Abstract
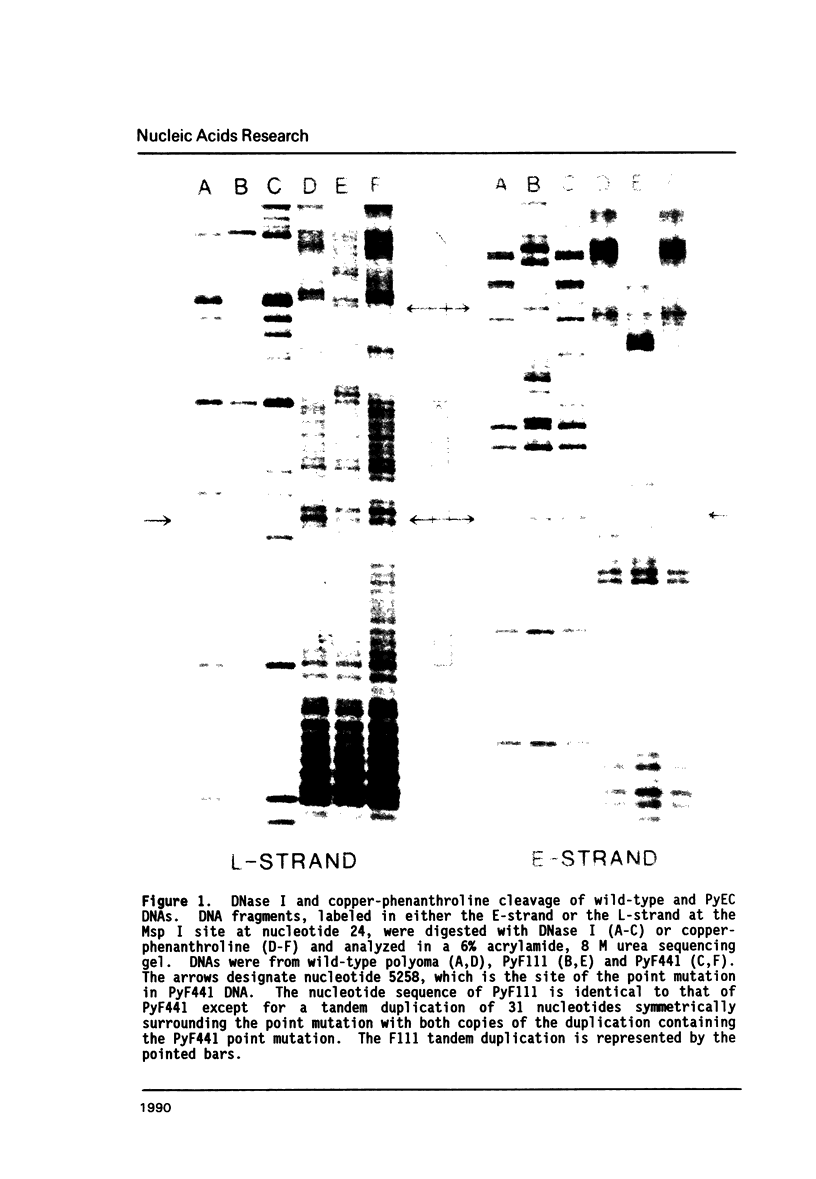
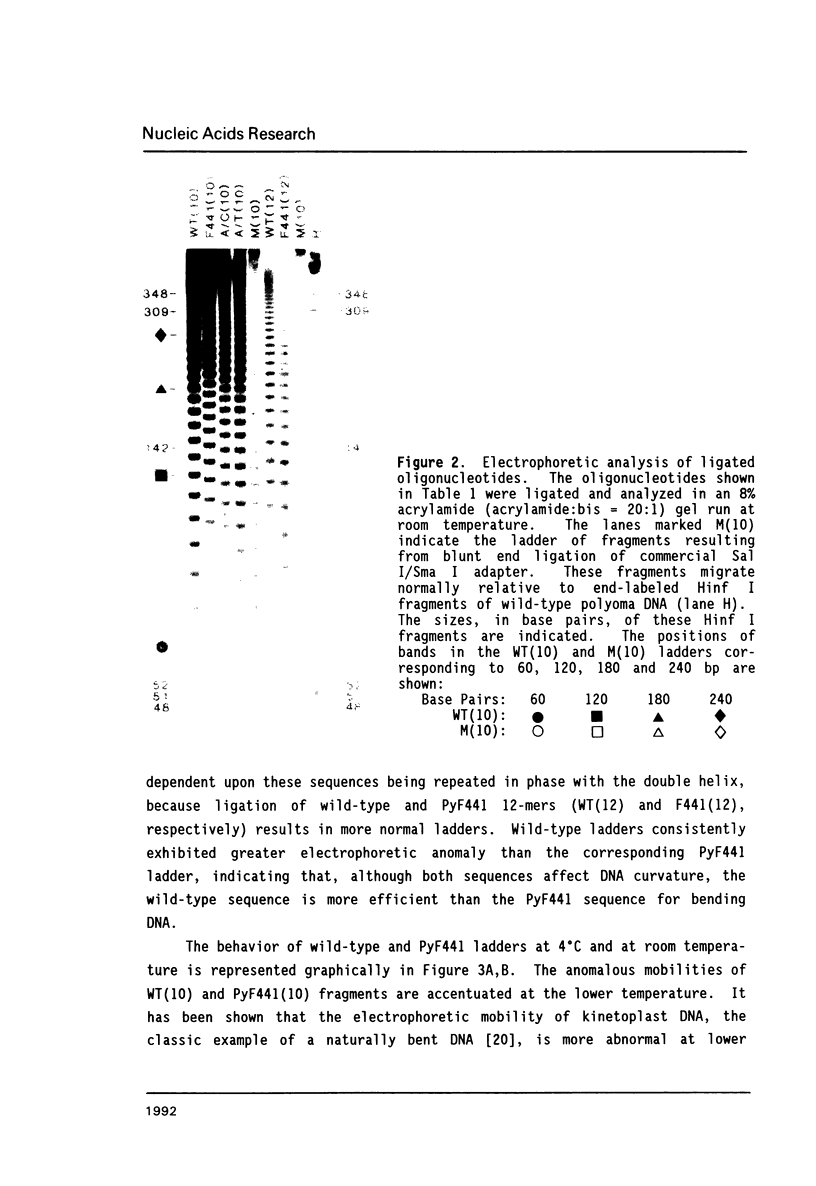
A point mutation in the enhancer of polyomavirus host range mutant, PyEC F441, permits productive infection of the murine embryonal carcinoma cell line, F9. This mutation at nucleotide position 5258 introduces a local conformational change in naked viral DNA. The effect of all four possible nucleotide sequences at position 5258 on local DNA conformation was analyzed by gel electrophoresis of fragments produced by ligation of synthetic oligonucleotides having these sequences. The results indicated that both the wild-type and the F441 sequences introduced local structural polymorphism that can lead to DNA bending. The wild-type sequence had a greater effect on DNA curvature than the F441 sequence. The two other sequences at nucleotide 5258 did not appear to introduce detectable amounts of DNA curvature.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Boccara M., Kelly F. Etude de la sensibilité au virus du polyome et à SV40 de plusieurs lignées cellulaires de tératocarcinome. Ann Microbiol (Paris) 1978 Feb-Mar;129(2):227–238. [PubMed] [Google Scholar]
- Bossi L., Smith D. M. Conformational change in the DNA associated with an unusual promoter mutation in a tRNA operon of Salmonella. Cell. 1984 Dec;39(3 Pt 2):643–652. doi: 10.1016/0092-8674(84)90471-9. [DOI] [PubMed] [Google Scholar]
- Cartwright I. L., Elgin S. C. Analysis of chromatin structure and DNA sequence organization: use of the 1,10-phenanthroline-cuprous complex. Nucleic Acids Res. 1982 Oct 11;10(19):5835–5852. doi: 10.1093/nar/10.19.5835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courey A. J., Plon S. E., Wang J. C. The use of psoralen-modified DNA to probe the mechanism of enhancer action. Cell. 1986 May 23;45(4):567–574. doi: 10.1016/0092-8674(86)90288-6. [DOI] [PubMed] [Google Scholar]
- Delli Bovi P., De Simone V., Giordano R., Amati P. Polyomavirus growth and persistence in Friend erythroleukemic cells. J Virol. 1984 Feb;49(2):566–571. doi: 10.1128/jvi.49.2.566-571.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deninger P. L., Esty A., LaPorte P., Hsu H., Friedmann T. The nucleotide sequence and restriction enzyme sites of the polyoma genome. Nucleic Acids Res. 1980 Feb 25;8(4):855–860. [PMC free article] [PubMed] [Google Scholar]
- Drew H. R., Travers A. A. DNA structural variations in the E. coli tyrT promoter. Cell. 1984 Jun;37(2):491–502. doi: 10.1016/0092-8674(84)90379-9. [DOI] [PubMed] [Google Scholar]
