Abstract
Objective
To investigate and refine the relationships among systemic lupus erythematosus (SLE) and related autoantibodies, interferon-α (IFN-α), and various ancestral backgrounds.
Methods
We investigated quantitatively defined genetic ancestry through principal component analysis in place of self-reported ancestry.
Results
African ancestry was found to be associated with presence of anti-RNP antibody (p = 0.0026), and anti-RNP was correlated with high levels of IFN-α (p = 2.8 × 10−5).
Conclusion
Our data support a model in which African ancestry increases the likelihood of SLE-associated autoantibody formation, which subsequently results in higher levels of serum IFN-α.
Key Indexing Terms: SYSTEMIC LUPUS ERYTHEMATOSUS, AUTOANTIBODIES, INTERFERONS, GENETICS
Systemic lupus erythematosus (SLE) is a heterogeneous disease characterized by different clinical and serological manifestations, which vary in prevalence across self-reported ancestral backgrounds1,2,3,4. Anti-Sm and anti-RNP antibodies are more prevalent in African American (AA) patients with SLE than in European (EA) and Hispanic American (HA) cohorts2,4. Moreover, AA and HA patients with SLE have a higher incidence of SLE-related renal disease than EA patients3. EA and HA patients tend to have more photosensitivity and oral ulcers compared to AA patients2,4. High levels of interferon-α (IFN-α), which plays an important pathogenic role in SLE, have been associated with positive serology and self-reported non-European ancestry5.
Using self-reported ancestry to categorize subjects can be problematic, as many systems for self-report are designed to indicate 1 ancestral background per subject, and proportional admixture is not taken into account. Assessment of genetic ancestry using ancestry-informative markers (AIM) provides a means to address incomplete data provided by self-report systems6,7,8. Principal component analysis using AIM can provide covariates designating genetic ancestry that can be used to control for admixture in diverse study populations6,7,8,9. We investigated the relationships among ancestry, serum IFN-α levels, and autoantibodies using quantitatively defined genetic ancestry.
MATERIALS AND METHODS
Patients, samples, and data
Serum samples were obtained from 220 patients with SLE (140 AA, 71 EA, and 9 HA by self-reports) from the Translational Research Initiative in the Department of Medicine at the University of Chicago Medical Center (UCMC). All the patients met the American College of Rheumatology criteria for SLE1,10. Autoantibodies to Ro, La, Sm, and RNP were measured by ELISA methods (Inova Diagnostics, San Diego, CA, USA) at UCMC, and standard clinical laboratory cutoff points were used to categorize them as positive or negative. Anti-dsDNA levels were measured using Crithidia luciliae immunofluorescence.
Serum IFN-α activity was measured through a reporter cell bioassay (WISH cells, ATCC no. CCL-25) that assesses the ability of patient sera to cause IFN-induced gene expression11. IFN-α activity was categorized into high versus low based on a cutoff point of 2 SD above the mean of healthy donors. A total of 374 AIM were genotyped in the context of a larger experiment12, and principal components derived from these AIM were used to designate genetic ancestry.
Statistical analysis
As a first-pass analysis, we performed multivariate logistic regression analysis on IFN-α and each autoantibody using principal components 1, 2, and 3 (PC1, PC2, PC3) as predictor variables. Sex and age were used as covariates in all regressions, and IFN-α was also included as a covariate in the autoantibody regressions because of the reported associations between IFN-α and autoantibodies5,13. We then carried forward variables with a p value < 0.20 for further analysis. Next, we performed backward stepwise regression modeling using each autoantibody and IFN-α as outcome variables with all the other variables as predictors for network analysis. Results with p < 0.05 in this overall model were considered significant, with OR to indicate the magnitude of the relationship.
