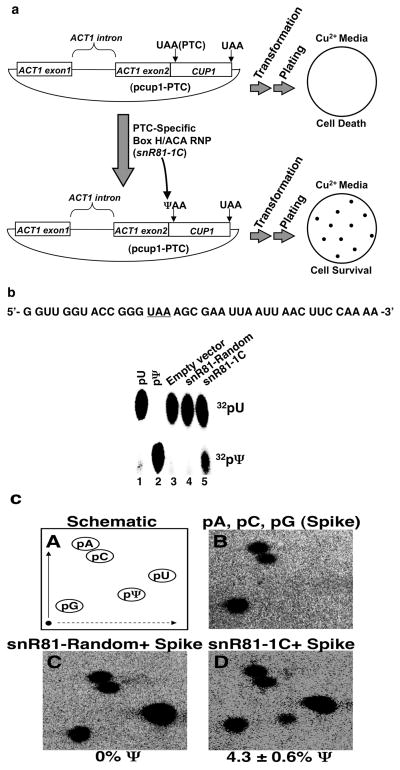
The in vitro results prompted us to investigate whether the pseudouridylation of a termination codon would elicit nonsense suppression in vivo. Taking advantage of the CUP1 reporter system6, we introduced a premature termination codon (PTC) at the second codon of the CUP1 gene, thus creating a new CUP1 reporter gene (termed cup1-PTC). Cup1p is a copper chelating protein that mediates resistance to copper sulfate (CuSO4)7. Thus, upon transformation of the cup1-PTC plasmid (pcup1-PTC) into a Saccharomyces cerevisiae cup1-Δ strain, one should be able to measure nonsense suppression by plating the cells on selective media containing CuSO4 (Fig. 2a).
Fig. 2.
Quantification of cup1-PTC pseudouridylation. (a) Schematic of the in vivo nonsense suppression assay. (b) In vitro pseudouridylation assay by thin layer chromatography. 5′ 32P-radiolabeled uridylate (pU) and pseudouridylate (pΨ) markers were run in parallel. The substrate—[α32P]UTP uniformly labeled RNA fragment—is shown. (c) Quantification of cup1-PTC pseudouridylation in vivo. The percentage of pseudouridylation was calculated [pΨ/(pΨ + pU)]. Adenosine 5′-monophosphate (pA), cytidine 5′-monophosphate (pC), guanosine 5′-monophosphate (pG), uridine 5′-monophosphate (pU), and pseudouridine 5′-monophosphate (pΨ) are indicated. (●) Origin. Error is given as the standard deviation of three independent experiments.
In order to direct site-specific Ψ formation in vivo, we took advantage of the H/ACA ribonucleoprotein (RNP) family. H/ACA RNPs are primarily responsible for the post-transcriptional isomerization of uridine to Ψ within RNA (Supplementary Fig. 2)8,9. In order to target the PTC within cup1-PTC we derived an H/ACA RNA, snR81-1C, from SNR81, a naturally occurring yeast H/ACA RNA. In addition, we also constructed a control H/ACA RNA, snR81-Random, which contained a random guide sequence.
To ensure that cup1-PTC was pseudouridylated in response to expression of snR81-1C, we measured Ψ formation within the PTC both in vitro and in vivo. To analyze Ψ formation in vitro, we monitored, by thin layer chromatography (TLC), Ψ formation on a 39 nucleotide fragment of RNA corresponding to the region of cup1-PTC containing the PTC (Fig. 2b). Incubation of the transcript in extracts prepared from cells expressing snR81-1C resulted in the formation of Ψ (Fig. 2b, lane 5), while extracts containing an empty vector or snR81-Random did not result in the formation of Ψ (lanes 3 and 4). These results indicate that snR81-1C is capable of directing pseudouridylation within the RNA fragment, most likely at the target site, i.e., the uridine of the PTC.