- Enver T., Patient R. Importance of helical periodicity. Nature. 1986 Jan 9;319(6049):99–100. doi: 10.1038/319099b0. [DOI] [PubMed] [Google Scholar]
- Fujimura F. K., Deininger P. L., Friedmann T., Linney E. Mutation near the polyoma DNA replication origin permits productive infection of F9 embryonal carcinoma cells. Cell. 1981 Mar;23(3):809–814. doi: 10.1016/0092-8674(81)90445-1. [DOI] [PubMed] [Google Scholar]
- Fujimura F. K. Nuclear activity from F9 embryonal carcinoma cells binding specifically to the enhancers of wild-type polyoma virus and PyEC mutant DNAs. Nucleic Acids Res. 1986 Apr 11;14(7):2845–2861. doi: 10.1093/nar/14.7.2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimura F. K., Silbert P. E., Eckhart W., Linney E. Polyoma virus infection of retinoic acid-induced differentiated teratocarcinoma cells. J Virol. 1981 Jul;39(1):306–312. doi: 10.1128/jvi.39.1.306-312.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galas D. J., Schmitz A. DNAse footprinting: a simple method for the detection of protein-DNA binding specificity. Nucleic Acids Res. 1978 Sep;5(9):3157–3170. doi: 10.1093/nar/5.9.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagerman P. J. Sequence dependence of the curvature of DNA: a test of the phasing hypothesis. Biochemistry. 1985 Dec 3;24(25):7033–7037. doi: 10.1021/bi00346a001. [DOI] [PubMed] [Google Scholar]
- Hagerman P. J. Sequence-directed curvature of DNA. Nature. 1986 May 22;321(6068):449–450. doi: 10.1038/321449a0. [DOI] [PubMed] [Google Scholar]
- Hochschild A., Ptashne M. Cooperative binding of lambda repressors to sites separated by integral turns of the DNA helix. Cell. 1986 Mar 14;44(5):681–687. doi: 10.1016/0092-8674(86)90833-0. [DOI] [PubMed] [Google Scholar]
- Jones K. A., Kadonaga J. T., Rosenfeld P. J., Kelly T. J., Tjian R. A cellular DNA-binding protein that activates eukaryotic transcription and DNA replication. Cell. 1987 Jan 16;48(1):79–89. doi: 10.1016/0092-8674(87)90358-8. [DOI] [PubMed] [Google Scholar]
- Katinka M., Vasseur M., Montreau N., Yaniv M., Blangy D. Polyoma DNA sequences involved in control of viral gene expression in murine embryonal carcinoma cells. Nature. 1981 Apr 23;290(5808):720–722. doi: 10.1038/290720a0. [DOI] [PubMed] [Google Scholar]
- Katinka M., Yaniv M., Vasseur M., Blangy D. Expression of polyoma early functions in mouse embryonal carcinoma cells depends on sequence rearrangements in the beginning of the late region. Cell. 1980 Jun;20(2):393–399. doi: 10.1016/0092-8674(80)90625-x. [DOI] [PubMed] [Google Scholar]