RESULTS
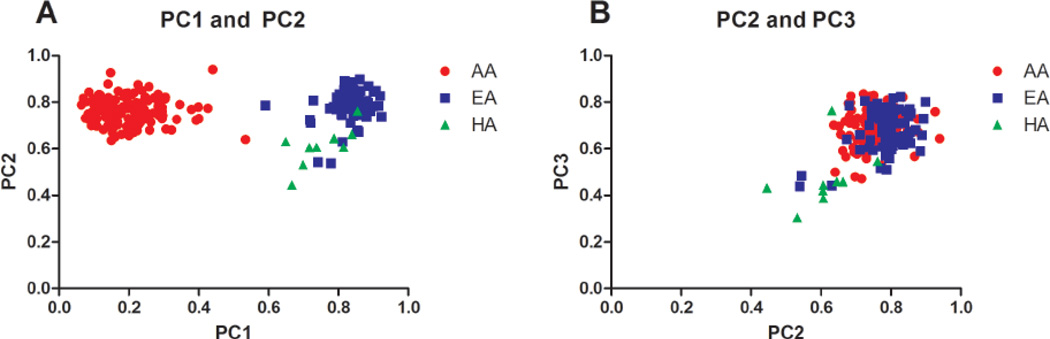
The average age of subjects was 45 ± 14.1 years and 85.5% were women. Figure 1 shows plots of the first 3 principal components derived from the AIM, with each subject color-coded by self-reported ancestry. PC1 separated African from non-African ancestry, and the second and third components corresponded to some degree with self-reported Hispanic ancestry. First, we detected associations between genetic ancestry and each antibody and IFN-α separately (Table 1). Lower values of PC1 were correlated with high IFN-α activity and positive anti-Ro, anti-Sm, and anti-RNP (p = 0.0025, 0.0194, 0.0112, and 0.0002, respectively), thus linking them to African ancestry. Lower values of PC2 were associated with the presence of anti-La (p = 0.0397), suggesting a positive connection between Hispanic ancestry and anti-La.
Figure 1.
Principal component (PC) analysis of ancestry-informative genetic markers in patients with systemic lupus erythematosus of various self-reported ancestral backgrounds. PC1, PC2, and PC3 correspond to the first, second, and third principal components from this analysis, and each dot represents 1 subject who is color-coded by self-reported ancestral background. A. PC1 vs PC2; B. PC2 vs PC3. AA: African-American; EA: European-American; HA: Hispanic American.
Table 1.
Associations between genetic ancestry and autoantibodies with age, sex, and interferon-α (IFN-α) as covariates.
| IFN-α | Anti-RNP | |||||
|---|---|---|---|---|---|---|
| β | SE | p | β | SE | p | |
| Sex | −0.6465 | 0.431 | 0.1336 | −0.0362 | 0.4689 | 0.9385 |
| Age | −0.0408 | 0.0111 | 0.0002 | −0.0162 | 0.0122 | 0.1848 |
| IFN-α* | NA | NA | NA | 1.4942 | 0.3303 | 6.1 × 10−6 |
| PC1 | −1.4764 | 0.4889 | 0.0025 | −2.1128 | 0.5576 | 0.0002 |
| PC2** | −3.6297 | 2.3468 | 0.1219 | — | — | — |
| PC3** | — | — | — | — | — | — |
| Anti-RO | Anti-SM | |||||
| β | SE | p | β | SE | p | |
| Sex | −0.1825 | 0.4459 | 0.6823 | 0.0313 | 0.5299 | 0.9529 |
| Age | 0.0072 | 0.5235 | 0.5235 | −0.0263 | 0.0132 | 0.0466 |
| IFN-α | 0.7732 | 0.3179 | 0.0150 | 0.7354 | 0.3605 | 0.0413 |
| PC1 | −1.2298 | 0.5261 | 0.0194 | −1.6780 | 0.6612 | 0.0112 |
| PC2** | — | — | — | −4.3808 | 2.4314 | 0.0716 |
| PC3** | 1.9206 | 1.7241 | 0.2653 | — | — | — |
| Anti-La | Anti-dsDNA | |||||
| β | SE | p | β | SE | p | |
| Sex | 0.1040 | 0.686 | 0.8795 | 0.2188 | 0.4638 | 0.6372 |
| Age | 0.0129 | 0.0155 | 0.4038 | −0.0273 | 0.012 | 0.023 |
| IFN-α | 1.0511 | 0.494 | 0.0334 | 0.8224 | 0.3168 | 0.0094 |
| PC1** | −1.5001 | 0.8949 | 0.0937 | — | — | — |
| PC2** | −6.1517 | 2.991 | 0.0397 | — | — | — |
| PC3** | — | — | — | — | — | — |
IFN-α was the dependent variable, so it was not used as a covariate.
Values were omitted if components did not meet the first-pass criterion (p < 0.20), indicated with a dash. PC: principal component; NA: not applicable.