To determine the pseudouridylation status of cup1-PTC in vivo, we analyzed the PTC of cellularly derived cup1-PTC mRNA using a site-specific and quantitative pseudouridylation assay, namely site-specific cleavage and radiolabeling followed by nuclease digestion and 2D-TLC10. To help locate the uridine and Ψ spots on the TLC plate, we spiked each reaction with 32P-radiolabeled 32pA, 32pC, and 32pG (Fig. 2c, panel B). Consistent with the results of our in vitro analysis (Fig. 2b), only CUP1 mRNA isolated from cells expressing snR81-1C produced a spot corresponding to Ψ (Fig. 2c, compare panel C with panel D). Quantification indicated that approximately 5% of the cup1-PTC transcript was pseudouridylated. Thus, our in vivo pseudouridylation results would predict that, upon expression of snR81-1C, a functional Cup1p (although in small amount) would be translated from the cup1-PTC mRNA.
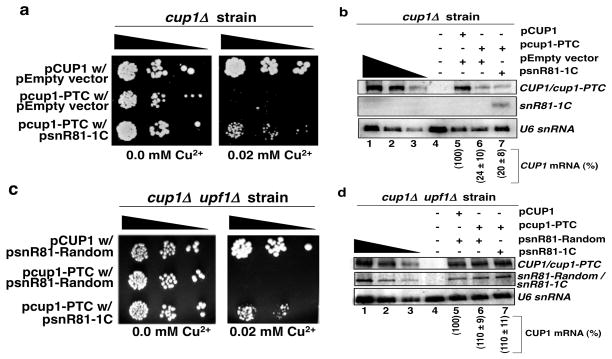
Figure 3a shows the results of the in vivo nonsense suppression assay (see Fig. 2a for illustration). As expected, when transformed with wild-type pCUP1, cup1Δ cells grew healthy on media containing 0.0 mM or 0.02 mM CuSO4 (top row). However, when transformed with pcup1-PTC, only cells co-transformed with psnR81-1C were able to survive on media containing 0.02 mM CuSO4 (compare the middle row with the bottom row). Partial restoration of growth appears to be consistent with the previous quantitative analysis demonstrating a low level of pseudouridylation (~5%) at the PTC (Fig. 2c).
Fig. 3.
Expression of an H/ACA RNA targeting the PTC of cup-PTC for pseudouridylation promotes nonsense suppression. (a) pCUP1 or pcup1-PTC along with either an empty vector or psnR81-1C were transformed into a cup1Δ strain. Cell growth was assessed on solid synthetic medium (-URA -LEU) containing either 0.0 mM or 0.02 mM CuSO4, as indicated. (b) Northern blot analysis of RNA extracted from cells described in (a). Normalized levels of CUP1 mRNA (lane 5) and cup1-PTC mRNA (lanes 6 and 7) are indicated in parentheses under each lane. Error is given as the standard deviation of three independent experiments. (c) cup1Δ upf1Δ strain was transformed with either pCUP1 or pcup1-PTC along with either psnR81-Random or psnR81-1C. Cell growth was assessed on solid synthetic medium (-HIS -URA -LEU) with or without CuSO4, as indicated. (d) Northern blot analysis of RNA extracted from cells described in (c). Normalized levels of CUP1 mRNA (lane 5) and cup1-PTC mRNA (lanes 6 and 7) are indicated in parentheses under each lane. Error is given as the standard deviation of three independent experiments.