- Koepsel R. R., Khan S. A. Static and initiator protein-enhanced bending of DNA at a replication origin. Science. 1986 Sep 19;233(4770):1316–1318. doi: 10.1126/science.3749879. [DOI] [PubMed] [Google Scholar]
- Koo H. S., Wu H. M., Crothers D. M. DNA bending at adenine . thymine tracts. Nature. 1986 Apr 10;320(6062):501–506. doi: 10.1038/320501a0. [DOI] [PubMed] [Google Scholar]
- Kovesdi I., Satake M., Furukawa K., Reichel R., Ito Y., Nevins J. R. A factor discriminating between the wild-type and a mutant polyomavirus enhancer. Nature. 1987 Jul 2;328(6125):87–89. doi: 10.1038/328087a0. [DOI] [PubMed] [Google Scholar]
- Maione R., Passananti C., De Simone V., Delli-Bovi P., Augusti-Tocco G., Amati P. Selection of mouse neuroblastoma cell-specific polyoma virus mutants with stage differentiative advantages of replication. EMBO J. 1985 Dec 1;4(12):3215–3221. doi: 10.1002/j.1460-2075.1985.tb04068.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marini J. C., Effron P. N., Goodman T. C., Singleton C. K., Wells R. D., Wartell R. M., Englund P. T. Physical characterization of a kinetoplast DNA fragment with unusual properties. J Biol Chem. 1984 Jul 25;259(14):8974–8979. [PubMed] [Google Scholar]
- Marini J. C., Levene S. D., Crothers D. M., Englund P. T. Bent helical structure in kinetoplast DNA. Proc Natl Acad Sci U S A. 1982 Dec;79(24):7664–7668. doi: 10.1073/pnas.79.24.7664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee S., Patel I., Bastia D. Conformational changes in a replication origin induced by an initiator protein. Cell. 1985 Nov;43(1):189–197. doi: 10.1016/0092-8674(85)90023-6. [DOI] [PubMed] [Google Scholar]
- Plon S. E., Wang J. C. Transcription of the human beta-globin gene is stimulated by an SV40 enhancer to which it is physically linked but topologically uncoupled. Cell. 1986 May 23;45(4):575–580. doi: 10.1016/0092-8674(86)90289-8. [DOI] [PubMed] [Google Scholar]
- Ryder K., Silver S., DeLucia A. L., Fanning E., Tegtmeyer P. An altered DNA conformation in origin region I is a determinant for the binding of SV40 large T antigen. Cell. 1986 Mar 14;44(5):719–725. doi: 10.1016/0092-8674(86)90838-x. [DOI] [PubMed] [Google Scholar]
- Sekikawa K., Levine A. J. Isolation and characterization of polyoma host range mutants that replicate in nullipotential embryonal carcinoma cells. Proc Natl Acad Sci U S A. 1981 Feb;78(2):1100–1104. doi: 10.1073/pnas.78.2.1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder M., Buchman A. R., Davis R. W. Bent DNA at a yeast autonomously replicating sequence. Nature. 1986 Nov 6;324(6092):87–89. doi: 10.1038/324087a0. [DOI] [PubMed] [Google Scholar]
