Abstract
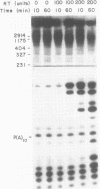
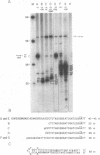
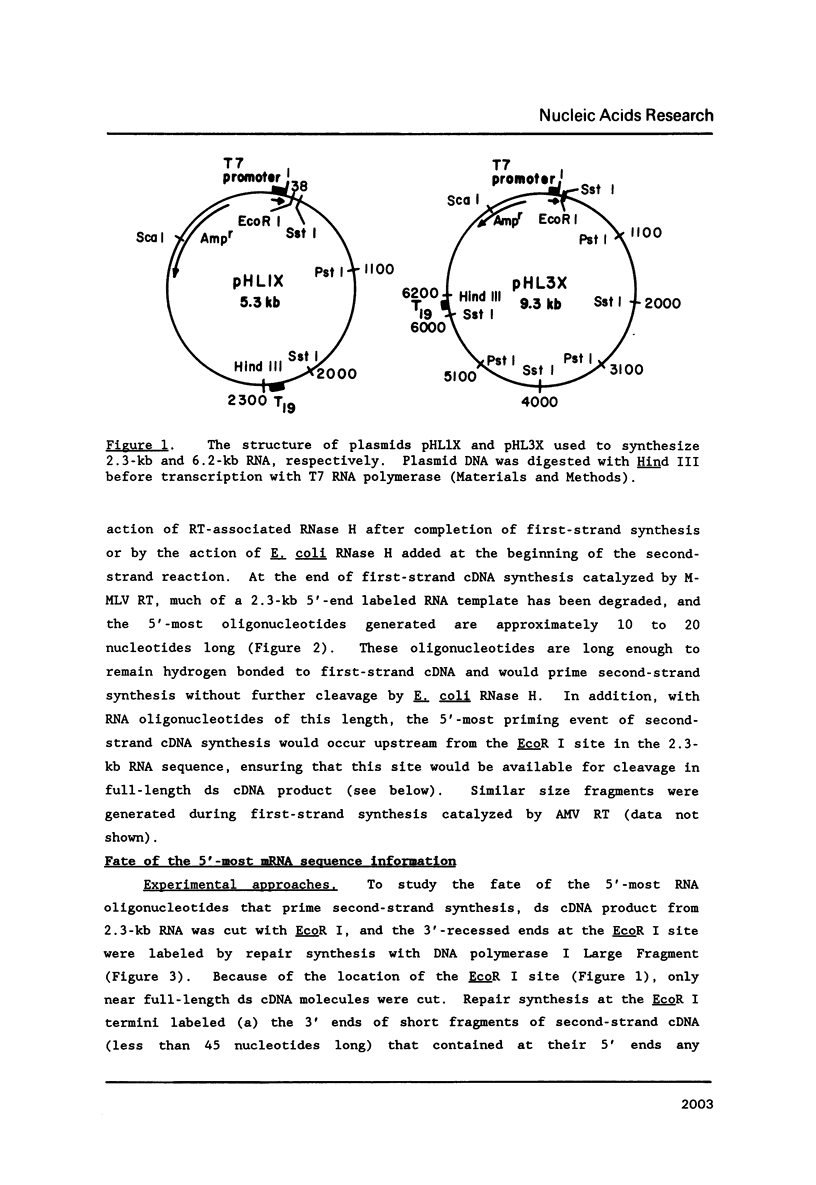
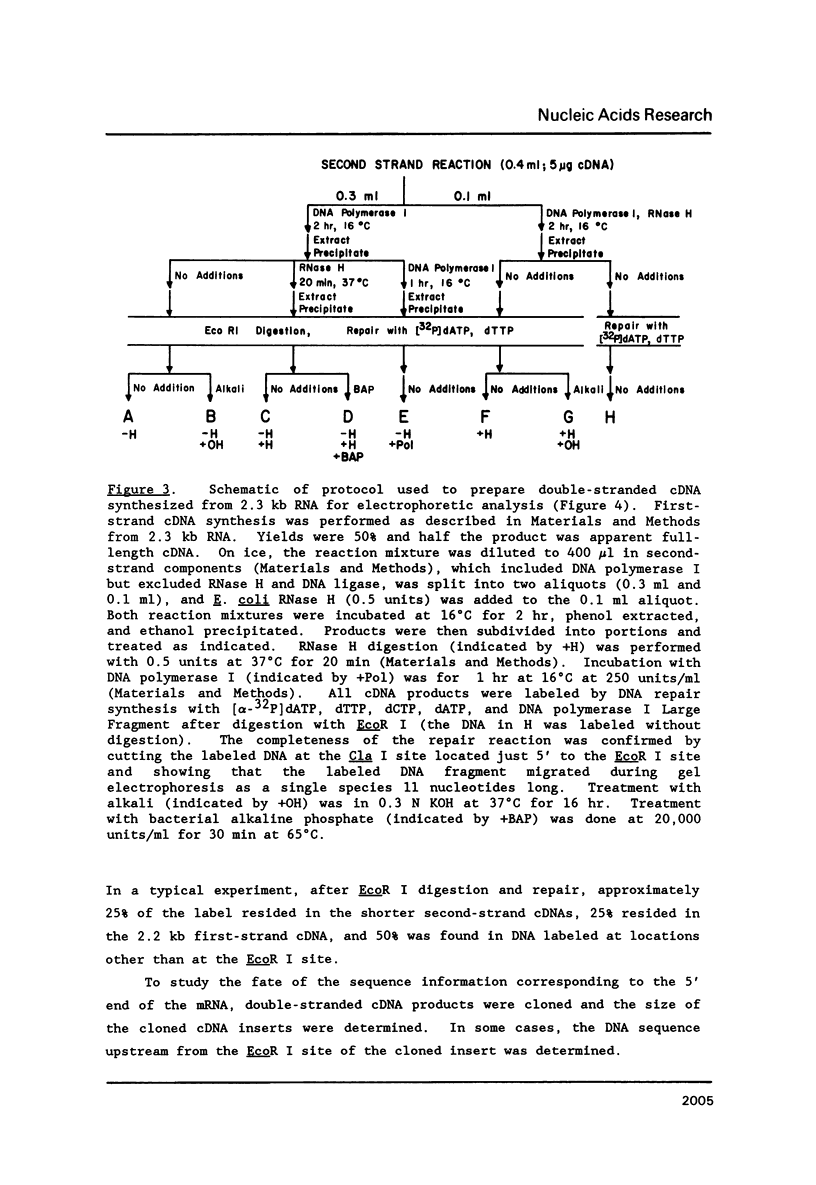
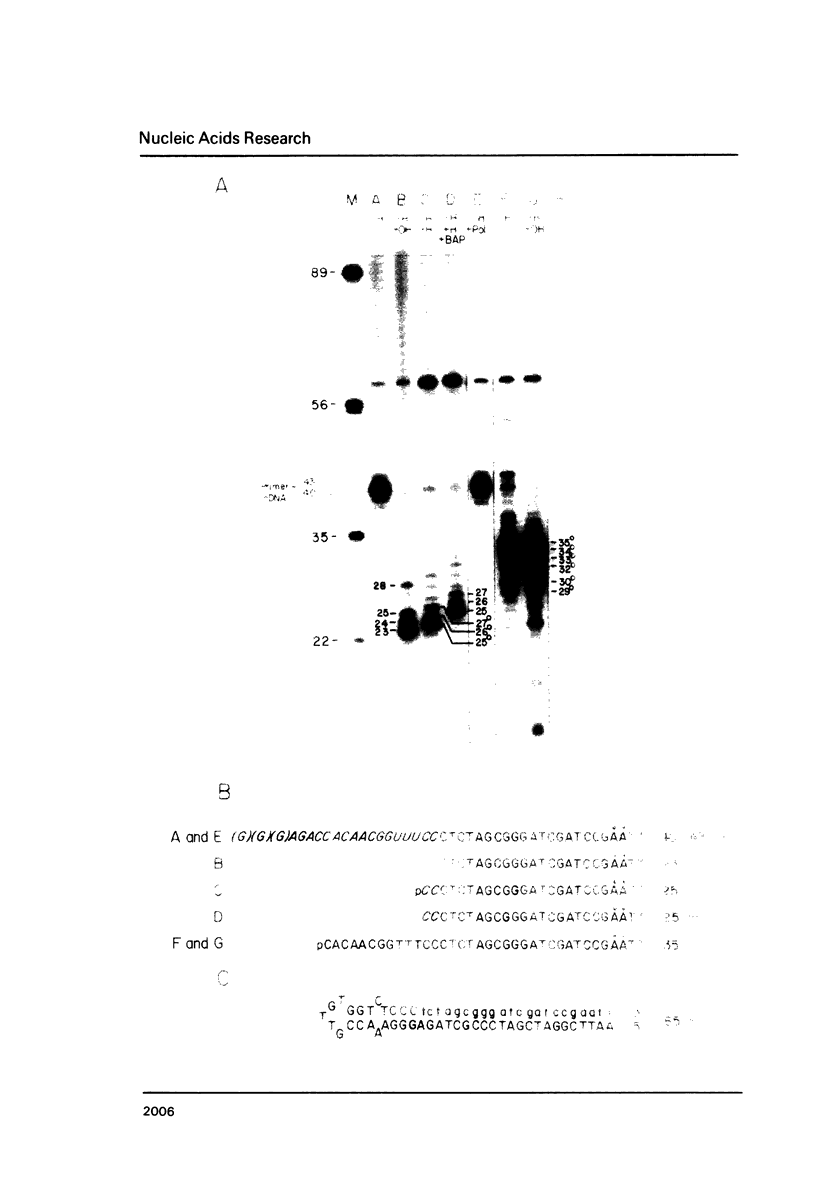
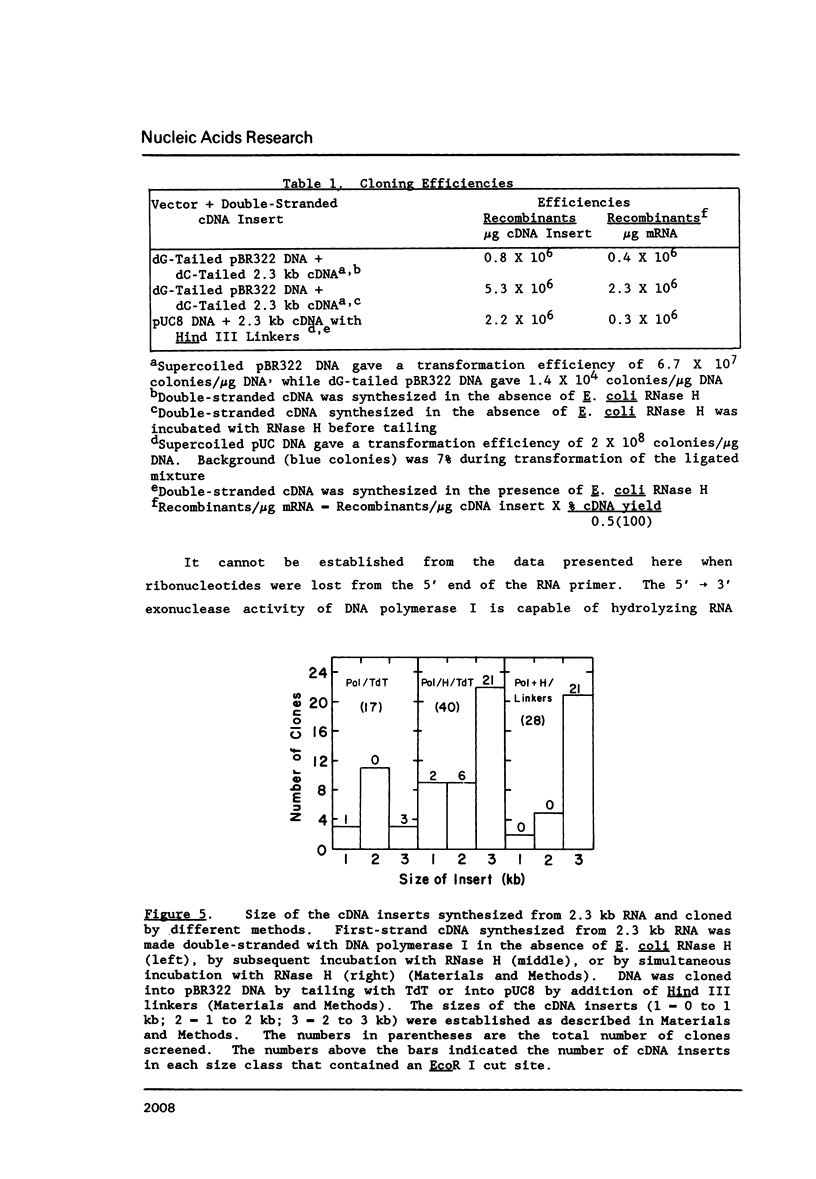
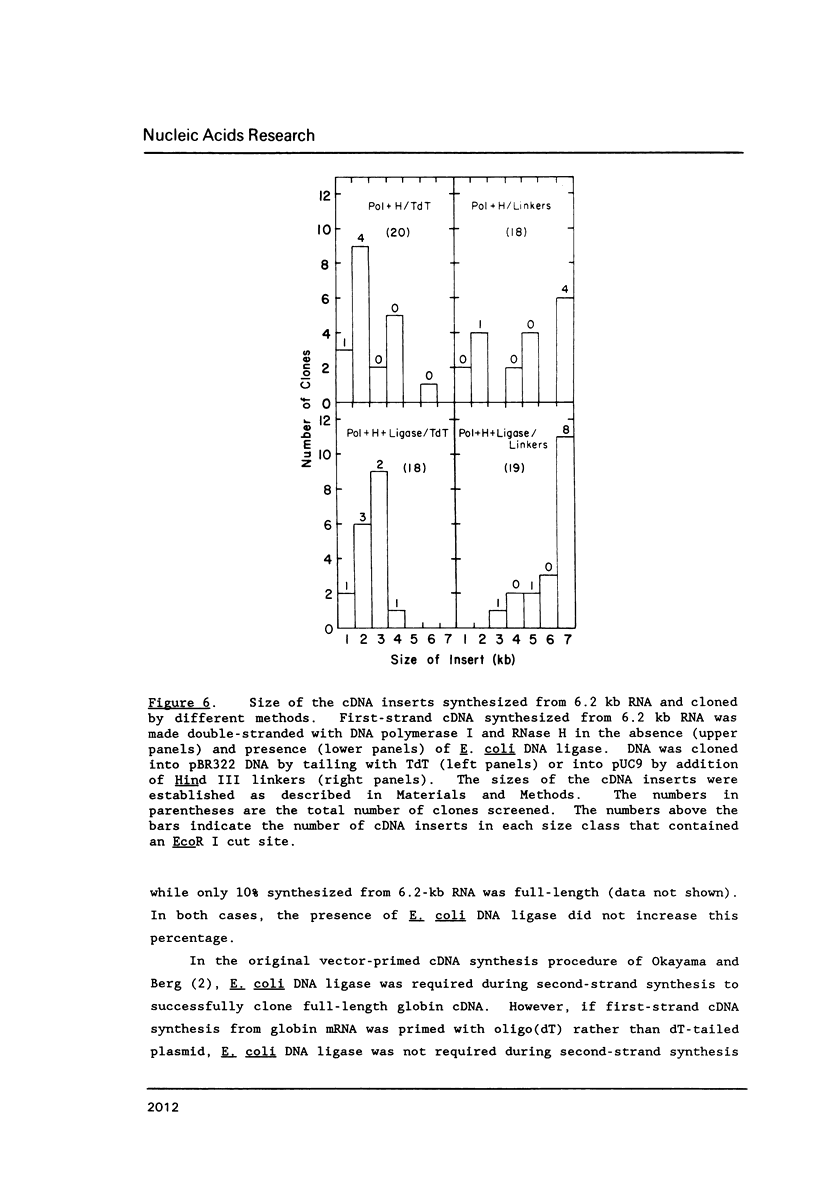
A simple method for generating cDNA libraries has been described (1) in which RNase H-DNA polymerase I-mediated second-strand cDNA synthesis primes from an RNA oligonucleotide derived from the 5' (capped) end of mRNA. The size of this oligonucleotide and the fate of the information corresponding to the RNA during subsequent cloning have not been established. We show here that the 5'-most RNA primer varies in length from 8 to 21 nucleotides, and that information corresponding to the length of the RNA primer is normally lost during cloning. A modification of the second-strand cDNA synthesis procedure is described which allows cloning of all, or almost all, of the primer sequence information. In addition, we show that the presence of E. coli DNA ligase during second-strand cDNA synthesis can increase the length of the cDNA clones obtained from long RNAs. Cloning by addition of linkers provides the greatest chance of obtaining near full-length cDNA clones from long mRNAs.
