Abstract
Although lung cancer remains the leading cancer killer in the United States, recently a number of developments indicate future clinical benefit. These include evidence that computed tomography–based screening decreases lung cancer mortality, the use of stereotactic radiation for early-stage tumors, the development of molecular methods to predict chemotherapy sensitivity, and genome-wide expression and mutation analysis data that have uncovered oncogene “addictions” as important therapeutic targets. Perhaps the most significant advance in the treatment of this challenging disease is the introduction of molecularly targeted therapies, a term that currently includes monoclonal antibodies and small-molecule tyrosine kinase inhibitors. The development of effective targeted therapeutics requires knowledge of the genes and pathways involved and how they relate to the biologic behavior of lung cancer. Drugs targeting the epidermal growth factor receptor, anaplastic lymphoma kinase, and vascular endothelial growth factor are now U.S. Food and Drug Administration approved for the treatment of advanced non-small cell lung cancer. These agents are generally better tolerated than conventional chemotherapy and show dramatic efficacy when their use is coupled with a clear understanding of clinical data, mechanism, patient selection, drug interactions, and toxicities. Integrating genome-wide tumor analysis with drug- and targeted agent-responsive phenotypes will provide a wealth of new possibilities for lung cancer–targeted therapeutics. Ongoing research efforts in these areas as well as a discussion of emerging targeted agents being evaluated in clinical trials are the subjects of this review.
Keywords: Lung cancer, targeted therapies, genome-wide tumor analysis, tyrosine kinase inhibitor, monoclonal antibody, EGFR, VEGF, ALK
Lung cancer is a heterogeneous disease clinically, histologically, biologically, and molecularly.1 The 2 main types of lung cancer, non-small cell lung cancer (NSCLC) (representing 80%–85% of cases) and small cell lung cancer (SCLC) (representing 15%–20%) are identified based on histological, clinical, and immunohistochemical characteristics and also differ molecularly with many genetic alterations exhibiting subtype specificity. Non-small cell lung cancer can be further histologically subdivided into adenocarcinoma (including bronchoalveolar carcinoma), squamous cell carcinoma, large cell carcinoma (including large cell neuroendocrine lung cancers), and mixed histologic types (e.g., adenosquamous carcinoma). In fact, these histologic subdivisions provide important information for NSCLC therapy selection such as the use of pemetrexed and bevacizumab in adenocarcinoma but not squamous lung cancers.
Surgical resection of the primary tumor remains the best chance of cure for NSCLC. Early stages I and II NSCLCs are generally treated with surgery (or radiation if the patient is not a surgical candidate) with or without adjuvant chemotherapy for stages IB and II. Locally advanced stages IIIA and IIIB disease is typically treated with a combination of chemotherapy and radiation if the patient is not a surgical candidate. Patients with metastatic disease (stage IV) are generally treated with chemotherapy to improve quality of life, palliate symptoms, and improve survival. Second- and third-line treatment for advanced or recurrent disease is chemotherapy and, more recently, epidermal growth factor receptor (EGFR) tyrosine kinase inhibitors (TKIs) if harboring an EGFR mutation or amplification. Patients with SCLC are generally treated with chemotherapy and concurrent radiation (if staged as limited disease) or chemotherapy alone (if staged as extensive disease).
TARGETED THERAPIES
The term targeted therapy potentially applies to all cancer treatments. Conventional cytotoxic chemotherapy is targeted against DNA replication (alkylating agents, topoisomerase inhibitors, anthracyclines, antimetabolites) or the mitotic microtubule apparatus (taxanes, vinca alkaloids). Recently, tumor expression of molecules relevant to the mechanism of conventional chemotherapeutics (such as excision repair cross-complementation group 1, ribonucleotide reductase M1, and thymidylate synthase) has been associated with response to certain cytotoxic agents, thus providing further evidence of their “targeted” nature. However, currently in general clinical usage, “targeted therapy” refers to 2 classes of cancer drugs: monoclonal antibodies (mAbs) and small-molecule TKIs.
Usually, to achieve a favorable efficacy-to-toxicity profile where a therapy kills tumor but not normal cells, a molecular target should be either unique to, overexpressed in, or mutated in tumors, when compared with normal tissues. The majority of molecular targets are expressed on or within cancer cells themselves. However, some targets—in particular, vascular endothelial growth factor (VEGF) and VEGF receptor (VEGFR)—are expressed in the tumor microenvironment or stroma.
Compared with the dramatic benefit of imatinib therapy for chronic myeloid leukemia (CML),2 the overall effect of targeted therapy on lung cancer outcomes has been modest to date. Only a minority of patients have tumors highly sensitive to these treatments. These cases generally develop resistance within months rather than years. In unselected populations, survival gains are measured in weeks. Molecular complexity and heterogeneity may account for the limitations of targeted therapy in lung cancer and, indeed, in most malignancies. In contrast to the single, well-described chromosomal aberration of CML, lung cancer cells are characterized by multiple molecular abnormalities.3 Furthermore, tumor genetic profiles differ markedly among patients, highlighting the importance of personalized or tailored therapy for this disease.
MOLECULAR TARGETING OF NSCLC
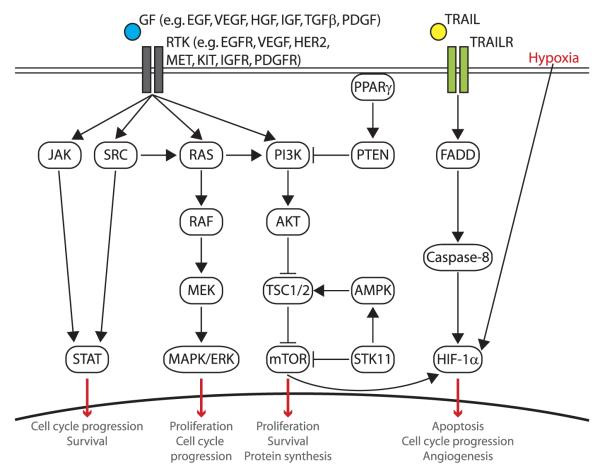
Oncogene activation (by gene amplification, point mutation, or DNA rearrangements) or loss of tumor suppressor gene (TSG) function (by loss of heterozygosity inactivating one allele and point mutation, epigenetic or transcription silencing inactivating the second allele)4,5 occurs in probably all lung cancers. This can result in dysregulation of signaling pathways, leading the cell to exhibit the “hallmarks of cancer” (including self-sufficiency of growth signals, insensitivity to growth-inhibitory (anti-growth) signals, evasion of programmed cell death (apoptosis), limitless replicative potential, sustained angiogenesis, and tissue invasion and metastasis).6,7 Research over the past decade has made a significant step forward by discovering “oncogene addiction” whereby the cell becomes dependent on this aberrant oncogenic signaling for survival.3,8–17 These “driver” oncogenes or oncogene “addictions” represent acquired conditional (on the oncogene) vulnerabilities in lung cancer cells and present as significant therapeutic targets by offering specificity of killing tumor but not normal cells. Thus, the cancer cell needs (is “addicted” to) the continued function of the oncogene (probably because of the associated acquired mutations that are required for the tumor cell to “tolerate” the oncogene), whereas normal cells do not need the continued function. This difference provides the therapeutic window. In lung cancer, commonly activated oncogenes include EGFR/HER1/ERBB1, HER2/ERBB2, MYC, KRAS, MET, CCND1, CDK4, EML4-ALK fusion, and BCL2. Oncogenic signaling pathways commonly dysregulated in lung cancer are summarized in Figure 1. For the most part, these pathways involve activation of a receptor tyrosine kinase (TK) at the cell surface by ligand binding and receptor homodimerization or heterodimerization leading to autophosphorylation of the intracellular tyrosine kinase domain, which in turn triggers multiple signal transduction cascades including the RAS/RAF/MEK (mitogen-activated and extracellular signal–regulated kinase kinase), PI3K (phosphatidylinositol-3-kinase)/AKT/mammalian target of rapamycin (mTOR), and STAT (signal transducer and activator of transcription) pathways.
FIGURE 1.
Signaling pathways and their targeted therapies in NSCLC. Aberrant signaling resulting in activation of growth stimulatory pathways or interference of growth inhibitory pathways has been implicated in lung cancer pathogenesis. Activation of RTKs through ligand binding leads to up-regulation of multiple signaling pathways including the RAS/RAF/MEK pathway, the PI3K/AKT/mTOR pathway, and the STAT. GF, growth factor.
Currently, 1 mAb and 2 TKIs are U.S. Food and Drug Administration (FDA)–approved for the treatment of NSCLC: bevacizumab (Avastin), crizotinib (Xalkori), and erlotinib (Tarceva). Although targeted therapies are generally better tolerated than conventional chemotherapy, they still exhibit toxicities. However, in the appropriate patient population, these drugs may provide greater benefit than conventional chemotherapy. Ongoing developments in targeted therapies for lung cancer have been the topic of several recent reviews,18–22 and selected clinical trials are outlined in detail in the supplementary material for this review (Supplemental Digital Content, http://links.lww.com/PPO/A4). Table 1 gives a summary of available agents that have shown efficacy in lung cancer. Detailed information on clinical trials for targeted agents in lung cancer is available in supplementary tables.
TABLE 1.
Targeted Therapies Approved, in Clinical Trial or in Preclinical Study for Lung Cancer
| Pathway | Target | Clinically Approved for Lung Cancer |
In Clinical Trials for Lung Cancer |
Novel Agents in Preclinical Study |
|---|---|---|---|---|
| RTKs | EGFR | Erlotinib, cetuximab* | Afatinib (BIBW 2992), BMS-690514, canertinib, CUDC-101, EKB-569, gefitinib†, icotinib, lapatinib, matuzumab, necitumumab, neratinib (HKI-272), nimotuzumab, panitumumab, pelitinib, PF0299804, vandetanib, XL647, zalutumumab |
AEE 788, AV-412, BMS-599626 |
| VEGFR | Axitinib, BMS-690514, brivanib alaninate (BMS-582664), cediranib, E7080, foretinib, linifanib, motesanib (AMG-706), neovastat (AE-941), pazopanib, ramucirumab, regorafenib, sorafenib, sunitinib, tivozanib, vandetanib, vargatef (BIBF 1120), vatalanib, XL184, XL647, XL999 |
Adnectin, AEE 788, TKI-258, TSU-68 |
||
| ALK | Crizotinib | GSK1838705A, nVP-TAE684 | ||
| HER2 | Afatinib (BIBW 2992), BMS-690514, CI-1033, CUDC-101, EKB-569, lapatinib, PF0299804, neratinib (HKI-272), pertuzumab, trastuzumab, XL647 |
AEE 788, AV-412, BMS-599626 | ||
| c-MET | Crizotinib | AMG 102, AV-299 (SCH-900105), foretinib, GSK1363089, MetMAb, tivantinib (ARQ197), XL184 |
AMG 208, PF-04217903, PHA-665752, SGX523, SU11274 |
|
| PDGFR | Axitinib, cediranib, dasatinib (BMS-354825), E7080, imatinib, IMC-3G3, linifanib, motesanib (AMG-706), pazopanib, ramucirumab, regorafenib, sorafenib, sunitinib, vargatef (BIBF 1120), vatalanib, XL999 |
TKI-258, TSU-68 | ||
| IGF-1R | AMG 479, BIIB022, cixutumumab (IMC-A12), figitumumab (CP751,851), MK-0646, OSI906 |
BMS-754807 | ||
| FGFR | Brivanib alaninate (BMS-582664), E-7080, regorafenib, vargatef (BIBF 1120), XL999 |
FP-1039, PD-173074, TKI-258, TSU-68 |
||
| c-KIT | Axitinib, cediranib, dasatinib (BMS-354825), imatinib, motesanib (AMG-706), pazopanib, regorafenib, sorafenib, sunitinib, vatalanib |
|||
| FLT-3 | MK0457, sorafenib, sunitinib, XL999 | |||
| SRC/BCR-ABL | AZD0530, dasatinib (BMS-354825), imatinib, XL999 |
KX2-391 | ||
| DDR2 | Dasatinib (BMS-354825) | |||
| Angiogenesis | VEGF | Bevacizumab | Aflibercept (AVE005), AMG706, cediranib (AZD2171) |
|
| RAS/RAF/ MEK/ERK |
RAS | Lonafarnib, tipifarnib | ISIS 2503 (H-ras) | |
| RAF | GSK2118436, regorafenib, sorafenib | AZ628, ISIS 5132, XL281 (BMS-908662) |
||
| MEK | GSK1120212, PD325901, selumetinib (AZD6244), sorafenib |
AS 703026, AZD8330, GDC-0973, RDEA119 |
||
| PI3K/AKT/mTOR | PI3K | BKM120, GDC-0941, PX-866, XL147, XL765 |
BEZ235, BGT226, LY294002 | |
| AKT | Nelfinavir, MK-2206, perifosine | |||
| mTOR | Everolimus (RAD001), PX-866, ridaforolimus, sirolimus/rapamycin, temsirolimus (CCI-779) |
AZD8055, BEZ235, OSI-027 | ||
| Apoptosis | IAPs | HGS1029 | ||
| TRAIL | Conatumumab (AMG 655), dulanermin (AMG 951), mapatumumab |
Apomab, lexatumumab | ||
| BCL2 | Gossypol, navitoclax (ABT-263), oblimersen, obatoclax |
ABT-737 | ||
| PARP | Iniparib (SAR240550), veliparbi |
AG014699, olaparib FUS1 Fus1 liposome complex |
||
| HSPs | HSP90 | Ganetespib (STA-9090), retaspimycin (IPI-504), SNX-5422 (PF04929113), tanespimycin |
17-AAG, alvespimycin | |
| HDACs | HDACs | Belinostat, CI-994 (tacedinaline), CUDC-101, entinostat, panobinostat (LBH589), Pivanex, romidepsin, vorinostat |
SB939 | |
| Proteosome | Proteosome | Carfilzomib, bortezomib, salinosporamide A (NPI-0052) |
CEP-18770, MLN9708 | |
| Stem cell pathways |
Hh (SMO) | RO4929097, vismodegib (GDC-0449), XL139 (BMS-833923) |
Cyclopamine, IPI-926, LDE225 | |
| Notch (γ-secretase) |
MK0752, MRK-003, PF03084014 | |||
| Telomerase | Telomerase | Imetelstat (GRN-163L), KML-001 (sodium meta-arsenite) |
Sodium meta-arsenite | |
| Cell cycle/cell proliferation |
p53 | p53 peptide vaccine, PRIMA-1 | ||
| MDM2 | JNJ-26854165, RO5045337 | |||
| Aurora kinase | AZD1152, alisertib (MLN8237), MK0457, MK5108 |
|||
| CDK | Purvalanol | |||
| Inflammation | COX-2 | Celecoxib | ||
| TGF-β | Trabedersen | |||
| PPARγ | Iniparib (BSI-201), CS 7017 | Olaparib (AZD2281) | ||
| Hypoxia | HIF1 | Oncothyreon |
Detailed information on clinical trials for these agents is available in supplementary material (Supplemental Digital Content, http://links.lww.com/PPO/A4).