Because nonsense suppression could be achieved both at the level of translation (termination suppression) and the level of mRNA decay where PTC-containing mRNA is usually the target of NMD (nonsense-mediated decay), it remained possible that the suppression we observed in Fig. 3a was a result of NMD suppression rather than suppression of translation termination. To test this possibility, we measured the levels of CUP1 mRNA using northern blot analysis (Fig. 3b). As expected, steady-state levels of cup1-PTC mRNA dropped significantly when compared with the level of wild-type CUP1 mRNA (compare lanes 6 and 7 with lane 5). However, expression of the PTC-specific guide RNA (snR81-1C) had no effect on steady state levels of cup1-PTC mRNA; nearly identical levels of cup1-PTC mRNA were detected in cells transformed with either psnR81-1C or with empty vector (compare lane 6 with lane 7). These results suggested that the observed suppression was a result of nonsense codon suppression rather than a result of suppressing NMD. To completely eliminate the potential complications of NMD, we deleted UPF1, a gene required for NMD11, and then repeated the nonsense suppression assay and northern blotting. As expected, deletion of UPF1 resulted in the stabilization of cup1-PTC (Figs 3d). Consistent with previously published results12, 13, we also observed a small degree of nonsense suppression as evidenced by a low (but above background) level of growth on media containing a low concentration (0.013 mM) of CuSO4 (Supplementary Fig. 3, compare row 5 with row 2). However, no growth was observed when CuSO4 concentration was raised to 0.02 mM (Fig. 3c, middle row). Apparently, the nonsense suppression phenotype conferred by deletion of UPF1 is not sufficient to promote growth on 0.02 mM CuSO4. To assess nonsense suppression induced by PTC pseudouridylation, we plated cells on media containing either 0.02 mM or 0.013 mM CuSO4 (Fig. 3c, and Supplementary Fig. 3). Under both conditions, expression of snR81-1C provided a growth advantage; the level of growth rescue is similar to that observed when UPF1 was intact (compare Fig. 3c with Fig. 3a, and Supplementary Fig. 3). These results further support the notion that expression of snR81-1C or pseudouridylation of the PTC resulted in suppression of translation termination rather than suppression of NMD. Given that the control, where snR81-Random was similarly expressed (Fig. 3d), showed no suppression (Fig. 3c), our results also indicate that the observed suppression of translation termination is guide-RNA specific.
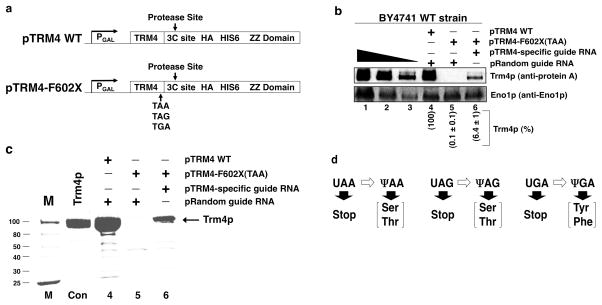
To further determine whetherΨ-mediated nonsense suppression can be generalized as well as which amino acids are incorporated at Ψ-substituted nonsense codons, we took advantage of a plasmid containing a C-terminally tagged TRM4 gene, pTRM4-WT (Fig. 4a). Through site-directed mutagenesis the codon for phenylalanine at position 602 (F602) of the TRM4 gene was changed to a nonsense codon (TAA, TAG, or TGA), creating three variants of the plasmid [pTRM4-F602X(TAA), pTRM4-F602X(TAG), and pTRM4-F602X(TGA)] (Fig. 4a).
Fig 4.
Generalization of Ψ-mediated nonsense suppression and determination of amino acids coded for by pseudouridylated nonsense codons. (a) Schematic representation of the constructs used for protein purification (also see text). (b) Western blot analysis was carried out using extracts prepared from wild-type cells transformed with either pTRM4 WT and a plasmid containing a random guide RNA gene (pRandom guide RNA) (lane 4), pTRM4-F602X(TAA) and pRandom guide RNA (lane 5), or pTRM4-F602X(TAA) and a plasmid containing a guide RNA gene that targets the nonsense codon (UAA 602) of TRM4-F602X(TAA) (lane 6). Enolase was probed as a loading control. The normalized levels of Trm4p are indicated in parentheses under each lane. Error is represented as the standard deviation from three independent experiments. (c) Cell cultures described in (b) were scaled up, and Trm4 proteins were purified and analyzed on a SDS-PAGE gel (stained with coomassie blue); lanes correspond to those in (b). In the control lane (Con), a known amount (6 μg) of purified Trm4p was loaded. (d) Identification of amino acids incorporated at Ψ-containing termination codons (also see Supplementary Figs. 4–8).
Figure 4b shows the western blot analysis of extracts prepared from wild-type cells expressing wild-type TRM4 (nonsense-free) or TRM4-F602X(TAA). When cells were transformed with the wild-type TRM4 plasmid, a strong protein A-tag signal was detected (Fig 4b, lane 4). However, when cells were transformed with pTRM4-F602X(TAA), a protein A signal was only detected when co-transformed with a TRM4-F602X(TAA)-specific guide RNA, indicating nonsense suppression (Fig 4b, compare lane 5 with lane 6).