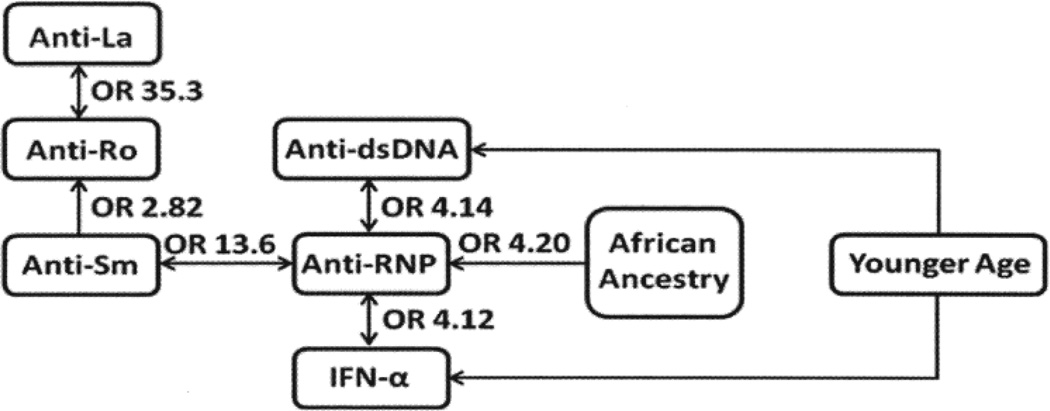
We next considered the significant variables in an overall logistic model to account for between-variable relationships. Significant results from this overall regression (p < 0.05) are shown in Figure 2. Interestingly, African ancestry was associated only with anti-RNP (p = 0.0026), suggesting that other serologic relationships with African genetic ancestry may be secondary to anti-RNP. Additionally, IFN-α was not directly associated with African ancestry, but instead was correlated with anti-RNP (p = 2.8 × 10−5).
Figure 2.
Network diagram shows relationships among genetic ancestry, age, autoantibodies, and interferon-α (IFN-α), with OR indicating the magnitude of the relationship.
DISCUSSION
A study by Weckerle, et al5 showed that self-reported non-European ancestry was associated with high serum IFN-α activity and positive autoantibodies in SLE, and after control for autoantibodies, a residual association remained between non-European ancestry and high IFN-α. We used genetic assessment of ancestry to investigate this question in detail, and this allowed us to resolve these overlapping associations to some degree. Our study supports a model in which African ancestry increases the likelihood of SLE-associated autoantibody formation, which subsequently results in higher levels of serum IFN-α. This finding is supported by previous in vitro work showing that SLE autoantibody immune complexes were able to induce IFN-α production, likely through Toll-like receptors14. We also replicated the relationship between higher IFN-α and younger age in SLE, and this finding continues to be robust across multiple analyses and cohorts5,15.
Previous studies have shown other autoantibodies such as anti-Sm, anti-Ro, and anti-DNA also were associated with AA patients2,3,5. That anti-RNP was the only independently linked antibody in our study could be related to the high prevalence of this antibody in AA patients with SLE and reduced statistical power to detect associations with other less prevalent autoantibodies. It is also possible that the careful designation of genetic ancestry increased our ability to distinguish associations particular to African ancestry, eliminating potential residual effects from genetic admixture. Future larger-scale efforts will be able to answer this question more definitively.
IFN-α is an important pathogenic mediator in SLE5,13, and improved understanding of the factors related to high IFN-α levels in humans is critical to our understanding of SLE. Our data support a model in which African genetic ancestry is linked to autoantibody production, leading to higher IFN-α activity.
Acknowledgments
Supported by Wake Forest University Health Sciences Center for Public Health Genomics (C.D. Langefeld); research grants from US National Institutes of Health (NIH; AR62277, AR42460, AI53747, AI31584, DE15223, RR20143, AI24717, AI62629, AR48940, AI83194, and AR49084), and from US Department of Veterans Affairs, Alliance for Lupus Research, and Rheuminations Inc. (J.B. Harley); NIH R01 AR060861, NIH K08 AI083790, NIH P30 DK42086, National Institute of Allergy and Infectious Diseases Clinical Research Loan Repayment AI071651, NIH CTSA Core Subsidy Grant, and CTSA Pilot Grants from UL1 RR024999, Lupus Research Institute Novel Research Grant, Alliance for Lupus Research Target Identification in Lupus Grant, Arthritis National Research, and a Foundation Eng Tan Scholar Award (T.B. Niewold).
K. Ko, MD; B.S. Franek, BS, Section of Rheumatology and Gwen Knapp Center for Lupus and Immunology Research, University of Chicago; M. Marion, MA, Wake Forest University Health Sciences; K.M. Kaufman, PhD, Center for Autoimmune Genomics and Etiology, Cincinnati Children’s Hospital; C.D. Langefeld, PhD, Wake Forest University Health Sciences; J.B. Harley, MD, PhD, Center for Autoimmune Genomics and Etiology, Cincinnati Children’s Hospital; T.B. Niewold, MD, Section of Rheumatology and Gwen Knapp Center for Lupus and Immunology Research, University of Chicago.