Although not currently U.S. FDA-approved for NSCLC, cetuximab is a recommended treatment in several practice guidelines, including those of the American Society of Clinical Oncology (ASCO) and the National Comprehensive Cancer Network (NCCN).
Previously approved in the U.S. and still approved elsewhere.
FGFR indicates FGF receptor.
Epidermal Growth Factor Receptor
Epidermal growth factor receptor (also termed HER1/ErbB1) has an extracellular ligand-binding domain, a transmembrane anchoring region, and an intracellular tyrosine kinase. The receptor subunits form homodimers and heterodimers to bind different ligands, leading to autophosphorylation of tyrosine residues on the intracellular tyrosine kinase domain.23 This action creates docking sites for numerous intracellular effector proteins activating multiple signal transduction cascades including the RAS/RAF/MEK, PI3K/AKT/mTOR, and STAT pathways (Fig. 1). These molecular signals ultimately result in cellular proliferation, resistance to apoptosis, invasion, metastasis, and angiogenesis.24–28 Independent of kinase-dependent signal transduction pathways, the EGFR complex may also be internalized and translocated to the nucleus, where it modifies gene transcription and contributes to DNA repair mechanisms.29,30
Epidermal growth factor receptor exhibits overexpression or aberrant activation in 50% to 90% of NSCLCs.31–33 Epidermal growth factor receptor protein expression has been associated with more aggressive phenotype and worse prognosis in NSCLC, bladder, breast, and head and neck cancers.33,34 Epidermal growth factor receptor is also expressed on normal epithelial cells, such as skin and gastrointestinal mucosa—a distribution that underlies the common toxicities of EGFR inhibitors.
In 2004, a significant advancement was made in the treatment of NSCLC following the observation that somatic mutations in the kinase domain of EGFR strongly correlated with sensitivity to EGFR TKIs.10,12 Exquisite sensitivity and marked tumor response have since been shown with EGFR TKIs (such as erlotinib and gefitinib) in EGFR mutant tumors10–12,35,36—an example of oncogene addiction in lung cancer where tumors initiated through EGFR mutation-activation of EGF signaling rely on continued EGF signaling for survival. Epidermal growth factor receptor mutation by either exon 19 deletion or exon 21 L858R mutation (termed “classic” EGFR mutations) each accounts for approximately 45% of EGFR mutations and shows an increased amount and duration of EGFR activation compared with wild-type receptors10 and has preferential activation of the PI3K/AKT/mTOR and STAT3/STAT5 pathways rather than the RAS/RAF/MEK pathway.36 Exon 19 mutations, most commonly in-frame deletions of amino acids 747–750, are clustered around the EGFR catalytic domain and flank the ATP-binding site. In addition to activating EGFR TK signaling, these structural changes enhance drug binding, resulting in complete blockade of mutated EGFR signaling at relatively low EGFR TKI doses.37 Higher response rates usually are seen with exon 19 mutations than with exon 21 mutations.38 Exon 21 mutations, which lie within the TK activation loop, are characteristically L858R substitutions.
Non-small cell lung cancer tumors harboring classic EGFR mutations have response rates greater than 60% with EGFR TKIs—compared with response rates of approximately 10% in wild-type EGFR cases—with progression-free survival (PFS) exceeding 1 year and overall survival (OS) exceeding 2 years.38 Whereas EGFR mutations do not appear to predict response to anti-EGFR mAbs,39,40 response rates to conventional chemotherapy in this population may be up to twice those in EGFR wild-type patients.41 Classic EGFR mutations are particularly prevalent in certain patient subgroups: female sex, East Asian ethnicity, never-smoking status, and adenocarcinoma histology.11,31,38,42–51 Approximately 10% of NSCLC cases in North America and Western Europe harbor EGFR mutations, compared with 30% to 50% of cases in East Asia.
The remaining 10% of EGFR TK mutations, in exons 18 and 20, do not confer sensitivity to EGFR TKIs and in some cases are associated with resistance. In exon 20, a threonine-to-methionine substitution at codon 790 (T790M) accounts for approximately 50% of cases of acquired resistance to EGFR TKIs. It appears that, in such cases, a small population of cancer cells harboring T790M mutations is present at diagnosis and selected for during EGFR TKI therapy. Proposed mechanisms of the therapeutic resistance caused by the T790M mutation include (1) a conformational change resulting in steric hindrance to EGFR TKI binding (analogous to the T315I mutation in CML)52 and (2) increased EGFR affinity for ATP.53 Resistance to TKI therapy has also been associated with EGFR exon 20 insertions, KRAS mutation, or amplification of the MET proto-oncogene,54–59 where MET activates the PI3K pathway through phosphorylation of ERBB3, independent of EGFR and ERBB2.59 A key finding has been the detection of a small (often <1%) subpopulation of resistant tumor cells in the primary tumor indicating the need to use EGFR-targeted therapy combined with therapy to also hit the small subpopulation of resistant tumor cells.
EGFR-INHIBITING DRUGS: CLINICALLY APPROVED FOR NSCLC
Currently, erlotinib (Tarceva) is the only EGFR-targeting agent FDA approved for the treatment of NSCLC based on results of the BR.21 phase III trial.60 Cetuximab, an anti-EGFR mAb approved for use in colorectal carcinoma and head and neck squamous cell carcinoma, is not currently FDA approved for NSCLC but is included in a number of NSCLC treatment guidelines, including those of the American Society of Clinical Oncology and the National Comprehensive Cancer Network. Epidermal growth factor receptor TKIs and mAbs exert their effects through separate mechanisms: EGFR TKIs bind competitively to the adenosine triphosphate pocket of EGFR, inhibiting EGFR phosphorylation and downstream signal transduction, whereas anti-EGFR mAbs antagonize ligand–receptor binding, thereby preventing receptor subunit dimerization, EGFR autophosphorylation, and signal transduction. In addition to erlotinib, gefitinib is another first-generation EGFR TKI that was previously approved for use in NSCLC based on phase II data, but when the phase III Iressa Survival Evaluation in Lung Cancer (ISEL) trial did not demonstrate a significant survival benefit compared with placebo,61 it was relabeled for use only in patients already benefiting from gefitinib or patients on clinical trials. Gefitinib remains approved for more general use in Asia. In Europe, it is approved for use in patients with tumors harboring activating EGFR mutations. Supplementary Table 1 summarizes selected phase III clinical trials in NSCLC using EGFR inhibitors (Supplemental Digital Content, http://links.lww.com/PPO/A4).
Molecular Biomarkers for Patient Selection
EGFR Expression
The relationship between expression of the EGFR protein and tumor sensitivity to EGFR inhibition is unclear. Numerous retrospective analyses of NSCLC patients treated with EGFR TKIs have found no relationship between tumor EGFR protein expression and objective response rate.45,62,63 However, in the BR.21 study of erlotinib and the ISEL study of gefitinib, patients with EGFR-positive tumors had significantly prolonged OS compared with EGFR-negative patients.63,64 The association between EGFR expression and outcomes in studies of anti-EGFR mAbs is also unclear, as most of these studies mandated EGFR positivity for study entry. Epidermal growth factor receptor protein expression can be assessed by immunohistochemistry (IHC), which is available in most medical centers; however, assessment is limited by reproducibility issues,65 lack of standardized scoring for EGFR IHC, and the heterogeneous expression of EGFR within tumor specimens.66,67
EGFR Copy Number
As in the case for EGFR expression, the association between the number of copies of the EGFR gene and response to EGFR inhibitors is unclear. In some clinical studies, there has been a positive correlation between increased EGFR copy number and increased response rate,62,68 time to progression,62,68 and OS.62,69 In the phase III BR.21 study of second-line erlotinib, increased EGFR copy number was associated with increased response rates but not increased survival.63 Increased EGFR copy number is highly correlated with the presence of EGFR mutations, but in Western populations, increased EGFR copy numberoccurs more frequently.62 Epidermal growth factor receptor copy number is most commonly assessed by fluorescence in situ hybridization (FISH), which is more readily standardized than IHC, although less widely available.
EGFR Mutations
Of any biomarker, EGFR mutations have the clearest association with tumor sensitivity to EGFR inhibitors, particularly TKIs. The current standard for EGFR mutational analysis is direct gene sequencing. There is ongoing debate as to whether and when tumor EGFR mutation testing should be performed. Although EGFR TKI as a first-line therapy could be extremely effective, it would require testing all patients at time of diagnosis. Furthermore, clinical data suggest that there is no difference in PFS with EGFR TKI administered as first-line—or second-line—or third-line therapy in patients with tumors harboring EGFR mutations.38 Yet patients with EGFR wild-type tumors have superior outcomes with conventional chemotherapy, rather than with EGFR TKIs in the first-line setting.41 Aided by their low toxicity, EGFR TKIs provide a survival benefit in second- and third-line therapy in unselected populations60; thus, EGFR TKIs could be included regardless of EGFR mutation status. Otherwise, selection of enriched populations (such as adenocarcinoma, female sex, and/or low smoking history) is a method to prioritize testing or select patients for therapy. However, in such populations, EGFR mutations still occur in only about 60% of patients,41 and EGFR mutations occur in up to 10% of patients lacking typical clinical predictors.70
K-ras Mutations
KRAS is mutated in approximately 20% to 30% of NSCLC and are mutually exclusive of EGFR mutations and ALK rearrangements.71,72 KRAS mutations in NSCLC are not clearly associated with resistance to the anti-EGFR mAb cetuximab,40 but they have been shown to be both prognostic (associated with worse OS) and predictive (associated with lack of benefit from adjuvant conventional chemotherapy in early-stage disease and with resistance to EGFR TKIs in advanced disease).60,68,73
Treatment Regimens and Results
Currently, insufficient evidence supports the use of EGFR inhibitors as adjuvant therapy for early-stage (stage I–III) NSCLC or during or following chemoradiation for locally advanced (stage III) NSCLC. Notably, a phase III trial in which patients with stage III NSCLC were randomized to gefitinib or placebo after completion of concurrent chemoradiation demonstrated significantly worse outcomes in the gefitinib arm (median OS, 23 vs 35 months; P = 0.01).74 This surprising result was not attributed to gefitinib-associated toxicity and remains largely unexplained.
In a clinically and histologically enriched patient population (East Asian, never-smokers, or former light smokers with adenocarcinoma NSCLC), gefitinib provides superior progression-free survival compared with carboplatin-paclitaxel as first-line monotherapy treatment of advanced (stage IV) NSCLC.41 However, even in this enriched population, for patients with tumors not harboring activating EGFR mutations, carboplatin-paclitaxel was superior to gefitinib (hazards ratio for progression or death, 0.48; 95% confidence interval, 0.36–0.64; P < 0.001). These findings have been replicated in other phase III trials of enriched populations.75–77
As a first-line treatment in combination with chemotherapy, the addition of an EGFR TKI (erlotinib or gefitinib) to platinum-based chemotherapy does not provide a survival benefit. The negative results of 4 large randomized trials (totaling >4000 patients) have been attributed to patient selection (not clinically or histologically enriched to benefit from EGFR inhibition due to EGFR mutation or amplification) and pharmacodynamic interference (EGFR TKIs induce G1 cell cycle arrest, potentially interfering with cell cycle–specific [S and G2/M phase] cytotoxicity of some chemotherapy drugs).45,78 In contrast, in the phase III FLEX (First-line in Lung cancer with ErbituX) trial, the addition of the anti-EGFR mAb cetuximab to cisplatin-vinorelbine chemotherapy provided a modest but statistically significant OS benefit as first-line therapy for patients with EGFR-positive NSCLC.