Next, we carried out large-scale immunoprecipitations to purify full length Trm4p produced as a consequence of Ψ-mediated nonsense suppression. Figure 4c shows an example [pTRM4-F602X(TAA)] of such experiments. The bands corresponding to full length Trm4p produced as a consequence of Ψ-mediated nonsense suppression were excised and sequenced by LC-MS/MS (Fig. 4c, and Supplementary Figs. 4–7). Remarkably, LC-MS/MS analysis indicated that pseudouridylated UAA and UAG (ΨAA and ΨAG) both directed the incorporation of either serine or threonine (Fig 4d, and Supplementary Figs. 4–6). Taking into account that the third base of a codon is usually non-specific (the wobble base), it makes sense that both ΨAA and ΨAG code for the same amino acids. With respect to targeted pseudouridylation of UGA (ΨGA), it directed the incorporation of tyrosine and phenylalanine (Fig. 4d, Supplementary Fig. 7). As all three termination codons directed the incorporation of two amino acids, we quantified their frequency of incorporation (Supplementary Fig. 8). Interestingly, although ΨAA and ΨAG both code for serine and threonine, serine is predominantly incorporated atΨAG, whereas serine and threonine are incorporated at a roughly similar frequency forΨAA. Furthermore, ΨGA primarily directs the incorporation of tyrosine.
Interestingly, however, given that the anticodons of the tRNASer and tRNAThr families do not look alike (Supplementary Fig. 9), our experimental data results raise an important question: how is the same pseudouridylated nonsense codon (e.g., ΨAA or ΨAG) recognized by the different families of tRNA? While it is possible that the presence of Ψ in mRNA-tRNA duplexes acts to stabilize interactions between the mRNA and near- or non-cognate tRNAs14, 15, an alternative explanation is that the presence of Ψ within the A-site may disorder the local RNA (or ribosome) structure, somehow allowing for the binding or accommodation of near- or non-cognate tRNAs, possibly through altering the hydration state of the nonsense codon16. It is also possible that unique RNA modifications in the anti-codon loop of tRNASer, tRNAThr, tRNAPhe, or tRNATyr, contribute to the recognition of pseudouridylated nonsense codons, thus allowing them to be decoded. In fact, modifications within the anticodon loop of tRNA have previously been demonstrated to impact recoding17. Perhaps more interestingly, it has not escaped our attention that the amino acids incorporated at each termination codon are biochemically and structurally similar. Specifically, serine and threonine, which are coded for by ΨAA and ΨAG, are both hydroxylated short chain amino acids. Likewise, tyrosine and phenylalanine, which are coded for by ΨGA, both contain an aromatic ring. Although the decoding center is ~75 Å away from the peptidyl transferase center18, whether there is a role for the amino acid in the decoding of Ψ-containing termination codons is an interesting idea that requires further analysis. If true, such a mechanism would represent a completely novel mode of decoding. It is interesting to note that frameshifting at sense codons also shows a strong preference for utilizing tRNASer and tRNAThr 19. Although detailed mechanisms are still unclear, the similarities in using similar polar amino acids (serine and threonine) in frameshifting and in decoding of pseudouridylated nonsense codons certainly deserve further attention.
Our data demonstrates that artificial H/ACA guide RNAs are able to direct the pseudouridylation of nonsense codons of mRNA, thus leading to nonsense suppression. It should be noted that artificial guide RNAs may have an unintended target(s), thus raising concerns about substrate specificity. We did, however, realize this concern when designing sense-to-nonsense mutations and their corresponding guide RNAs, and purposely avoided the sites and their guide sequences that could target other endogenous mRNAs. In fact, BLAST search against the yeast genome did not generate any other potential targets that appear to be suitable substrates for our artificial H/ACA RNAs. Thus, it is unlikely that the observed effects are due to the nonspecific effect of modifications of unintended off-targets.