Full text
PDF










Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aviv H., Leder P. Purification of biologically active globin messenger RNA by chromatography on oligothymidylic acid-cellulose. Proc Natl Acad Sci U S A. 1972 Jun;69(6):1408–1412. doi: 10.1073/pnas.69.6.1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmichael G. G., McMaster G. K. The analysis of nucleic acids in gels using glyoxal and acridine orange. Methods Enzymol. 1980;65(1):380–391. doi: 10.1016/s0076-6879(80)65049-6. [DOI] [PubMed] [Google Scholar]
- Challberg M. D., Englund P. T. Specific labeling of 3' termini with T4 DNA polymerase. Methods Enzymol. 1980;65(1):39–43. doi: 10.1016/s0076-6879(80)65008-3. [DOI] [PubMed] [Google Scholar]
- Deng G., Wu R. An improved procedure for utilizing terminal transferase to add homopolymers to the 3' termini of DNA. Nucleic Acids Res. 1981 Aug 25;9(16):4173–4188. doi: 10.1093/nar/9.16.4173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng G., Wu R. Terminal transferase: use of the tailing of DNA and for in vitro mutagenesis. Methods Enzymol. 1983;100:96–116. doi: 10.1016/0076-6879(83)00047-6. [DOI] [PubMed] [Google Scholar]
- Gubler U., Hoffman B. J. A simple and very efficient method for generating cDNA libraries. Gene. 1983 Nov;25(2-3):263–269. doi: 10.1016/0378-1119(83)90230-5. [DOI] [PubMed] [Google Scholar]
- Keller W., Crouch R. Degradation of DNA RNA hybrids by ribonuclease H and DNA polymerases of cellular and viral origin. Proc Natl Acad Sci U S A. 1972 Nov;69(11):3360–3364. doi: 10.1073/pnas.69.11.3360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leis J. P., Berkower I., Hurwitz J. Mechanism of action of ribonuclease H isolated from avian myeloblastosis virus and Escherichia coli. Proc Natl Acad Sci U S A. 1973 Feb;70(2):466–470. doi: 10.1073/pnas.70.2.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Okayama H., Berg P. High-efficiency cloning of full-length cDNA. Mol Cell Biol. 1982 Feb;2(2):161–170. doi: 10.1128/mcb.2.2.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omer C. A., Faras A. J. Mechanism of release of the avian rotavirus tRNATrp primer molecule from viral DNA by ribonuclease H during reverse transcription. Cell. 1982 Oct;30(3):797–805. doi: 10.1016/0092-8674(82)90284-7. [DOI] [PubMed] [Google Scholar]
- Resnick R., Omer C. A., Faras A. J. Involvement of retrovirus reverse transcriptase-associated RNase H in the initiation of strong-stop (+) DNA synthesis and the generation of the long terminal repeat. J Virol. 1984 Sep;51(3):813–821. doi: 10.1128/jvi.51.3.813-821.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]