Erlotinib is approved for maintenance treatment of patients with advanced NSCLC whose disease has not progressed after 4 cycles of platinum-based first-line chemotherapy. Similar to second-line treatment, maintenance treatment is administered after completion of first-line chemotherapy. In a phase III clinical trial, 889 patients (with approximately 70% of patients’ tumors EGFR positive by IHC) whose disease did not progress during first-line platinum-based chemotherapy were randomized to erlotinib or placebo. Erlotinib-treated patients had a significant increase in PFS and OS.79
Epidermal growth factor receptor TKIs also show benefit as second-line therapy where erlotinib prolongs survival in patients with previously treated advanced NSCLC. In the phase III National Cancer Institute Canada BR.21 (“BR” designates “bronchus”) trial, patients randomized to erlotinib had a median OS of 6.7 months, compared with 4.7 months for placebo (P < 0.001). In contrast, in the phase III ISEL trial, gefitinib did not provide a significant survival advantage over placebo (median, 5.6 vs. 5.1 months; P = 0.09).
EGFR-INHIBITING AGENTS: NOVEL EGFR mABS
Supplementary Table 1 summarizes selected phase III clinical trials in NSCLC using EGFR inhibitors (Supplemental Digital Content, http://links.lww.com/PPO/A4). Necitumumab (IMC-11F8) is a fully human IgG1 mAb directed against EGFR currently in 2 phase III clinical trials.80 The INSPIRE trial is evaluating necitumumab in combination with pemetrexed-cisplatin as first-line therapy in advanced NSCLC. Enrollment was stopped early in 2011 because of concerns of thromboembolism, and the trial will continue with existing patients. A second phase III trial, SQUIRE, is testing the efficacy of necitumumab in combination with gemcitabine and cisplatin as first-line therapy in stage IV squamous NSCLC. No safety concerns have been reported, and enrolment continues.
MET, HER2, AND EGFR TKI RESISTANCE
Although EGFR mutant tumors are initially exquisitely sensitive to EGFR TKIs, disease progression eventually develops in most patients within a year,81 usually because of secondary mutations that arise.81,82 Resistance-associated mutations include a T790M mutation in the EGFR TK domain (which has been reported in approximately 50%–60% of EGFR TKI resistant tumors),83 EGFR exon 20 insertions, KRAS mutation, or amplification of the MET proto-oncogene.54–59 In addition, recently, a subgroup of EGFR mutant, EGFR TKI resistant tumors has been identified that have developed features of small cell lung carcinoma (while retaining the EGFR mutation).84 Interestingly, these tumors then respond to SCLC-like chemotherapy. Subsequently when they are resistant to this chemotherapy they revert to an adenocarcinoma phenotype. This transition strongly suggests resistance developing by some epigenetic mechanism. Therapeutic resistance caused by the T790M mutation could be due to a conformational change resulting in steric hindrance to EGFR TKI binding52 and/or increased EGFR affinity for ATP.53 Amplification of MET is thought to provide a “bypass” mechanism whereby signaling continues independently of EGFR activation.
Similar to EGFR, MET is a receptor tyrosine kinase capable of driving RAS/RAF/MEK and PI3K/AKT/mTOR pathway signaling.85 MET is activated upon binding of the hepatocyte growth factor. Independent of therapeutic resistance incurred through the EGFR T790M mutation, amplification of the MET proto-oncogene is also thought to mediate resistance to EGFR TKIs,54–59 where MET activates the PI3K/AKT/mTOR pathway through phosphorylation of HER3, independent of EGFR and HER2.59 Inhibition of MET is being approached with mAbs (such as MetMAb) and small-molecule MET inhibitors (tivantinib/ARQ-197). Phase II clinical trials in EGFRTKI–naive advanced NSCLCs found a trend for better PFS with erlotinib and tivantinib compared with erlotinib and placebo, particularly in patients with KRAS mutant tumors.86 Phase III trials comparing erlotinib and tivantinib with erlotinib and placebo in non-squamous and EGFR wild-type advanced-stage lung cancer are currently underway.80 A phase II trial of advanced NSCLC comparing MetMAb, a monovalent mAb that specifically binds the Met receptor, plus erlotinib to placebo plus erlotinib as second- or third-line therapy found the combination of MetMAb and erlotinib had significant benefit to PFS and OS in tumors with high MET IHC levels but conferred a worse OS in tumors with no MET amplification.87
Second-generation EGFR TKIs that bind irreversibly to EGFR tyrosine kinase are novel compounds that induce much less therapeutic resistance and appear to be effective against secondary resistance mutations such as T790M as well.88,89 Irreversible EGFR TKIs currently being evaluated in clinical trials, such as PF00299804 (PF-299), afatinib (BIBW 2992), and neratinib (HKI-272), have affinity for both EGFR and HER2, a receptor tyrosine kinase that can activate lung cancer signaling pathways such as RAS/RAF/MEK and PI3K/AKT/mTOR.85
The ligand for HER2 remains unknown, but it is activated following homodimerization or heterodimerization (with EGFR or HER3 preferentially).90 Although HER2 amplification or overexpression confers sensitivity to anti-HER2 mAbs (trastuzumab) or HER2 TKIs (lapatinib) in breast and gastric cancer, it does not in NSCLC.91,92 However, exon 20 mutations in HER2 (occurring in 3%–10% of lung adenocarcinomas) do confer sensitivity to lapatinib in NSCLC cell lines.93,94 HER2 mutations also confer resistance to EGFR TKIs, regardless of EGFR mutation status as HER2 replaces EGFR in driving growth signals. In addition to trastuzumab and lapatinib, other mAbs and TKIs in clinical trials include MGAH22 and PF00299804 (PF-299), afatinib (BIBW 2992), and neratinib (HKI-272). The LUX-LUNG 1 phase III study evaluated the benefit of afatinib with best supportive care in advanced NSCLC and found a clinical benefit in PFS.95 Two other phase III studies evaluating afatinib are underway.80 Following positive results from ongoing phase II studies showing a benefit of PF00299804, a pan-HER TKI, in advanced NSCLC patients,96,97 phase III studies of PF00299804 in advanced NSCLC are also underway comparing with either erlotinib (ARCHER 1009) or with placebo (NCT01000025).80 A phase III study evaluating neratinib, however, found low activity in all patients, with only 2% responding but a dramatic response (response rate = 75%) in a rare subset of patients with EGFR G719X tumors.98 Supplementary Table 1 summarizes selected phase III clinical trials in NSCLC using EGFR inhibitors (Supplemental Digital Content, http://links.lww.com/PPO/A4).
Anaplastic Lymphoma Kinase
In 2007, a novel fusion gene with transforming ability was reported in a small subset of NSCLC patients.99 Formed by the inversion of 2 closely located genes on chromosome 2p, fusion of protein tyrosine kinase echinoderm microtubule-associated protein–like 4 (EML4) with anaplastic lymphoma kinase (ALK), a transmembrane tyrosine kinase, yields the EML4-ALK fusion protein. The fusion results in constitutive oligomerization leading to persistent mitogenic signaling and malignant transformation, and a recent meta-analysis of 13 studies encompassing 2835 tumors reported the EML4-ALK fusion protein is present in 4% of NSCLCs.100 Despite the rarity of these aberrations in NSCLC, the vast number of lung cancer cases worldwide results in an estimated 40,000 such cases occurring annually.101 EML4-ALK fusions are, in nearly every case, found exclusive of EGFR and KRAS mutations and occur predominantly in adenocarcinomas, never or light smokers, younger age, and male sex.102,103 In contrast to EGFR mutations, ALK translocations do not appear to have a clear association with race/ethnicity. Anaplastic lymphoma kinase–positive patients have a significantly longer PFS with pemetrexed, whereas EGFR or KRAS mutant patients do not.104 Tumors with EML4-ALK fusions exhibit dramatic clinical responses to ALK-targeted therapy,105–109 and the ALK and MET inhibitor crizotinib (PF-02341066) is now approved for use for lung cancer treatment in patients harboring the fusion protein.
ALK-INHIBITING DRUGS: CLINICALLY APPROVED FOR NSCLC
Crizotinib (Xalkori) is an orally bioavailable adenosine triphosphate–competitive inhibitor of the ALK tyrosine kinase. Crizotinib is the only ALK inhibitor currently FDA approved, and its use is restricted to patients with advanced NSCLC harboring EML4-ALK fusions. Crizotinib also inhibits the MET tyrosine kinase, although it is not clear to what extent this mechanism contributes to therapeutic effect in ALK-positive NSCLC. Supplementary Table 2 summarizes selected phases I–III clinical trials of ALK inhibitors in NSCLC (Supplemental Digital Content, http://links.lww.com/PPO/A5).
Molecular Biomarkers for Patient Selection
The clinical development of ALK inhibitors is unique among targeted therapies for lung cancer in that all studies have been biomarker based, where only patients with tumors harboring EML4-ALK fusions have been enrolled. Anaplastic lymphoma kinase rearrangements can be identified by IHC, FISH, or polymerase chain reaction, where FISH is the most clinically applicable; however, dual IHC and FISH may increase sensitivity.103 Whether all NSCLC patients should be tested or a subset preferentially enriched for ALK fusions (e.g., adenocarcinomas) remains to be determined for cost and efficacy. The terms “ALK positivity,” “ALK rearrangement,” “ALK fusion,” and “ALK translocation” are generally synonymous and refer to the presence of the EML4-ALK translocation.
Treatment Regimens and Results
Crizotinib has not yet been studied as adjuvant (postoperative) therapy for early-stage (stage I-III) NSCLC or as therapy for locally advanced (stage III) disease. In first-line treatment for advanced (stage IV) NSCLC, results are awaiting for a randomized phase III trial (PROFILE 1014) comparing crizotinib with cisplatin-pemetrexed or carboplatin-pemetrexed in patients with previously untreated stage IV ALK-positive NSCLC. In second-line and beyond treatment for advanced disease, crizotinib showed efficacy in an expanded cohort of ALK-positive NSCLC from a multicenter phase I study.110 Tumor specimens from approximately 1500 patients with advanced NSCLC were screened for ALK translocations. A total of 82 patients, most of whom were treated previously with chemotherapy, were enrolled. The radiographic objective response rate was 57%, with stable disease 33%, yielding a clinical benefit rate of 90%. The estimated 6-month PFS rate was 72%. PROFILE 1007 is a randomized phase III clinical trial of crizotinib versus pemetrexed or docetaxel chemotherapy for patients who had progressed after platinum-based chemotherapy, whereas PROFILE 1005 is a phase II single-arm study that provides crizotinib to patients on PROFILE 1007 randomized to chemotherapy with disease progression or to patients not eligible for PROFILE 1007.
ALK-INHIBITING AGENTS: NOVEL AGENTS
Nearly all ongoing clinical trials aimed at inhibiting ALK activity in NSCLC use crizotinib, but despite remarkable initial responses, cancers eventually develop resistance to crizotinib, usually within a year,111,112 thereby limiting the potential clinical benefit. Secondary resistance mutations in the TK domain of ALK were identified in patients who had a disease progression on crizotinib. Thus, as with second-generation development of irreversible EGFR TKIs against EGFR T790M resistance mutations, efforts are being made to identify agents effective against crizotinib-resistant EML4-ALK–positive cancers. Two ALK inhibitors structurally different from crizotinib, NVP-TAE684 and AP26113, as well as the heat shock protein 90 (HSP90) inhibitors 17-AAG and ISI-504 have shown potency in vitro where crizotinib does not.112–114 A phase II trial is ongoing to evaluate efficacy of ISI-504 a phase II trial in ALK inhibitor– naive versus pretreated NSCLCs.80
VEGF and VEGFR
Angiogenesis, the growth of new blood vessels from pre-existing vasculature, is essential for tumor development as tumor cells require an ongoing source of oxygen and nutrients for proliferation,115 thus rendering angiogenesis a rational target for cancer therapy. The VEGF–VEGFR axis regulates endothelial cell survival, mitogenesis, migration, mobilization of endothelial progenitor cells from bone marrow, and vascular permeability.
Whereas VEGF is the principal mediator of angiogenesis, other angiogenic proteins include platelet-derived growth factor (PDGF), fibroblast growth factor (FGF), interleukin 8, and angiopoietins 1 and 2. Vascular endothelial growth factor stimulates proliferation and migration, inhibits apoptosis, promotes survival, and regulates endothelial cell permeability.116 VEGF signaling is stimulated by tumor hypoxia, growth factors and cytokines, and oncogenic activation.117 The VEGF-related gene family of growth factors includes VEGF-A, -B, -C, -D, and -E and placenta growth factors 1 and 2. Vascular endothelial growth factor A, commonly referred to as VEGF, binds to 3 receptors: VEGFR1, VEGFR2, and VEGFR3. Vascular endothelial growth factor receptors are receptor tyrosine kinases. Similar to EGFR, they convey proliferation signals via intracellular mediators. Vascular endothelial growth factor is highly expressed in both NSCLC and SCLC,118 and its expression is associated with poor prognosis in NSCLC71,119,120; therefore, inhibition of VEGF signaling in tumor cells is an important therapeutic target. One caveat to this therapy is VEGF signaling is also present in the stroma as well as the tumor cells. Supplementary Table 3 summarizes selected phase III clinical trials in NSCLC using antiangiogenic inhibitors (Supplemental Digital Content, http://links.lww.com/PPO/A6).
VEGF-INHIBITING DRUGS: CLINICALLY APPROVED FOR NSCLC
Currently, only 1 VEGF inhibitor is FDA approved for NSCLC, the mAb bevacizumab (Avastin). Bevacizumab blocks the binding of all VEGF-A isoforms to VEGFRs, inhibiting the biologic activities of VEGF. Because angiogenesis is relevant to the development and growth of multiple tumor types, bevacizumab is approved for multiple indications, including NSCLC (nonsquamous histology), colorectal cancer, breast cancer, glioblastoma multiforme, and renal cell carcinoma.