Our RNA-guided modification strategy is of significant clinical interest, given the current estimates that approximately 33% of genetic diseases can be attributed to the presence of a PTC20. On the other hand, since the artificial guide RNAs are derived from naturally occurring H/ACA RNAs (only the short guide sequence is changed), we predict that the nonsense codons of some mRNAs are naturally pseudouridylated by endogenous H/ACA RNAs as long as the guide sequence matches the target. Indeed, using computational algorithms to predict nonsense codons that may be natural targets of the endogenous H/ACA RNP machinery yields a number of potential candidates (Supplementary Fig. 10). In addition, our lab has recently demonstrated that an exact match between the H/ACA RNA guide sequence and the target sequence is not necessary for efficient modification under certain conditions. In fact, the mismatches are required for inducible pseudouridylation in response to cell stress21. Thus, there are likely a large number of pseudouridylation targets in mRNAs. While some of these potential targets are nonsense codons, a majority of them are expected to be sense codons. Given our surprising discovery that pseudouridylation of nonsense codons converts them into sense codons, it is not impossible that pseudouridylation of sense codons will alter their decoding, making mRNA pseudouridylation a novel mechanism of RNA editing. If such is true, the genetic code would expand considerably. We predict that targeted pseudouridylation of mRNA is a yet-to-be appreciated mechanism of generating protein diversity.
Methods Summary
Relevant properties of strains and growth conditions are described in text. Strain construction and additional growth conditions are described in the Methods. Standard procedures were used for all protein and RNA analyses and are described in the Methods. Mass spectrometry was performed at the University of Rochester Medical Center Proteomics Core and is described in the Methods.
Methods
Yeast strains, Transformation and growth assay
The cup1-Δ yeast strain was kindly provided by Dr. Christine Guthrie (University of California San Francisco)6. The UPF1 locus was deleted from a cup1-Δ strain using standard protocol as described previously22. For the analysis of CuSO4 resistance the appropriate plasmids were transformed into either cup1-Δ or cup1-Δ upf1-Δ yeast strains as previously described22, except that after heat shock cells were precipitated and resuspended in 100 μL of water rather than YPD. Single colonies were selected and grown to saturation in SGal drop-out media, cells were diluted to an O.D.600 0.001 and then a series of five-fold dilutions were spotted on to SGal drop-out media, with or without CuSO4. Growth phenotypes were assessed after cells were grown for 3–5d at 30°C.
Plasmids
The pCUP1 plasmid was a gift from Dr. David Mcpheeters (Case Western Reserve University), while pTRM4 WT was a gift from Dr. Eric Phizicky and Dr. Beth Grayhack (University of Rochester Medical Center). pcup1-PTC and pTRM4 F602X variants were created by site-directed mutagenesis using Pfu polymerase (Stratagene) and the appropriate oligonucleotides and plasmids. Novel H/ACA RNA genes were constructed by PCR using four overlapping oligonucleotide primers and were either cloned into 2 μm URA3 or 2 μm LEU2 vector [both gifts from Dr. Eric Phizicky (University of Rochester Medical Center)] as BAMHI/HINDIII fragments23.
In vitro transcription and translation
In order to generate mRNA transcripts for in vitro translation, DNA templates were synthesized through PCR using two overlapping DNA oligonucleotides. The double-stranded DNA templates thus synthesized contained either a TAA nonsense codon or a CAA codon in the middle, flanked by a 6HIS-coding sequence near the 5′ end and a FLAG-coding sequence at the 3′ end (Fig. 1b). For efficient in vitro translation, the templates also contained a Kozak sequence immediately upstream of the 6HIS coding sequence (Fig. 1b). In addition, a T7 promoter sequence was included at the 5′ terminus. Following in vitro T7 transcription24, 25, UAA- or CAA-containing mRNA transcripts were synthesized (see Fig. 1b). To create a similar mRNA, with the uridine of the nonsense codon (or the cytidine of the CAA codon) changed to Ψ, a two-piece splint ligation was employed24. The 5′ RNA was in vitro synthesized through T7 transcription, ending with CUC at its 3′ terminus (see Fig 1b), and the 3′ piece (5′ - GAG ΨAA GAC UAC AAG GAC GAC GAC GAC AAG AUC UAG - 3′) (see Fig. 1a) was chemically synthesized (Thermo Scientific). The 5′ and 3′ halves were ligated using a bridging oligonucleotide and T4 DNA ligase24. In vitro synthesized RNAs were gel-purified before being used in the in vitro translation reactions. In vitro translation reactions were carried out in 30 μL reactions of Red Nova Lysate as described by the supplier (Novagen). PCR of two overlapping oligodeoxynucleotides was also used to generate the template for in vitro transcription of the substrate used in the in vitro pseudouridylation assay (Fig. 2C).