- Swartzendruber D. E., Lehman J. M. Neoplastic differentiation: interaction of simian virus 40 and polyoma virus with murine teratocarcinoma cells in vitro. J Cell Physiol. 1975 Apr;85(2 Pt 1):179–187. doi: 10.1002/jcp.1040850204. [DOI] [PubMed] [Google Scholar]
- Takahashi K., Vigneron M., Matthes H., Wildeman A., Zenke M., Chambon P. Requirement of stereospecific alignments for initiation from the simian virus 40 early promoter. Nature. 1986 Jan 9;319(6049):121–126. doi: 10.1038/319121a0. [DOI] [PubMed] [Google Scholar]
- Tanaka K., Chowdhury K., Chang K. S., Israel M., Ito Y. Isolation and characterization of polyoma virus mutants which grow in murine embryonal carcinoma and trophoblast cells. EMBO J. 1982;1(12):1521–1527. doi: 10.1002/j.1460-2075.1982.tb01349.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trevor K., Lehman J. M. The interaction of polyoma virus with F9 embryonal carcinoma cells and chemically induced differentiated progeny: fate of the viral DNA and expression of viral antigens. J Cell Physiol Suppl. 1982;2:69–83. doi: 10.1002/jcp.1041130512. [DOI] [PubMed] [Google Scholar]
- Wu C. An exonuclease protection assay reveals heat-shock element and TATA box DNA-binding proteins in crude nuclear extracts. Nature. 1985 Sep 5;317(6032):84–87. doi: 10.1038/317084a0. [DOI] [PubMed] [Google Scholar]
- Wu H. M., Crothers D. M. The locus of sequence-directed and protein-induced DNA bending. Nature. 1984 Apr 5;308(5959):509–513. doi: 10.1038/308509a0. [DOI] [PubMed] [Google Scholar]
- Xiao J. H., Davidson I., Ferrandon D., Rosales R., Vigneron M., Macchi M., Ruffenach F., Chambon P. One cell-specific and three ubiquitous nuclear proteins bind in vitro to overlapping motifs in the domain B1 of the SV40 enhancer. EMBO J. 1987 Oct;6(10):3005–3013. doi: 10.1002/j.1460-2075.1987.tb02606.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahn K., Blattner F. R. Binding and bending of the lambda replication origin by the phage O protein. EMBO J. 1985 Dec 16;4(13A):3605–3616. doi: 10.1002/j.1460-2075.1985.tb04124.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahn K., Blattner F. R. Sequence-induced DNA curvature at the bacteriophage lambda origin of replication. Nature. 1985 Oct 3;317(6036):451–453. doi: 10.1038/317451a0. [DOI] [PubMed] [Google Scholar]
- de Villiers J., Schaffner W. A small segment of polyoma virus DNA enhances the expression of a cloned beta-globin gene over a distance of 1400 base pairs. Nucleic Acids Res. 1981 Dec 11;9(23):6251–6264. doi: 10.1093/nar/9.23.6251. [DOI] [PMC free article] [PubMed] [Google Scholar]