Molecular and Clinical Biomarkers for Patient Selection
In NSCLC, no biomarkers consistently predict response to antiangiogenic agents. Tumor and plasma VEGF levels are prognostic, but not reliably predictive.72 Vascular endothelial growth factor and VEGFR polymorphisms, which have been characterized in breast cancer patients treated with bevacizumab,121 are under study in NSCLC. Other candidate pharmacodynamic markers include multiple plasma cytokines and angiogenic factors, among them intracellular adhesion molecule 1, E selectin, soluble VEGFRs, matrix metalloproteinases, and interleukins.
Analogous to EGFR inhibitor–associated rash, treatment-associated hypertension has emerged as a possible predictor of benefit from bevacizumab. In Eastern Cooperative Oncology Group 4599, patients randomized to receive bevacizumab who developed hypertension had a median OS of 14.0 months, compared with 11.3 months for patients without hypertension. However, it should be noted that only 8% of bevacizumab-treated patients in this study developed hypertension and that this comparison did not reach statistical significance.122
Treatment Regimens and Results
Bevacizumab has shown efficacy in first-line treatment of advanced (stage IV) NSCLC. The addition of bevacizumab to carboplatin-paclitaxel for patients with advanced nonsquamous NSCLC significantly prolongs survival. In the phase III Eastern Cooperative Oncology Group 4599 clinical trial, patients receiving carboplatin-paclitaxel plus bevacizumab had a median survival of 12.3 months, compared with 10.3 months in the group receiving carboplatin-paclitaxel alone (P = 0.003). However, the addition of bevacizumab to cisplatin-gemcitabine does not provide a survival benefit. In the phase III AVAiL (AVAstin in Lung) trial, patients were randomized to cisplatin-gemcitabine alone, cisplatin-gemcitabine plus bevacizumab 7.5 mg/kg, or cisplatin-gemcitabine plus bevacizumab 15 mg/kg. Median OS in the 3 groups was, respectively, 13.1, 13.6 (P = 0.42 compared with chemotherapy alone), and 13.4 (P = 0.76 compared with chemotherapy alone) months.
Currently, insufficient evidence supports the use of bevacizumab as a component of adjuvant therapy for early-stage (stage I-III) NSCLC, and combining bevacizumab with concurrent chemoradiation for inoperable stage III NSCLC is not recommended because it is associated with increased incidence of tracheoesophageal fistulae.123
In addition to inhibiting the growth of new blood vessels in tumors, disruption of the VEGF pathway exerts several effects on normal vasculature. Thus, bevacizumab toxicities include thromboembolic events, bleeding, wound healing complications, hypertension, and proteinuria.124–127 In patients with lung cancer, specifically with squamous cell histology, a particular concern is hemoptysis, and squamous cell tumor histology remains an absolute contraindication to bevacizumab.
VEGF-INHIBITING AGENTS: NOVEL AGENTS
The anti-VEGF mAb bevacizumab is the only antiangiogenic drug currently approved for lung cancer treatment. Several antiangiogenic agents have been FDA-approved for the treatment of various cancers, including sunitinib (renal cell carcinoma [RCC], hepatocellular carcinoma, pancreatic neuroendocrine tumors), sorafenib (RCC, hepatocellular carcinoma), vandetanib (medullary thyroid cancer), and pazopanib (RCC). However, these drugs have not demonstrated survival benefits in NSCLC. Selected clinical trials of these inhibitors as well as additional novel agents in NSCLC are summarized in Supplementary Table 3 (Supplemental Digital Content, http://links.lww.com/PPO/A6).
VEGFR TKIs
Sunitinib and sorafenib are TKIs targeting multiple kinases, including VEGFR, PDGF receptor (PDGFR), cKIT, and FLT-3.128 Phase II trials of single-agent therapy with sunitinib in NSCLC reported activity129; however, toxicity issues were demonstrated in trials combining sunitinib with chemotherapies (SABRE-L).130,131 A phase II trial testing the benefit of sunitinib and pemetrexed alone or in combination in advanced pretreated NSCLCs as well as a phase III/IV trial evaluating sunitinib for maintenance therapy as a single agent is ongoing.80 Dual inhibition of EGFR and VEGF signaling is being evaluated in phase II (SUN 1058) and phase III (SUN 1087) trials studying the benefit of erlotinib alone or in combination with sunitinib in advanced NSCLC.80 Both trials have finished recruiting and await results.
Following promising results in a phase II study that found benefit with sorafenib therapy in patients with prior therapies,132 sorafenib was evaluated in phase III trials as a first-line therapy in combination with chemotherapies (ESCAPE128 and NExUS). The ESCAPE study found no improvement in OS with the addition of sorafenib to carboplatin-paclitaxel.128 The NExUS study comparing cisplatin-gemcitabine with and without sorafenib has completed enrollment and awaits results. Recent results from the phase II BATTLE trial in pretreated lung cancers found benefit for sorafenib in KRAS mutant tumors,79 illustrating the need for biomarkers to identify patient populations that will most benefit from targeted therapies.
Vandetanib (ZD6474) is a dual VEGFR2 and EGFR TKI, albeit with only moderate EGFR TKI activity. Phase II data found PFS benefit of vandetanib to docetaxel as second-line therapy in patients who had failed platinum-based chemotherapy.133 A phase III study (ZEAL) evaluating the benefit of adding vandetanib with pemetrexed found no OS or PFS benefit but improved objective RR and time to deterioration of symptoms with vandetanib.134 However, another phase III trial comparing erlotinib alone or with combination with vandetanib found no difference in PFS or OS in an unselected population of advanced NSCLC with treatment failure following at least 1 chemotherapy.135 Pazopanib, a multikinase TKI approved for use in RCC, targets VEGFR1/2/3, PDGFR>/A, and cKIT and is currently undergoing phase III trials as second-line monotherapy.
Two VEGFR TKIs undergoing clinical trials in lung cancer that have not been approved for use in any cancer are vargatef (BIBF 1120) and motesanib (AMG-706). Vargatef targets VEGFRs, PDGFRs, and FGF receptors and is currently in 2 phase III trials evaluating its efficacy in second-line therapy combined with either docetaxel or pemetrexed.80 Motesanib inhibits all known VEGFRs, PDGFR, and c-KIT and is currently being evaluated in a phase III trial in combination with paclitaxel and carboplatin for advanced NSCLC.
VEGF mAbs
Aflibercept is a fusion protein with high affinity for VEGF. A phase II clinical trial found aflibercept was well tolerated in nonsquamous, advanced population,136 which led to an ongoing phase III trial comparing the efficacy of aflibercept with docetaxel in advanced NSCLC.80
RAS/RAF/MEK Pathway
Activation of the RAS/RAF/MEK pathway occurs frequently in lung cancer, most commonly via activating mutations in KRAS (predominantly in codon 12), which occur in ~20% of lung cancers, particularly adenocarcinomas.137,138 Mutation results in constitutive activation of downstream signaling pathways such as PI3K/AKT/mTOR and MAPK, rendering KRAS mutant tumors independent of EGFR signaling and therefore resistant to EGFR TKIs as well as chemotherapy.35,56,139 KRAS mutations are mutually exclusive with EGFR and HER2 mutations and are primarily observed in lung adenocarcinomas of smokers.35,140 The prevalence and importance of KRAS in lung tumorigenesis make it an attractive therapeutic target; however, it remains “undruggable,” with 2 unsuccessful approaches being farnesyltransferase inhibitors (to inhibit posttranslational processing and membrane localization of RAS proteins) and antisense oligonucleotides against RAS.137 Recent efforts have focused on downstream effectors of RAS signaling: RAF kinase and mitogen-activated protein kinase (MAPK) kinase (MEK).137,141 BRAF is the direct effector of RAS, and although commonly mutated in melanoma (~70%), mutations are rare in lung cancer (~3%), predominantly in adenocarcinoma, and mutually exclusive to EGFR and KRAS mutations.142–145 Small-molecule kinase inhibitors have been designed to inhibit RAF kinase activity such as the multikinase inhibitor sorafenib (which inhibits VEGFR, PDGFR, FLT-3, RAF, MEK, and KIT) as well as some BRAF mutant-specific inhibitors such as vemurafenib (PLX-4032), PLX-4720, and GDC-0879146 Sorafenib was approved by the FDA in 2005 for the treatment of RCC. It is a relatively weak RAF inhibitor but showed efficacy in KRAS mutant NSCLC in the phase II BATTLE trial.79
Several potent and selective MEK inhibitors such as selumetinib (AZD6244) and GSK1120212 show potential in inhibiting RAS/RAF/MEK signaling. In an unselected population of advanced NSCLCs, a phase II trial comparing selumetinib with pemetrexed therapy found no benefit with selumetinib therapy,147 but 2 more phase II trials are either underway or completed including the BATTLE-2 trial comparing erlotinib, the AKT inhibitor MK-2206, selumetinib, and sorafenib. Evaluation of GSK1120212 is still in early phase I trials.
Supplementary Table 4 summarizes selected phases II and III clinical trials in NSCLC using RAF and MEK inhibitors (Supplemental Digital Content, http://links.lww.com/PPO/A7).
Insulin Growth Factor Pathway Inhibition
The insulin growth factor (IGF) pathway mediates the growth and differentiation of bone and skeletal muscle. It comprises 2 receptors, insulin receptor (IR) and insulin-like growth factor 1 receptor (IGF-1R), and 3 principal ligands, IGF-1, IGF-2, and insulin.148 Insulin-like growth factor 1 receptor is a receptor tyrosine kinase that forms homodimers and heterodimers with IR and HER2. Like HER2, IGF-1R does not appear to be mutated in cancers. Activation upon ligand binding results in up-regulation of various signaling pathways including the PI3K/AKT/mTOR and RAS/RAK/MEK pathways. Dysregulation of IGF signaling in lung cancer is evidenced by frequent (up to 70%) overexpression of IGF-1R in NSCLC,149,150 where increased signaling results in tumor growth and drug resistance.151 Furthermore, increased levels of IGF-1 are associated with increased risk of lung cancer.152,153
Several mAbs and small molecules have commenced efficacy testing in lung cancer (Supplementary Table 4, http://links.lww.com/PPO/A7), although progress in IGF-R1– targeted TKIs has been limited by the high degree of TK domain homology between IGF-1R and IR.154 Following promising phase II results of figitumumab (CP-751,871), a mAb against IGF-1R, in nonadenocarcinoma histology,155 a randomized phase III trial (ADVIGO 1018) commenced comparing the efficacy of figitumumab plus erlotinib versus erlotinib alone in refractory, advanced NSCLC but was terminated due to lack of effect in the primary endpoint of OS. Another phase III trial (ADVIGO 1016) evaluating the efficacy of combining paclitaxel, carboplatin, and figitumumab as first-line therapy in advanced NSCLC was also terminated because of lack of efficacy as well as adverse effects.156 Other IGF-1R–targeted mAbs such as cixutumumab and AMG479 as well as IGF-1R TKIs such as OSI906 have commenced preliminary phases I and II evaluation, but the lack of success found with figitumumab suggests that further progress of IGF-1R–targeted therapy will require identification of biomarkers and clinicopathologic factors—such as squamous cell histology—to select appropriate patient groups.157
PI3K/AKT/mTOR Pathway Inhibition
The PI3K/AKT/mTOR pathway is a downstream signaling pathway of several receptor tyrosine kinases, such as EGFR. Downstream targets of AKT (a serine/threonine kinase downstream from PI3K) are involved in cell growth, angiogenesis, cell metabolism, protein synthesis, and suppression of apoptosis directly or via the activation of mTOR. Activation can occur through the binding of the SH2 domains of p85, the regulatory subunit of PI3K, to phosphotyrosine residues of activated RTKs such as EGFR or via binding of PI3K to activated RAS.158 The pathway has 2 negative regulators: the tumor suppressor gene, PTEN, and TSC1/TUSC2 complex, which act upstream and downstream of AKT, respectively. In lung tumorigenesis, activation of the PI3K/AKT/mTOR pathway occurs early in pathogenesis, generally through mutations in PI3K; AKT; or PTEN as well as EGFR or KRAS; amplification of PIK3CA, which encodes the catalytic subunit of PI3K; PTEN loss; or activation of AKT and results in cell survival through inhibition of apoptosis.13,159–164
Several novel PI3K (BKM120 and GDC-0941) and AKT (MK-2206) inhibitors are currently being evaluated in phase II trials in patients with advanced NSCLC after promising phase I results (Supplementary Table 4, http://links.lww.com/PPO/A7). The results from 2 BKM120 trials will be of particular interest as patient selection includes molecular biomarker analysis, requiring either activated PI3K pathway (being tested in combination with chemotherapy) or tumors with mutations in KRAS, NRAS, and/or BRAF.