Northern blot analysis
Total RNA was isolated from yeast using Trizol essentially as described by the supplier (Invitrogen), except that cells were vortexed with acid-sterilized glass beads for 5 minutes. For northern blot analysis, 6 μg of total RNA was separated on 8% polyacrylamide-7M urea gels and electrotransferred at 4 °C to Amersham Hybond-N+ membranes in 0.5X TBE buffer for 16 h at 15 V. Hybridizations were preformed essentially as described23.
Protein purification and Immunoblot analysis
For the analysis of Trm4p protein sequence, BY4741 was transformed with the appropriate plasmids as described before22. Yeast whole cell extracts and IgG sepharose chromatography was preformed as previously described26. For analysis of in vitro translation products, membranes were probed with either a monoclonal Flag antibody (M2; Sigma-Aldrich, St. Louis, Mo.) or monoclonal His-probe (H-3; Santa Cruz Biotechnology, Santa Cruz, Calif.). Goat Anti-Mouse IgG (H+L)-AP conjugate (Bio-Rad) was then used as a secondary antibody. Proteins were visualized using 1-Step NBT/BCIP (Pierce). For the analysis of Trm4p, yeast crude extracts were separated on 4–15% Tris-HCl Ready gels (BioRad). Proteins were then transferred to 0.1 μm nitrocellulose membranes (Whatman) and probed with either monoclonal Protein A (Sigma-Aldrich) or anti-Eno1p (a gift from Dr. Mark Dumont, University of Rochester Medical Center). Goat anti-Mouse IgG (H+L)-AP conjugate (Bio-Rad) was used as a secondary antibody. Proteins were visualized using 1-Step NBT/BCIP (Pierce).
Pseudouridylation assays
In vitro pseudouridylation assays were performed using yeast whole cell extract. Cells were grown to mid log phase and pelleted. Pellets were resuspended in 200 μL of extraction buffer containing 20 mM HEPES at pH 7.9, 0.42 MNaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 0.5 mM DTT, 0.5 mM PMSF, and 25% glycerol. 400 μL of sterile acid-washed glass beads were added to the cell suspension, and cells were subsequently homogenized through vigorous vortexing (5 × 30 s) at 4°C. Following a 5-min microfugation (14,000g, 4°C), the supernatant was recovered, and used for the pseudouridylation assay. The substrate RNA was prepared by in vitro transcription in the presence of [α-P32]UTP. The substrate was gel purified and incubated in the extracts for 2 hr. The radiolabled substrate was recovered and digested with nuclease P1 and analyzed by 1D-TLC as previously described10. Pseudouridylation of cellularly derived cup1-PTC RNA was analyzed as previously described10, except that modifications were analyzed by 2D-TLC27.
Mass Spectrometry
Mass spectrometry was preformed at the University Of Rochester Proteomics Center. Coomassie stained gel bands corresponding to full length Trm4p were subjected to in gel trypsin digestion. An 80 minute LC-MS/MS run was performed in-line with a Finnigan LTQ Ion Trap mass spectrometer (Thermo Scientific), using a flow rate of 250ul/min. The data collected from the LTQ runs was searched using MASCOT (Matrix Science), initially against the full Saccharomyces database, secondly against a custom database which included the wild type Trm4p sequence, as well as a Trm4p sequence with an “X” in the amino acid position that corresponds to the stop codon. Peptides identified by Mascot with an ion score of 15 or greater were inspected further for MS/MS fragmentation patterns that map through most of the peptide sequence, especially on and through the mutant amino acid position. Peptides with Expect values greater than 0.05 were not accepted. To allow for relative quantitation, we repeated the LC-MS/MS experiments using dynamic inclusion for only the peptides of interest. Total spectral counts obtained by dynamic inclusion therefore represent the relative abundances of each respective peptide.