References
- 1.Hochberg MC. Updating the American College of Rheumatology revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum. 1997;40:1725. doi: 10.1002/art.1780400928. [DOI] [PubMed] [Google Scholar]
- 2.Molina JF, Molina J, Garcia C, Gharavi AE, Wilson WA, Espinoza LR. Ethnic differences in the clinical expression of systemic lupus erythematosus: A comparative study between African-Americans and Latin Americans. Lupus. 1997;6:63–67. doi: 10.1177/096120339700600109. [DOI] [PubMed] [Google Scholar]
- 3.Alarcon GS, Friedman AW, Straaton KV, Moulds JM, Lisse J, Bastian HM, et al. Systemic lupus erythematosus in three ethnic groups: III. A comparison of characteristics early in the natural history of the LUMINA cohort. LUpus in MInority populations: Nature vs. Nurture. Lupus. 1999;8:197–209. doi: 10.1191/096120399678847704. [DOI] [PubMed] [Google Scholar]
- 4.Cooper GS, Parks CG, Treadwell EL, St. Clair EW, Gilkeson GS, Cohen PL, et al. Differences by race, sex and age in the clinical and immunologic features of recently diagnosed systemic lupus erythematosus patients in the southeastern United States. Lupus. 2002;11:161–167. doi: 10.1191/0961203302lu161oa. [DOI] [PubMed] [Google Scholar]
- 5.Weckerle CE, Franek BS, Kelly JA, Kumabe M, Mikolaitis RA, Green SL, et al. Network analysis of associations between serum interferon-alpha activity, autoantibodies, and clinical features in systemic lupus erythematosus. Arthritis Rheum. 2011;63:1044–1053. doi: 10.1002/art.30187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bonilla C, Parra EJ, Pfaff CL, Dios S, Marshall JA, Hamman RF, et al. Admixture in the Hispanics of the San Luis Valley, Colorado, and its implications for complex trait gene mapping. Ann Hum Genet. 2004;68(Pt 2):139–153. doi: 10.1046/j.1529-8817.2003.00084.x. [DOI] [PubMed] [Google Scholar]
- 7.Fejerman L, Haiman CA, Reich D, Tandon A, Deo RC, John EM, et al. An admixture scan in 1,484 African American women with breast cancer. Cancer Epidemiol Biomarkers Prev. 2009;18:3110–3117. doi: 10.1158/1055-9965.EPI-09-0464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang H, Haiman CA, Kolonel LN, Henderson BE, Wilkens LR, Le Marchand L, et al. Self-reported ethnicity, genetic structure and the impact of population stratification in a multiethnic study. Hum Genet. 2010;128:165–177. doi: 10.1007/s00439-010-0841-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ringner M. What is principal component analysis? Nat Biotechnol. 2008;26:303–304. doi: 10.1038/nbt0308-303. [DOI] [PubMed] [Google Scholar]
- 10.Tan EM, Cohen AS, Fries JF, Masi AT, McShane DJ, Rothfield NF, et al. The 1982 revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum. 1982;25:1271–1277. doi: 10.1002/art.1780251101. [DOI] [PubMed] [Google Scholar]
- 11.Niewold TB, Hua J, Lehman TJ, Harley JB, Crow MK. High serum IFN-alpha activity is a heritable risk factor for systemic lupus erythematosus. Genes Immun. 2007;8:492–502. doi: 10.1038/sj.gene.6364408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lessard CJ, Adrianto I, Kelly JA, Kaufman KM, Grundahl KM, Adler A, et al. Identification of a systemic lupus erythematosus susceptibility locus at 11p13 between PDHX and CD44 in a multiethnic study. Am J Hum Genet. 2011;88:83–91. doi: 10.1016/j.ajhg.2010.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kirou KA, Lee C, George S, Louca K, Peterson MG, Crow MK. Activation of the interferon-alpha pathway identifies a subgroup of systemic lupus erythematosus patients with distinct serologic features and active disease. Arthritis Rheum. 2005;52:1491–1503. doi: 10.1002/art.21031. [DOI] [PubMed] [Google Scholar]
- 14.Lovgren T, Eloranta ML, Kastner B, Wahren-Herlenius M, Alm GV, Ronnblom L. Induction of interferon-alpha by immune complexes or liposomes containing systemic lupus erythematosus autoantigen- and Sjogren’s syndrome autoantigen-associated RNA. Arthritis Rheum. 2006;54:1917–1927. doi: 10.1002/art.21893. [DOI] [PubMed] [Google Scholar]
- 15.Niewold TB, Adler JE, Glenn SB, Lehman TJ, Harley JB, Crow MK. Age- and sex-related patterns of serum interferon-alpha activity in lupus families. Arthritis Rheum. 2008;58:2113–2119. doi: 10.1002/art.23619. [DOI] [PMC free article] [PubMed] [Google Scholar]