Rapamycin and its derivatives (everolimus, temsirolimus) block mTOR functions and yield antiproliferative activity in a variety of malignancies. Everolimus (RAD001) has demonstrated tolerability in advanced-stage NSCLC patients in a phase II study with some clinical activity.165 An analysis of molecular biomarkers was attempted in the patient cohort, but limited sample precluded any conclusions. Ongoing phase II trials are testing the efficacy of everolimus in combination with chemotherapies.80 The mTOR inhibitor temsirolimus is clinically approved for use in renal cell carcinoma. Studies in lung cancer are ongoing. In an attempt to enhance the effectiveness of temsirolimus and bypass resistance to mTOR inhibition, a phase I trial dually inhibited mTOR and IGF-1R with temsirolimus and cixutumumab in solid tumors.166 Temsirolimus combined with cixutumumab was well tolerated, but tumor reduction was observed only in Ewing sarcoma and adrenocortical carcinoma. As a single agent, temsirolimus was recently evaluated in the phase II setting in advanced NSCLC, and results are expected soon.80 The studies of mTOR inhibitors have all been conducted in unselected patient populations. Molecular characterization of PI3K/ AKT/mTOR pathway biomarkers is expected to allow a better selection of tumors responsive to mTOR, as well as AKT and PI3K inhibition. Loss of PTEN functions, for example, results in AKT activation and cell growth and proliferation, and targeted therapies to the PI3K/AKT/mTOR pathway (such as LY294002 and rapamycin) have shown significant efficacy in vitro in both NSCLC and SCLC cells with activated AKT signaling.167–169
Tumor Necrosis Factor–Related Apoptosis-Inducing Ligand Therapy
Tumor necrosis factor–related apoptosis-inducing ligand (TRAIL) is a membrane-bound protein constitutively expressed on macrophages, T cells, natural killer cells, and dendritic cells that selectively kills tumor cells while leaving most normal cells unharmed.170 It initiates apoptosis by binding to cell surface TRAIL death receptors (TRAIL-R1/R2), which trimerize for receptor activation leading to intracellular recruitment of FADD (Fas-associated death domain) and its sequestered procaspase 8 to form the death-inducing signaling complex. Both the extrinsic (direct) and intrinsic (indirect) apoptotic pathways can then be activated through the cleavage of procaspase 8.
Therapeutic targeting of TRAIL receptors is an attractive target, particularly because TRAIL-induced apoptosis is independent of p53, which is often inactivated in lung cancer cells. However, many lung cancer cell lines demonstrate intrinsic resistance to TRAIL-induced apoptosis, thought to be conferred, in part, through mutations in TRAIL death receptors and the death-inducing signaling complex, rendering them resistant to TRAIL therapy. Currently, TRAIL targeting molecules comprise either recombinant human TRAIL or agnostic mAbs targeted against either TRAIL-R1 and/or TRAIL-R2 (Supplementary Table 4, http://links.lww.com/PPO/A7). A phase IB trial using the recombinant human TRAIL dulanermin in combination with paclitaxel and carboplatin with and without bevacizumab showed potential as first-line therapy in advanced NSCLCs171 and is currently being evaluated in a phase II setting.80 Mapatumumab and conatumumab are 2 agonist mAbs being tested in clinical trials that target TRAIL-R1 and TRAIL-R2, respectively. As a single agent, mapatumumab was well tolerated in NSCLC patients with refractory disease,172 but combination with carboplatin-paclitaxel as first-line therapy showed little effect.173 A similar study using conatumumab with carboplatin-paclitaxel has been completed and is awaiting results80 following promising monotherapy results.174
Histone Deacetylase Inhibition
Epigenetic events can lead to changes in gene expression without any changes in DNA sequence and therefore, importantly, are potentially reversible.175 Histone modification is a mechanism for epigenetic control of gene transcription where histone deacetylation results in condensing of chromatin, resulting in transcriptionally inactive DNA. Inhibitors of histone deacetylases (HDACs) resulting in pharmacologic restoration of expression of epigenetically silenced genes—such as TSGs—are an exciting targeted therapeutic approach and show promise in lung cancer176,177 (Supplementary Table 4, http://links.lww.com/PPO/A7). A phase II/III trial found a significantly improved RR with the addition of vorinostat, an HDAC inhibitor, to carboplatin-paclitaxel for the treatment of advanced NSCLC; however, the study was terminated because the primary endpoint of OS was not reached.178 Similar studies plus a phase III trial testing the efficacy of another HDAC inhibitor, CI-994, in combination with gemcitabine as a second-line therapy are ongoing.80
Heat Shock Protein Inhibition
Heat shock proteins are molecular chaperones involved in posttranslational folding, stability, activation, and maturation of many proteins essential to signal transduction and cell cycle progression. They also chaperone oncogenic proteins, and inhibition of HSP90, the best studied HSP proteins, leads to the degradation of known oncogenes such as HER2, BRAF, and BCR-ABL, resulting in the inhibition of multiple oncogenic transduction pathways.
Geldanamycin is a natural compound HSP90 inhibitor from which several 17–amino acid derivatives have been developed including 17-AAG, SNX-5422, retaspimycin, and ganetespib (Supplementary Table 4, http://links.lww.com/PPO/A7).179 A phase I trial of SNX-5422 in refractory tumors was well tolerated,180 but development has been halted because of ocular toxicity observed in mouse models and another phase I study.181 A phase II study of retaspimycin (IPI-504) in advanced NSCLC with prior EGFR TKI therapy and molecular profiling found that patients with ALK rearrangements had improved response compared with patients with EGFR wild-type or mutant tumors.182 Consequently, a number of other phase II studies are underway in patients with tumors harboring KRAS mutations or ALK rearrangements80 to better define the benefit of molecular selection of patients.
Telomerase Inhibition
Activation of telomerase, the telomere-lengthening enzyme, in premalignant cells prevents loss of telomere ends beyond critical points and is essential for cell immortality. Although silenced in normal cells, telomerase is activated in greater than 80% of NSCLCs and almost uniformly in SCLCs.183–185 Inhibition of telomerase in cancer cells leads to telomere shortening and ultimately either cellular senescence or apoptosis.186,187 Targeted approaches to telomerase inhibition include using antisense oligonucleotides that bind to human telomerase RNA187 (such as imetelstat, which has started Phase II trials) (Supplementary Table 4, http://links.lww.com/PPO/A7).80
Targeting Lung Cancer Stem Cells
The cancer stem cell (CSC) model hypothesizes that there is a population of rare, stem-like tumor cells capable of self-renewing and undergoing asymmetric division, thereby giving rise to differentiated progeny that comprises the bulk of the tumor.188–190 Although the first evidence for CSCs (also termed tumor-initiating cells) was reported in acute myeloid leukemia,191 support for their existence in solid tumors, including lung cancer, is increasing.105,107,192–196 Regulation of CSCs in lung cancer is likely by the hedgehog (Hh), Wnt, and Notch stem cell signaling pathways as persistent dysregulation of these pathways is found in both SCLC and NSCLC.197–210
Cancer stem cells are thought to have higher resistance to cytotoxic therapies and radiotherapy than the bulk of differentiated tumor cells. Thus, whereas conventional treatment strategies may initially “debulk” the primary tumor through elimination of differentiated tumor cells, the small population of CSCs eventually regenerates the tumor, giving rise to recurrence. Approaches to specifically treating the CSC population include selective targeting using CSC detection molecules; sensitization of CSCs to conventional therapies and differentiation therapies; inhibition of signaling pathways important to CSCs such as Hh, Wnt, and Notch signaling pathways; and inhibition of telomerase. Inhibition of the Hh pathway has been demonstrated with cyclopamine, a naturally occurring inhibitor of SMO, which has led to the development of synthetic oral inhibitors that show clinical activity in basal cell carcinoma.211 Inhibition of the Notch signaling pathway shows potential with γ-secretase inhibitors. Several inhibitors have shown efficacy in NSCLC,212,213 and 2 phase II trials using the γ-secretase inhibitor RO4929097 as second-line therapy in advanced NSCLC with prior chemotherapy have commenced.80
MOLECULAR TARGETING OF SCLC
No targeted therapies have been presently approved for the treatment of SCLC. A number of agents have either been tested or are ongoing in clinical trials (Supplementary Table 5, http://links.lww.com/PPO/A8), although the limited success of the former suggests we need better understanding of the molecular biology of SCLC to identify better therapeutic targets. As with other cancers, SCLS exhibits dysregulation of signaling pathways regulating various cellular processes such as cell cycle, cell proliferation, apoptosis, and angiogenesis, but agents targeting these pathways have little activity. Clinical studies of targeted therapy in SCLC have been reviewed recently.18,19 Limited access to tumor samples has hindered understanding of the SCLC genome, but recent whole-genome sequencing efforts214,215 will identify somatic “driver” mutations that provide good targets for therapeutic development. This will allow a more systemic approach to clinical trials that is more likely to generate improved results.
CONCLUSIONS
Immense effort in basic research now affords clinicians a considerable selection of targeted agents for therapy. However, one overall message that has emerged from clinical trials of targeted therapies in NSCLC to date is that the heterogeneous nature of NSCLC significantly limits the ability to detect therapeutic benefit of a specific agent in unselected patient populations. Thus, the current dilemma in clinical trial design is how to select patient populations that will derive the most benefit. This requires identifying patients who exhibit dramatic results from these agents and subsequently identifying clinicopathologic factors or molecular biomarkers associated with response. Two multi-institutional examples of coordinated efforts to accelerate our understanding of the lung cancer genome are The Cancer Genome Atlas216 and the Lung Cancer Mutation Consortium (LCMC).217 The LCMC has completed sequencing of tumor DNA for mutations in common and potentially “actionable” oncogene using CLIA-certified methods. The LCMC then couples the patients with particular mutations to clinical trials with agent(s) targeting that mutant oncoprotein or activated pathway.
Both in vitro studies and clinical trials of targeted agents have also shown that inhibition of a single target or pathway is an ineffective approach to cancer therapy. Experience with EGFR and ALK TKIs has shown that resistance mechanisms will continue to emerge in tumor cells. Combination targeted therapies, such as erlotinib and bevacizumab, are now being evaluated in clinical trials that are directed at multiple targets, pathways, or cellular processes.
Finally, although traditional chemotherapy approaches to lung cancer treatment may not provide any further significant advancement to disease response and OS in lung cancer, it is apparent that their efficacy can be marked and diverse when combined with knowledge of molecular biomarkers or use with targeted therapies. Elucidating these patient subsets will guide clinicians in the context of personalized medicine.
Supplementary Material
Acknowledgments
Sources of Funding: This research was supported by the National Cancer Institute Lung Cancer Specialized Program of Research Excellence (P50CA70907), Department of Defense PROSPECT (W81XWH0710306), Lung Cancer Mutation Consortium RC2 CA148394-01, TSANZ Allen & Hanburys Respiratory Fellowship. Dr Heymach has received research funding from AstraZeneca and GlaxoSmithKline, and Dr Minna has received research funding from AstraZeneca and Geron. Dr Heymach has consulted or advised AstraZeneca, Boehringer Ingelheim, Genentech, and GlaxoSmithKline, and Dr Minna has consulted or advised Amgen and Genentech.
Footnotes
Conflicts of Interest For the remaining authors none were declared.
Supplemental digital contents are available for this article. Direct URL citations appear in the printed text and are provided in the HTML and PDF versions of this article on the journal’s Web site (www.journalppo.com).