Supplementary Material
Acknowledgments
We thank Fred Hagen and the Proteomics Core at the University of Rochester for performing the mass spectrometry analysis. We also thank Eric Phizicky and Beth Grayhack for the wildtype TRM4 construct, Christine Guthrie for the cup1Δ yeast strain, David Mcpheeters for the wildtype CUP1 construct, and Mark Dumont for the anti-Eno1p antibody. Lastly, we would like to thank members of the Yu laboratory, especially Dr. Xinliang Zhao, for helpful discussions.
Footnotes
Contributions
JK and YTY designed and interpreted the experiments. Mass spectrometry was performed at the Proteomics Core at the University of Rochester Medical Center. JK performed all other experiments.
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Brenner S, Barnett L, Katz ER, Crick FH. UGA: a third nonsense triplet in the genetic code. Nature. 1967;213:449–50. doi: 10.1038/213449a0. [DOI] [PubMed] [Google Scholar]
- 2.Brenner S, Stretton AO, Kaplan S. Genetic code: the ‘nonsense’ triplets for chain termination and their suppression. Nature. 1965;206:994–8. doi: 10.1038/206994a0. [DOI] [PubMed] [Google Scholar]
- 3.Weigert MG, Garen A. Base composition of nonsense codons in E. coli. Evidence from amino-acid substitutions at a tryptophan site in alkaline phosphatase. Nature. 1965;206:992–4. doi: 10.1038/206992a0. [DOI] [PubMed] [Google Scholar]
- 4.Cohn WE. 5-Ribosyl uracil, a carbon-carbon ribofuranosyl nucleoside in ribonucleic acids. Biochim Biophys Acta. 1959;32:569–71. doi: 10.1016/0006-3002(59)90644-4. [DOI] [PubMed] [Google Scholar]
- 5.Charette M, Gray MW. Pseudouridine in RNA: what, where, how, and why. IUBMB Life. 2000;49:341051. doi: 10.1080/152165400410182. [DOI] [PubMed] [Google Scholar]
- 6.Lesser CF, Guthrie C. Mutational analysis of pre-mRNA splicing in Saccharomyces cerevisiae using a sensitive new reporter gene, CUP1. Genetics. 1993;133:851–63. doi: 10.1093/genetics/133.4.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hamer DH, Thiele DJ, Lemontt JE. Function and autoregulation of yeast copperthionein. Science. 1985;228:685–90. doi: 10.1126/science.3887570. [DOI] [PubMed] [Google Scholar]
- 8.Ganot P, Bortolin ML, Kiss T. Site-specific pseudouridine formation in preribosomal RNA is guided by small nucleolar RNAs. Cell. 1997;89:799–809. doi: 10.1016/s0092-8674(00)80263-9. [DOI] [PubMed] [Google Scholar]
- 9.Ni J, Tien AL, Fournier MJ. Small nucleolar RNAs direct site-specific synthesis of pseudouridine in ribosomal RNA. Cell. 1997;89:565–73. doi: 10.1016/s0092-8674(00)80238-x. [DOI] [PubMed] [Google Scholar]
- 10.Zhao X, Yu YT. Detection and quantitation of RNA base modifications. RNA. 2004;10:996–1002. doi: 10.1261/rna.7110804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leeds P, Peltz SW, Jacobson A, Culbertson MR. The product of the yeast UPF1 gene is required for rapid turnover of mRNAs containing a premature translational termination codon. Genes Dev. 1991;5:2303–14. doi: 10.1101/gad.5.12a.2303. [DOI] [PubMed] [Google Scholar]
- 12.Leeds P, Wood JM, Lee BS, Culbertson MR. Gene products that promote mRNA turnover in Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:2165–77. doi: 10.1128/mcb.12.5.2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wilusz CJ, Wang W, Peltz SW. Curbing the nonsense: the activation and regulation of mRNA surveillance. Genes Dev. 2001;15:1781–5. doi: 10.1101/gad.943701. [DOI] [PubMed] [Google Scholar]