REFERENCES
- 1.Gazdar AF. Should we continue to use the term non-small-cell lung cancer? Ann Oncol. 2010;21(suppl 7):vii225–vii229. doi: 10.1093/annonc/mdq372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Druker BJ, Guilhot F, O’Brien SG, et al. Five-year follow-up of patients receiving imatinib for chronic myeloid leukemia. N Engl J Med. 2006;355:2408–2417. doi: 10.1056/NEJMoa062867. [DOI] [PubMed] [Google Scholar]
- 3.Weir BA, Woo MS, Getz G, et al. Characterizing the cancer genome in lung adenocarcinoma. Nature. 2007;450:893–898. doi: 10.1038/nature06358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Knudson AG., Jr The ninth Gordon Hamilton-Fairley memorial lecture. Hereditary cancers: clues to mechanisms of carcinogenesis. Br J Cancer. 1989;59:661–666. doi: 10.1038/bjc.1989.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Breuer RH, Postmus PE, Smit EF. Molecular pathology of non-smallcell lung cancer. Respiration. 2005;72:313–330. doi: 10.1159/000085376. [DOI] [PubMed] [Google Scholar]
- 6.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 7.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 8.Kwei KA, Kim YH, Girard L, et al. Genomic profiling identifies TITF1 as a lineage-specific oncogene amplified in lung cancer. Oncogene. 2008;27:3635–3640. doi: 10.1038/sj.onc.1211012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Merlino GT, Xu YH, Ishii S, et al. Amplification and enhanced expression of the epidermal growth factor receptor gene in A431 human carcinoma cells. Science. 1984;224:417–419. doi: 10.1126/science.6200934. [DOI] [PubMed] [Google Scholar]
- 10.Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 11.Pao W, Miller V, Zakowski M, et al. EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci U S A. 2004;101:13306–13311. doi: 10.1073/pnas.0405220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Paez JG, Janne PA, Lee JC, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 13.Massion PP, Kuo WL, Stokoe D, et al. Genomic copy number analysis of non-small cell lung cancer using array comparative genomichybridization: implications of the phosphatidylinositol 3-kinase pathway. Cancer Res. 2002;62:3636–3640. [PubMed] [Google Scholar]
- 14.Friend SH, Bernards R, Rogelj S, et al. A human DNA segment with properties of the gene that predisposes to retinoblastoma and osteosarcoma. Nature. 1986;323:643–646. doi: 10.1038/323643a0. [DOI] [PubMed] [Google Scholar]
- 15.Nobori T, Miura K, Wu DJ, et al. Deletions of the cyclin-dependent kinase-4 inhibitor gene in multiple human cancers. Nature. 1994;368:753–756. doi: 10.1038/368753a0. [DOI] [PubMed] [Google Scholar]
- 16.Steck PA, Pershouse MA, Jasser SA, et al. Identification of a candidate tumour suppressor gene, MMAC1, at chromosome 10q23. 3 that is mutated in multiple advanced cancers. Nat Genet. 1997;15:356–362. doi: 10.1038/ng0497-356. [DOI] [PubMed] [Google Scholar]
- 17.Weinstein IB. Cancer. Addiction to oncogenes—the Achilles heal of cancer. Science. 2002;297:63–64. doi: 10.1126/science.1073096. [DOI] [PubMed] [Google Scholar]
- 18.William WN, Jr, Glisson BS. Novel strategies for the treatment of small-cell lung carcinoma. Nat Rev Clin Oncol. 2011;8:611–619. doi: 10.1038/nrclinonc.2011.90. [DOI] [PubMed] [Google Scholar]
- 19.Abidin AZ, Garassino MC, Califano R, et al. Targeted therapies in small cell lung cancer: a review. Ther Adv Med Oncol. 2010;2:25–37. doi: 10.1177/1758834009356014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pal SK, Figlin RA, Reckamp K. Targeted therapies for non-small cell lung cancer: an evolving landscape. Mol Cancer Ther. 2010;9:1931–1944. doi: 10.1158/1535-7163.MCT-10-0239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ray MR, Jablons D, He B. Lung cancer therapeutics that target signaling pathways: an update. Exp Rev Respir Med. 2010;4:631–645. doi: 10.1586/ers.10.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Janku F, Garrido-Laguna I, Petruzelka LB, et al. Novel therapeutic targets in non-small cell lung cancer. J Thorac Oncol. 2011;6:1601–1612. doi: 10.1097/JTO.0b013e31822944b3. [DOI] [PubMed] [Google Scholar]
- 23.Normanno N, Bianco C, Strizzi L, et al. The ErbB receptors and their ligands in cancer: an overview. Curr Drug Targets. 2005;6:243–257. doi: 10.2174/1389450053765879. [DOI] [PubMed] [Google Scholar]
- 24.Schreiber AB, Winkler ME, Derynck R. Transforming growth factor-alpha: a more potent angiogenic mediator than epidermal growth factor. Science. 1986;232:1250–1253. doi: 10.1126/science.2422759. [DOI] [PubMed] [Google Scholar]
- 25.Petit AM, Rak J, Hung MC, et al. Neutralizing antibodies against epidermal growth factor and ErbB-2/neu receptor tyrosine kinases down-regulate vascular endothelial growth factor production by tumor cells in vitro and in vivo: angiogenic implications for signal transduction therapy of solid tumors. Am J Pathol. 1997;151:1523–1530. [PMC free article] [PubMed] [Google Scholar]
- 26.Gibson S, Tu S, Oyer R, et al. Epidermal growth factor protects epithelial cells against Fas-induced apoptosis. Requirement for Akt activation. J Biol Chem. 1999;274:17612–17618. doi: 10.1074/jbc.274.25.17612. [DOI] [PubMed] [Google Scholar]
- 27.O-charoenrat P, Modjtahedi H, Rhys-Evans P, et al. Epidermal growth factor-like ligands differentially up-regulate matrix metalloproteinase 9 in head and neck squamous carcinoma cells. Cancer Res. 2000;60:1121–1128. [PubMed] [Google Scholar]
- 28.Harari PM, Allen GW, Bonner JA. Biology of interactions: antiepidermal growth factor receptor agents. J Clin Oncol. 2007;25:4057–4065. doi: 10.1200/JCO.2007.11.8984. [DOI] [PubMed] [Google Scholar]
- 29.Lin SY, Makino K, Xia W, et al. Nuclear localization of EGF receptor and its potential new role as a transcription factor. Nat Cell Biol. 2001;3:802–808. doi: 10.1038/ncb0901-802. [DOI] [PubMed] [Google Scholar]
- 30.Dittmann K, Mayer C, Fehrenbacher B, et al. Radiation-induced epidermal growth factor receptor nuclear import is linked to activation of DNA-dependent protein kinase. J Biol Chem. 2005;280:31182–31189. doi: 10.1074/jbc.M506591200. [DOI] [PubMed] [Google Scholar]
- 31.Rusch V, Baselga J, Cordon-Cardo C, et al. Differential expression of the epidermal growth factor receptor and its ligands in primary non-small cell lung cancers and adjacent benign lung. Cancer Res. 1993;53(suppl 10):2379–2385. [PubMed] [Google Scholar]
- 32.Dutu T, Michiels S, Fouret P, et al. Differential expression of biomarkers in lung adenocarcinoma: a comparative study between smokers and never-smokers. Ann Oncol. 2005;16:1906–1914. doi: 10.1093/annonc/mdi408. [DOI] [PubMed] [Google Scholar]
- 33.Hirsch FR, Varella-Garcia M, Bunn PA, Jr, et al. Epidermal growth factor receptor in non-small-cell lung carcinomas: correlation between gene copy number and protein expression and impact on prognosis. J Clin Oncol. 2003;21:3798–3807. doi: 10.1200/JCO.2003.11.069. [DOI] [PubMed] [Google Scholar]
- 34.Nicholson RI, Gee JM, Harper ME. EGFR and cancer prognosis. Eur J Cancer. 2001;37(suppl 4):S9–S15. doi: 10.1016/s0959-8049(01)00231-3. [DOI] [PubMed] [Google Scholar]
- 35.Eberhard DA, Johnson BE, Amler LC, et al. Mutations in the epidermal growth factor receptor and in KRAS are predictive and prognostic indicators in patients with non-small-cell lung cancer treated with chemotherapy alone and in combination with erlotinib. J Clin Oncol. 2005;23:5900–5909. doi: 10.1200/JCO.2005.02.857. [DOI] [PubMed] [Google Scholar]
- 36.Sordella R, Bell DW, Haber DA, et al. Gefitinib-sensitizing EGFR mutations in lung cancer activate anti-apoptotic pathways. Science. 2004;305:1163–1167. doi: 10.1126/science.1101637. [DOI] [PubMed] [Google Scholar]
- 37.Yun CH, Boggon TJ, Li Y, et al. Structures of lung cancer-derived EGFR mutants and inhibitor complexes: mechanism of activation and insights into differential inhibitor sensitivity. Cancer Cell. 2007;11:217–227. doi: 10.1016/j.ccr.2006.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rosell R, Moran T, Queralt C, et al. Screening for epidermal growth factor receptor mutations in lung cancer. N Engl J Med. 2009;361:958–967. doi: 10.1056/NEJMoa0904554. [DOI] [PubMed] [Google Scholar]
- 39.Hanna N, Lilenbaum R, Ansari R, et al. Phase II trial of cetuximab in patients with previously treated non-small-cell lung cancer. J Clin Oncol. 2006;24:5253–5258. doi: 10.1200/JCO.2006.08.2263. [DOI] [PubMed] [Google Scholar]
- 40.Gatzemeier U, Paz-Ares L, Pereira J Rodrigues, et al. Molecular and clinical biomarkers of cetuximab efficacy: data from the phase III FLEX study in non-small cell lung cancer (NSCLC) J Thorac Oncol. 2009;4:S324. Abstract B322.323. [Google Scholar]
- 41.Mok TS, Wu YL, Thongprasert S, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med. 2009;361:947–957. doi: 10.1056/NEJMoa0810699. [DOI] [PubMed] [Google Scholar]
- 42.Fukuoka M, Yano S, Giaccone G, et al. Multi-institutional randomized phase II trial of gefitinib for previously treated patients with advanced non-small-cell lung cancer (the IDEAL 1 trial) [corrected] J Clin Oncol. 2003;21:2237–2246. doi: 10.1200/JCO.2003.10.038. [DOI] [PubMed] [Google Scholar]
- 43.Kris MG, Natale RB, Herbst RS, et al. Efficacy of gefitinib, an inhibitor of the epidermal growth factor receptor tyrosine kinase, in symptomatic patients with non-small cell lung cancer: a randomized trial. JAMA. 2003;290:2149–2158. doi: 10.1001/jama.290.16.2149. [DOI] [PubMed] [Google Scholar]
- 44.Janne PA, Gurubhagavatula S, Yeap BY, et al. Outcomes of patients with advanced non-small cell lung cancer treated with gefitinib (ZD1839, “Iressa”) on an expanded access study. Lung Cancer. 2004;44:221–230. doi: 10.1016/j.lungcan.2003.12.014. [DOI] [PubMed] [Google Scholar]
- 45.Perez-Soler R, Chachoua A, Hammond LA, et al. Determinants of tumor response and survival with erlotinib in patients with non-small-cell lung cancer. J Clin Oncol. 2004;22:3238–3247. doi: 10.1200/JCO.2004.11.057. [DOI] [PubMed] [Google Scholar]
- 46.Kim KS, Jeong JY, Kim YC, et al. Predictors of the response to gefitinib in refractory non-small cell lung cancer. Clin Cancer Res. 2005;11:2244–2251. doi: 10.1158/1078-0432.CCR-04-2081. [DOI] [PubMed] [Google Scholar]
- 47.Veronese ML, Algazy K, Bearn L, et al. Gefitinib in patients with advanced non-small cell lung cancer (NSCLC): the expanded access protocol experience at the University of Pennsylvania. Cancer Invest. 2005;23:296–302. doi: 10.1081/cnv-61528. [DOI] [PubMed] [Google Scholar]
- 48.Shigematsu H, Lin L, Takahashi T, et al. Clinical and biological features associated with epidermal growth factor receptor gene mutations in lung cancers. J Natl Cancer Inst. 2005;97:339–346. doi: 10.1093/jnci/dji055. [DOI] [PubMed] [Google Scholar]
- 49.Franklin WA, Veve R, Hirsch FR, et al. Epidermal growth factor receptor family in lung cancer and premalignancy. Semin Oncol. 2002;29(1 suppl 4):3–14. doi: 10.1053/sonc.2002.31520. [DOI] [PubMed] [Google Scholar]
- 50.Herbst RS. Review of epidermal growth factor receptor biology. Int J Radiat Oncol Biol Phys. 2004;59(suppl 2):21–26. doi: 10.1016/j.ijrobp.2003.11.041. [DOI] [PubMed] [Google Scholar]
- 51.Fujino S, Enokibori T, Tezuka N, et al. A comparison of epidermal growth factor receptor levels and other prognostic parameters in non-small cell lung cancer. Eur J Cancer. 1996;32A:2070–2074. doi: 10.1016/s0959-8049(96)00243-2. [DOI] [PubMed] [Google Scholar]
- 52.Greulich H, Chen TH, Feng W, et al. Oncogenic transformation by inhibitor-sensitive and -resistant EGFR mutants. PLoS Med. 2005;2:e313. doi: 10.1371/journal.pmed.0020313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yun CH, Mengwasser KE, Toms AV, et al. The T790M mutation in EGFR kinase causes drug resistance by increasing the affinity for ATP. Proc Natl Acad Sci U S A. 2008;105:2070–2075. doi: 10.1073/pnas.0709662105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gazdar AF. Activating and resistance mutations of EGFR in non-small-cell lung cancer: role in clinical response to EGFR tyrosine kinase inhibitors. Oncogene. 2009;28(suppl 1):S24–S31. doi: 10.1038/onc.2009.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Thomas RK, Greulich H, Yuza Y, et al. Detection of oncogenic mutations in the EGFR gene in lung adenocarcinoma with differential sensitivity to EGFR tyrosine kinase inhibitors. Cold Spring Harb Symp Quant Biol. 2005;70:73–81. doi: 10.1101/sqb.2005.70.056. [DOI] [PubMed] [Google Scholar]
- 56.Pao W, Wang TY, Riely GJ, et al. KRAS mutations and primary resistance of lung adenocarcinomas to gefitinib or erlotinib. PLoS Med. 2005;2:e17. doi: 10.1371/journal.pmed.0020017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pao W, Miller VA, Politi KA, et al. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med. 2005;2:e73. doi: 10.1371/journal.pmed.0020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bean J, Brennan C, Shih JY, et al. MET amplification occurs with or without T790M mutations in EGFR mutant lung tumors with acquired resistance to gefitinib or erlotinib. Proc Natl Acad Sci U S A. 2007;104:20932–20937. doi: 10.1073/pnas.0710370104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Engelman JA, Zejnullahu K, Mitsudomi T, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 2007;316:1039–1043. doi: 10.1126/science.1141478. [DOI] [PubMed] [Google Scholar]
- 60.Shepherd FA, Pereira J Rodrigues, Ciuleanu T, et al. Erlotinib in previously treated non-small-cell lung cancer. N Engl J Med. 2005;353:123–132. doi: 10.1056/NEJMoa050753. [DOI] [PubMed] [Google Scholar]
- 61.Thatcher N, Chang A, Parikh P, et al. Gefitinib plus best supportive care in previously treated patients with refractory advanced non-small-cell lung cancer: results from a randomised, placebo-controlled, multicentre study (Iressa Survival Evaluation in Lung Cancer) Lancet. 2005;366:1527–1537. doi: 10.1016/S0140-6736(05)67625-8. [DOI] [PubMed] [Google Scholar]
- 62.Cappuzzo F, Hirsch FR, Rossi E, et al. Epidermal growth factor receptor gene and protein and gefitinib sensitivity in non-small-cell lung cancer. J Natl Cancer Inst. 2005;97:643–655. doi: 10.1093/jnci/dji112. [DOI] [PubMed] [Google Scholar]
- 63.Tsao MS, Sakurada A, Cutz JC, et al. Erlotinib in lung cancer - molecular and clinical predictors of outcome. N Engl J Med. 2005;353:133–144. doi: 10.1056/NEJMoa050736. [DOI] [PubMed] [Google Scholar]
- 64.Hirsch FR, Varella-Garcia M, Bunn PA, Jr, et al. Molecular predictors of outcome with gefitinib in a phase III placebo-controlled study in advanced non-small-cell lung cancer. J Clin Oncol. 2006;24:5034–5042. doi: 10.1200/JCO.2006.06.3958. [DOI] [PubMed] [Google Scholar]
- 65.Eberhard DA, Giaccone G, Johnson BE. Biomarkers of response to epidermal growth factor receptor inhibitors in Non-Small-Cell Lung Cancer Working Group: standardization for use in the clinical trial setting. J Clin Oncol. 2008;26:983–994. doi: 10.1200/JCO.2007.12.9858. [DOI] [PubMed] [Google Scholar]
- 66.Italiano A, Vandenbos FB, Otto J, et al. Comparison of the epidermal growth factor receptor gene and protein in primary non-small-cell-lung cancer and metastatic sites: implications for treatment with EGFR-inhibitors. Ann Oncol. 2006;17:981–985. doi: 10.1093/annonc/mdl038. [DOI] [PubMed] [Google Scholar]
- 67.Suzuki S, Dobashi Y, Sakurai H, et al. Protein overexpression and gene amplification of epidermal growth factor receptor in non-small cell lung carcinomas. An immunohistochemical and fluorescence in situ hybridization study. Cancer. 2005;103:1265–1273. doi: 10.1002/cncr.20909. [DOI] [PubMed] [Google Scholar]
- 68.Miller VA, Riely GJ, Zakowski MF, et al. Molecular characteristics of bronchioloalveolar carcinoma and adenocarcinoma, bronchioloalveolar carcinoma subtype, predict response to erlotinib. J Clin Oncol. 2008;26:1472–1478. doi: 10.1200/JCO.2007.13.0062. [DOI] [PubMed] [Google Scholar]
- 69.Hirsch FR, Varella-Garcia M, McCoy J, et al. Increased epidermal growth factor receptor gene copy number detected by fluorescence in situ hybridization associates with increased sensitivity to gefitinib in patients with bronchioloalveolar carcinoma subtypes: a Southwest Oncology Group Study. J Clin Oncol. 2005;23:6838–6845. doi: 10.1200/JCO.2005.01.2823. [DOI] [PubMed] [Google Scholar]
- 70.Matsuo K, Ito H, Yatabe Y, et al. Risk factors differ for non-small-cell lung cancers with and without EGFR mutation: assessment of smoking and sex by a case-control study in Japanese. Cancer Sci. 2007;98:96–101. doi: 10.1111/j.1349-7006.2006.00347.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jantus-Lewintre E, Sanmartin E, Sirera R, et al. Combined VEGF-A and VEGFR-2 concentrations in plasma: diagnostic and prognostic implications in patients with advanced NSCLC. Lung Cancer. 2011;74:326–331. doi: 10.1016/j.lungcan.2011.02.016. [DOI] [PubMed] [Google Scholar]
- 72.Longo R, Gasparini G. Challenges for patient selection with VEGF inhibitors. Cancer Chemother Pharmacol. 2007;60:151–170. doi: 10.1007/s00280-006-0403-6. [DOI] [PubMed] [Google Scholar]
- 73.Coate LE, John T, Tsao MS, et al. Molecular predictive and prognostic markers in non-small-cell lung cancer. Lancet Oncol. 2009;10:1001–1010. doi: 10.1016/S1470-2045(09)70155-X. [DOI] [PubMed] [Google Scholar]
- 74.Kelly K, Chansky K, Gaspar LE, et al. Phase III trial of maintenance gefitinib or placebo after concurrent chemoradiotherapy and docetaxel consolidation in inoperable stage III non-small-cell lung cancer: SWOG S0023. J Clin Oncol. 2008;26:2450–2456. doi: 10.1200/JCO.2007.14.4824. [DOI] [PubMed] [Google Scholar]
- 75.Inoue A, Kobayashi D, Maemondo M, et al. A randomized phase III study comparing gefitinib with carboplatin (CBDCA) plus paclitaxel (TXL) for the first-line treatment of non-small cell lung cancer (NSCLC) with sensitive EGFR mutations: NEJ002 study. Eur J Cancer. 2009;45(suppl 1):6. Abstract 9LBA. [Google Scholar]
- 76.Lee JS, Park K, Kim S-W, et al. A randomized phase III study of gefitinib (Iressa) versus standard chemotherapy (gemcitabine plus cisplatin) as a first-line treatment for never-smokers with advanced or metastatic adenocarcinoma of the lung. J Thorac Oncol. 2009;4(9 suppl 1):S283. Abstract PRS.284. [Google Scholar]
- 77.Mitsudomi T, Morita S, Yatabe Y, et al. Gefitinib versus cisplatin plus docetaxel in patients with non-small-cell lung cancer harbouring mutations of the epidermal growth factor receptor (WJTOG3405): an open label, randomised phase 3 trial. Lancet Oncol. 2010;11:121–128. doi: 10.1016/S1470-2045(09)70364-X. [DOI] [PubMed] [Google Scholar]
- 78.Kimura TM, Mahaffey CM, Pryde BJ, et al. Apoptotic effects of the docetaxel->OSI-774 combination in non-small cell lung carcinoma (NSCLC) cells. Proc Am Soc Clin Oncol. 2004;23:649. Abstract 7026. [Google Scholar]
- 79.Kim E, Herbst R, Wistuba I, et al. The BATTLE trial (Biomarker-integrated Approaches of Targeted Therapy for Lung Cancer Elimination): personalizing therapy for lung cancer. Cancer Discov. 2011;1:43–51. doi: 10.1158/2159-8274.CD-10-0010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. [Accessed October 18, 2011];Clinicaltrials.gov. Available at: www.clinicaltrials.gov.
- 81.Maemondo M, Inoue A, Kobayashi K, et al. Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N Engl J Med. 2010;362:2380–2388. doi: 10.1056/NEJMoa0909530. [DOI] [PubMed] [Google Scholar]
- 82.Engelman JA, Janne PA. Mechanisms of acquired resistance to epidermal growth factor receptor tyrosine kinase inhibitors in non-small cell lung cancer. Clin Cancer Res. 2008;14:2895–2899. doi: 10.1158/1078-0432.CCR-07-2248. [DOI] [PubMed] [Google Scholar]
- 83.Balak MN, Gong Y, Riely GJ, et al. Novel D761Y and common secondary T790M mutations in epidermal growth factor receptor-mutant lung adenocarcinomas with acquired resistance to kinase inhibitors. Clin Cancer Res. 2006;12:6494–6501. doi: 10.1158/1078-0432.CCR-06-1570. [DOI] [PubMed] [Google Scholar]
- 84.Sequist LV, Waltman BA, Dias-Santagata D, et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci Transl Med. 2011;3:75ra26. doi: 10.1126/scitranslmed.3002003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yarden Y, Sliwkowski MX. Untangling the ErbB signalling network. Nat Rev Mol Cell Biol. 2001;2:127–137. doi: 10.1038/35052073. [DOI] [PubMed] [Google Scholar]
- 86.Sequist LV, von Pawel J, Garmey EG, et al. Randomized phase II study of erlotinib plus tivantinib versus erlotinib plus placebo in previously treated non-small-cell lung cancer. J Clin Oncol. 2011;29:3307–3315. doi: 10.1200/JCO.2010.34.0570. [DOI] [PubMed] [Google Scholar]
- 87.Spigel DR, Ervin TJ, Ramlau R, et al. Final efficacy results from OAM4558g, a randomized phase II study evaluating MetMAb or placebo in combination with erlotinib in advanced NSCLC. J Clin Oncol. 2011;29(suppl 15):7505. [Google Scholar]
- 88.Kwak EL, Sordella R, Bell DW, et al. Irreversible inhibitors of the EGF receptor may circumvent acquired resistance to gefitinib. Proc Natl Acad Sci U S A. 2005;102:7665–7670. doi: 10.1073/pnas.0502860102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Engelman JA, Zejnullahu K, Gale CM, et al. PF00299804, an irreversible pan-ERBB inhibitor, is effective in lung cancer models with EGFR and ERBB2 mutations that are resistant to gefitinib. Cancer Res. 2007;67:11924–11932. doi: 10.1158/0008-5472.CAN-07-1885. [DOI] [PubMed] [Google Scholar]
- 90.Garrett TP, McKern NM, Lou M, et al. The crystal structure of a truncated ErbB2 ectodomain reveals an active conformation, poised to interact with other ErbB receptors. Mol Cell. 2003;11:495–505. doi: 10.1016/s1097-2765(03)00048-0. [DOI] [PubMed] [Google Scholar]
- 91.Mathew J, Perez EA. Trastuzumab emtansine in human epidermal growth factor receptor 2-positive breast cancer: a review. Curr Opin Oncol. 2011;23:594–600. doi: 10.1097/CCO.0b013e32834b895c. [DOI] [PubMed] [Google Scholar]
- 92.Mackenzie M, Spithoff K, Jonker D. Systemic therapy for advanced gastric cancer: a clinical practice guideline. Curr Oncol. 2011;18:e202–e209. doi: 10.3747/co.v18i4.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Shigematsu H, Takahashi T, Nomura M, et al. Somatic mutations of the HER2 kinase domain in lung adenocarcinomas. Cancer Res. 2005;65:1642–1646. doi: 10.1158/0008-5472.CAN-04-4235. [DOI] [PubMed] [Google Scholar]
- 94.Stephens P, Hunter C, Bignell G, et al. Lung cancer: intragenic ERBB2 kinase mutations in tumours. Nature. 2004;431:525–526. doi: 10.1038/431525b. [DOI] [PubMed] [Google Scholar]
- 95.Miller VA, Hirsh V, Cadranel J, et al. Phase IIB/III double-blind randomized trial of afatinib (BIBW 2992, an irreverisible inhibitor of EGFR/HER1 and HER2) and best supportive care (BSC) versus placebo and BSC in patients with NSCLC failing 1-2 lines of chemotherapy and erlotinib or gefitinib (LUX-LUNG 1) Ann Oncol. 2010;21(suppl 8):viii1. [Google Scholar]
- 96.Campbell A, Reckamp KL, Camidge DR, et al. PF-00299804 (PF299) patient (pt)-reported outcomes (PROs) and efficacy in adenocarcinoma (adeno) and nonadeno non-small cell lung cancer (NSCLC): a phase (P) II trial in advanced NSCLC after failure of chemotherapy (CT) and erlotinib (E) J Clin Oncol. 2010;28(suppl 15):7596. [Google Scholar]
- 97.Boyer MJ, Blackhall FH, Park K, et al. Efficacy and safety of PF299804 versus erlotinib (E): a global, randomized phase II trial in patients (pts) with advanced non-small cell lung cancer (NSCLC) after failure of chemotherapy (CT) J Clin Oncol. 2010;28(suppl 18):LBA7523. [Google Scholar]
- 98.Sequist LV, Besse B, Lynch TJ, et al. Neratinib, an irreversible pan-ErbB receptor tyrosine kinase inhibitor: results of a phase II trial in patients with advanced non-small-cell lung cancer. J Clin Oncol. 2010;28:3076–3083. doi: 10.1200/JCO.2009.27.9414. [DOI] [PubMed] [Google Scholar]
- 99.Soda M, Choi YL, Enomoto M, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448:561–566. doi: 10.1038/nature05945. [DOI] [PubMed] [Google Scholar]
- 100.Solomon B, Varella-Garcia M, Camidge DR. ALK gene rearrangements: a new therapeutic target in a molecularly defined subset of non-small cell lung cancer. J Thorac Oncol. 2009;4:1450–1454. doi: 10.1097/JTO.0b013e3181c4dedb. [DOI] [PubMed] [Google Scholar]
- 101.Palmer RH, Vernersson E, Grabbe C, et al. Anaplastic lymphoma kinase: signalling in development and disease. Biochem J. 2009;420:345–361. doi: 10.1042/BJ20090387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Shaw AT, Yeap BY, Mino-Kenudson M, et al. Clinical features and outcome of patients with non-small-cell lung cancer who harbor EML4-ALK. J Clin Oncol. 2009;27:4247–4253. doi: 10.1200/JCO.2009.22.6993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Rodig SJ, Mino-Kenudson M, Dacic S, et al. Unique clinicopathologic features characterize ALK-rearranged lung adenocarcinoma in the western population. Clin Cancer Res. 2009;15:5216–5223. doi: 10.1158/1078-0432.CCR-09-0802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Camidge DR, Kono SA, Lu X, et al. Anaplastic lymphoma kinase gene rearrangements in non-small cell lung cancer are associated with prolonged progression-free survival on pemetrexed. J Thorac Oncol. 2011;6:774–780. doi: 10.1097/JTO.0b013e31820cf053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Bertolini G, Roz L, Perego P, et al. Highly tumorigenic lung cancer CD133+ cells display stem-like features and are spared by cisplatin treatment. Proc Natl Acad Sci U S A. 2009;106:16281–16286. doi: 10.1073/pnas.0905653106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.De Francesco F, Tirino V, Desiderio V, et al. Human CD34/CD90 ASCs are capable of growing as sphere clusters, producing high levels of VEGF and forming capillaries. PLoS One. 2009;4:e6537. doi: 10.1371/journal.pone.0006537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Tirino V, Camerlingo R, Franco R, et al. The role of CD133 in the identification and characterisation of tumour-initiating cells in non-small-cell lung cancer. Eur J Cardiothorac Surg. 2009;36:446–453. doi: 10.1016/j.ejcts.2009.03.063. [DOI] [PubMed] [Google Scholar]