- 14.Agris PF. The importance of being modified: roles of modified nucleosides and Mg2+ in RNA structure and function. Prog Nucleic Acid Res Mol Biol. 1996;53:79–129. doi: 10.1016/s0079-6603(08)60143-9. [DOI] [PubMed] [Google Scholar]
- 15.Davis DR. Stabilization of RNA stacking by pseudouridine. Nucleic Acids Res. 1995;23:5020–6. doi: 10.1093/nar/23.24.5020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Auffinger P, Westhof E. RNA hydration: three nanoseconds of multiple molecular dynamics simulations of the solvated tRNA(Asp) anticodon hairpin. J Mol Biol. 1997;269:326–41. doi: 10.1006/jmbi.1997.1022. [DOI] [PubMed] [Google Scholar]
- 17.Agris PF. Decoding the genome: a modified view. Nucleic Acids Res. 2004;32:223–38. doi: 10.1093/nar/gkh185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Song H, et al. The crystal structure of human eukaryotic release factor eRF1--mechanism of stop codon recognition and peptidyl-tRNA hydrolysis. Cell. 2000;100:311–21. doi: 10.1016/s0092-8674(00)80667-4. [DOI] [PubMed] [Google Scholar]
- 19.Atkins JF, Gesteland RF, Reid BR, Anderson CW. Normal tRNAs promote ribosomal frameshifting. Cell. 1979;18:1119–31. doi: 10.1016/0092-8674(79)90225-3. [DOI] [PubMed] [Google Scholar]
- 20.Frischmeyer PA, Dietz HC. Nonsense-mediated mRNA decay in health and disease. Hum Mol Genet. 1999;8:1893–900. doi: 10.1093/hmg/8.10.1893. [DOI] [PubMed] [Google Scholar]
- 21.Wu G, Xiao M, Yang C, Yu YT. U2 snRNA is inducibly pseudouridylated at novel sites by Pus7p and snR81 RNP. EMBO J. 2010;30:79–89. doi: 10.1038/emboj.2010.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ma X, et al. Pseudouridylation of yeast U2 snRNA is catalyzed by either an RNA-guided or RNA-independent mechanism. EMBO J. 2005;24:2403–13. doi: 10.1038/sj.emboj.7600718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chernyakov I, Whipple JM, Kotelawala L, Grayhack EJ, Phizicky EM. Degradation of several hypomodified mature tRNA species in Saccharomyces cerevisiae is mediated by Met22 and the 5′-3′ exonucleases Rat1 and Xrn1. Genes Dev. 2008;22:1369–80. doi: 10.1101/gad.1654308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu YT. Site-specific 4-thiouridine incorporation into RNA molecules. Methods Enzymol. 2000;318:71–88. doi: 10.1016/s0076-6879(00)18045-0. [DOI] [PubMed] [Google Scholar]
- 25.Zhao X, Yu YT. Pseudouridines in and near the branch site recognition region of U2 snRNA are required for snRNP biogenesis and pre-mRNA splicing in Xenopus oocytes. RNA. 2004;10:681–90. doi: 10.1261/rna.5159504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gelperin DM, et al. Biochemical and genetic analysis of the yeast proteome with a movable ORF collection. Genes Dev. 2005;19:2816–26. doi: 10.1101/gad.1362105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zebarjadian Y, King T, Fournier MJ, Clarke L, Carbon J. Point mutations in yeast CBF5 can abolish in vivo pseudouridylation of rRNA. Mol Cell Biol. 1999;19:7461–72. doi: 10.1128/mcb.19.11.7461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reichow SL, Hamma T, Ferré-D’Amaré AR, Varani G. The structure and function of small nucleolar ribonucleoproteins. Nucleic Acids Res. 2007;35:1452–64. doi: 10.1093/nar/gkl1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.