- 108.De Rosa A, De Francesco F, Tirino V, et al. A new method for the cryopreserving ASCs: an attractive and suitable large-scale and long-term cell banking technology. Tissue Eng Part C Methods. 2009;15:659–667. doi: 10.1089/ten.TEC.2008.0674. [DOI] [PubMed] [Google Scholar]
- 109.Costantino E, Maddalena F, Calise S, et al. TRAP1, a novel mitochondrial chaperone responsible for multi-drug resistance and protection from apoptotis in human colorectal carcinoma cells. Cancer Lett. 2009;279:39–46. doi: 10.1016/j.canlet.2009.01.018. [DOI] [PubMed] [Google Scholar]
- 110.Kwak EL, Bang YJ, Camidge DR, et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med. 2010;363:1693–1703. doi: 10.1056/NEJMoa1006448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Choi YL, Soda M, Yamashita Y, et al. EML4-ALK mutations in lung cancer that confer resistance to ALK inhibitors. N Engl J Med. 2010;363:1734–1739. doi: 10.1056/NEJMoa1007478. [DOI] [PubMed] [Google Scholar]
- 112.Sasaki T, Okuda K, Zheng W, et al. The neuroblastoma-associated F1174L ALK mutation causes resistance to an ALK kinase inhibitor in ALK-translocated cancers. Cancer Res. 2010;70:10038–10043. doi: 10.1158/0008-5472.CAN-10-2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Katayama R, Khan TM, Benes C, et al. Therapeutic strategies to overcome crizotinib resistance in non-small cell lung cancers harboring the fusion oncogene EML4-ALK. Proc Natl Acad Sci U S A. 2011;108:7535–7540. doi: 10.1073/pnas.1019559108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Normant E, Paez G, West KA, et al. The Hsp90 inhibitor IPI-504 rapidly lowers EML4-ALK levels and induces tumor regression in ALK-driven NSCLC models. Oncogene. 2011;30:2581–2586. doi: 10.1038/onc.2010.625. [DOI] [PubMed] [Google Scholar]
- 115.Cardones AR, Banez LL. VEGF inhibitors in cancer therapy. Curr Pharm Des. 2006;12:387–394. doi: 10.2174/138161206775201910. [DOI] [PubMed] [Google Scholar]
- 116.Korpanty G, Smyth E, Sullivan LA, et al. Antiangiogenic therapy in lung cancer: focus on vascular endothelial growth factor pathway. Exp Biol Med. 2010;235:3–9. doi: 10.1258/ebm.2009.009191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ferrara N. Vascular endothelial growth factor: basic science and clinical progress. Endocr Rev. 2004;25:581–611. doi: 10.1210/er.2003-0027. [DOI] [PubMed] [Google Scholar]
- 118.Stefanou D, Batistatou A, Arkoumani E, et al. Expression of vascular endothelial growth factor (VEGF) and association with microvessel density in small-cell and non-small-cell lung carcinomas. Histol Histopathol. 2004;19:37–42. doi: 10.14670/HH-19.37. [DOI] [PubMed] [Google Scholar]
- 119.Kaya A, Ciledag A, Gulbay BE, et al. The prognostic significance of vascular endothelial growth factor levels in sera of non-small cell lung cancer patients. Respir Med. 2004;98:632–636. doi: 10.1016/j.rmed.2003.12.017. [DOI] [PubMed] [Google Scholar]
- 120.Dudek AZ, Mahaseth H. Circulating angiogenic cytokines in patients with advanced non-small cell lung cancer: correlation with treatment response and survival. Cancer Invest. 2005;23:193–200. doi: 10.1081/cnv-200055949. [DOI] [PubMed] [Google Scholar]
- 121.Schneider BP, Wang M, Radovich M, et al. Association of vascular endothelial growth factor and vascular endothelial growth factor receptor-2 genetic polymorphisms with outcome in a trial of paclitaxel compared with paclitaxel plus bevacizumab in advanced breast cancer: ECOG 2100. J Clin Oncol. 2008;26:4672–4678. doi: 10.1200/JCO.2008.16.1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Dahlberg SE, Sandler AB, Brahmer JR, et al. Clinical course of advanced non-small cell lung cancer patients experiencing hypertension during treatment with bevacizumab in combination with carboplatin and paclitaxel on E4599. J Clin Oncol. 2010;8:949–954. doi: 10.1200/JCO.2009.25.4482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Spigel DR, Hainsworth JD, Yardley DA, et al. Tracheoesophageal fistula formation in patients with lung cancer treated with chemoradiation and bevacizumab. J Clin Oncol. 2009;28:43–48. doi: 10.1200/JCO.2009.24.7353. [DOI] [PubMed] [Google Scholar]
- 124.Ferrara N. VEGF: an update on biological and therapeutic aspects. Curr Opin Biotechnol. 2000;11:617–624. doi: 10.1016/s0958-1669(00)00153-1. [DOI] [PubMed] [Google Scholar]
- 125.Hicklin DJ, Ellis LM. Role of the vascular endothelial growth factor pathway in tumor growth and angiogenesis. J Clin Oncol. 2005;23:1011–1027. doi: 10.1200/JCO.2005.06.081. [DOI] [PubMed] [Google Scholar]
- 126.Kamba T, McDonald DM. Mechanisms of adverse effects of anti-VEGF therapy for cancer. Br J Cancer. 2007;96:1788–1795. doi: 10.1038/sj.bjc.6603813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Kitamoto Y, Tokunaga H, Miyamoto K, et al. VEGF is an essential molecule for glomerular structuring. Nephrol Dial Transplant. 2002;17(suppl 9):25–27. doi: 10.1093/ndt/17.suppl_9.25. [DOI] [PubMed] [Google Scholar]
- 128.Scagliotti G, Govindan R. Targeting angiogenesis with multitargeted tyrosine kinase inhibitors in the treatment of non-small cell lung cancer. Oncologist. 2010;15:436–446. doi: 10.1634/theoncologist.2009-0225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Socinski MA, Novello S, Brahmer JR, et al. Multicenter, phase II trial of sunitinib in previously treated, advanced non-small-cell lung cancer. J Clin Oncol. 2008;26:650–656. doi: 10.1200/JCO.2007.13.9303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Herbst RS, O’Neill VJ, Fehrenbacher L, et al. Phase II study of efficacy and safety of bevacizumab in combination with chemotherapy or erlotinib compared with chemotherapy alone for treatment of recurrent or refractory non small-cell lung cancer. J Clin Oncol. 2007;25:4743–4750. doi: 10.1200/JCO.2007.12.3026. [DOI] [PubMed] [Google Scholar]
- 131.Herbst RS. Toxicities of antiangiogenic therapy in non-small-cell lung cancer. Clin Lung Cancer. 2006;8(suppl 1):S23–S30. doi: 10.3816/clc.2006.s.010. [DOI] [PubMed] [Google Scholar]
- 132.Schiller JH, Lee JW, Hanna NH, et al. A randomized discontinuation phase II study of sorafenib versus placebo in patients with non-small cell lung cancer who have failed at least two prior chemotherapy regimens: E2501. J Clin Oncol. 2008;26(suppl 15):8014. [Google Scholar]
- 133.Heymach J, Paz-Ares L, De Braud F, et al. Randomized phase II study of vandetanib (VAN) alone or in combination with carboplatin and paclitaxel (CP) as first-line treatment for advanced non-small cell lung cancer (NSCLC) J Clin Oncol. 2007;25(suppl 18):7544. doi: 10.1200/JCO.2008.17.3138. [DOI] [PubMed] [Google Scholar]
- 134.De Boer R, Arrieta O, Gottfried FH. Vandetanib plus pemetrexed versus pemetrexed as second-line therapy in patients with advanced non-small cell lung cancer (NSCLC): a randomized, double-blind phase III trial (ZEAL) J Clin Oncol. 2011;9:1067–1074. doi: 10.1200/JCO.2010.29.5717. [DOI] [PubMed] [Google Scholar]
- 135.Natale RB, Thongprasert S, Greco FA, et al. Phase III trial of vandetanib compared with erlotinib in patients with previously treated advanced non-small-cell lung cancer. J Clin Oncol. 2011;29:1059–1066. doi: 10.1200/JCO.2010.28.5981. [DOI] [PubMed] [Google Scholar]
- 136.Massarelli E, Miller VA, Leighl NB, et al. Phase II study of the efficacy and safety of intravenous (IV) AVE0005 (VEGF Trap) givenevery 2 weeks in patients (Pts) with platinum- and erlotinib-resistant adenocarcinoma of the lung (NSCLA) J Clin Oncol. 2007;25(suppl 18):7627. [Google Scholar]
- 137.Downward J. Targeting RAS signalling pathways in cancer therapy. Nat Rev Cancer. 2003;3:11–22. doi: 10.1038/nrc969. [DOI] [PubMed] [Google Scholar]
- 138.Karnoub AE, Weinberg RA. Ras oncogenes: split personalities. Nat Rev Mol Cell Biol. 2008;9:517–531. doi: 10.1038/nrm2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Linardou H, Dahabreh IJ, Kanaloupiti D, et al. Assessment of somatic k-RAS mutations as a mechanism associated with resistance to EGFR-targeted agents: a systematic review and meta-analysis of studies in advanced non-small-cell lung cancer and metastatic colorectal cancer. Lancet Oncol. 2008;9:962–972. doi: 10.1016/S1470-2045(08)70206-7. [DOI] [PubMed] [Google Scholar]
- 140.Riely GJ, Kris MG, Rosenbaum D, et al. Frequency and distinctive spectrum of KRAS mutations in never smokers with lung adenocarcinoma. Clin Cancer Res. 2008;14:5731–5734. doi: 10.1158/1078-0432.CCR-08-0646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Adjei AA. K-ras as a target for lung cancer therapy. J Thorac Oncol. 2008;3(6 suppl 2):S160–S163. doi: 10.1097/JTO.0b013e318174dbf9. [DOI] [PubMed] [Google Scholar]
- 142.Ding L, Getz G, Wheeler DA, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008;455:1069–1075. doi: 10.1038/nature07423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Davies H, Bignell GR, Cox C, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 144.Naoki K, Chen TH, Richards WG, et al. Missense mutations of the BRAF gene in human lung adenocarcinoma. Cancer Res. 2002;62:7001–7003. [PubMed] [Google Scholar]
- 145.Sasaki H, Kawano O, Endo K, et al. Uncommon V599E BRAF mutations in Japanese patients with lung cancer. J Surg Res. 2006;133:203–206. doi: 10.1016/j.jss.2005.10.022. [DOI] [PubMed] [Google Scholar]
- 146.Bollag G, Hirth P, Tsai J, et al. Clinical efficacy of a RAF inhibitor needs broad target blockade in BRAF-mutant melanoma. Nature. 2010;467:596–599. doi: 10.1038/nature09454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Hainsworth JD, Cebotaru CL, Kanarev V, et al. A phase II, open-label, randomized study to assess the efficacy and safety of AZD6244 (ARRY-142886) versus pemetrexed in patients with non-small cell lung cancer who have failed one or two prior chemotherapeutic regimens. J Thorac Oncol. 2010;5:1630–1636. doi: 10.1097/JTO.0b013e3181e8b3a3. [DOI] [PubMed] [Google Scholar]
- 148.Maki RG. Small is beautiful: insulin-like growth factors and their role in growth, development, and cancer. J Clin Oncol. 2010;28:4985–4995. doi: 10.1200/JCO.2009.27.5040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Gong Y, Yao E, Shen R, et al. High expression levels of total IGF-1R and sensitivity of NSCLC cells in vitro to an anti-IGF-1R antibody (R1507) PLoS One. 2009;4:e7273. doi: 10.1371/journal.pone.0007273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Nahta R, Yu D, Hung MC, et al. Mechanisms of disease: understanding resistance to HER2-targeted therapy in human breast cancer. Nat Clin Pract Oncol. 2006;3:269–280. doi: 10.1038/ncponc0509. [DOI] [PubMed] [Google Scholar]
- 151.Morgillo F, Kim WY, Kim ES, et al. Implication of the insulin-like growth factor-IR pathway in the resistance of non-small cell lung cancer cells to treatment with gefitinib. Clin Cancer Res. 2007;13:2795–2803. doi: 10.1158/1078-0432.CCR-06-2077. [DOI] [PubMed] [Google Scholar]
- 152.Yu H, Spitz MR, Mistry J, et al. Plasma levels of insulin-like growth factor-I and lung cancer risk: a case-control analysis. J Natl Cancer Inst. 1999;91:151–156. doi: 10.1093/jnci/91.2.151. [DOI] [PubMed] [Google Scholar]
- 153.Han JY, Choi BG, Choi JY, et al. The prognostic significance of pre-treatment plasma levels of insulin-like growth factor (IGF)-1, IGF-2, and IGF binding protein-3 in patients with advanced non-small cell lung cancer. Lung Cancer. 2006;54:227–234. doi: 10.1016/j.lungcan.2006.07.014. [DOI] [PubMed] [Google Scholar]
- 154.Ullrich A, Gray A, Tam AW, et al. Insulin-like growth factor I receptor primary structure: comparison with insulin receptor suggests structural determinants that define functional specificity. EMBO J. 1986;5:2503–2512. doi: 10.1002/j.1460-2075.1986.tb04528.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Karp DD, Paz-Ares LG, Novello S, et al. Phase II study of the anti-insulin-like growth factor type 1 receptor antibody CP-751,871 in combination with paclitaxel and carboplatin in previously untreated, locally advanced, or metastatic non-small-cell lung cancer. J Clin Oncol. 2009;27:2516–2522. doi: 10.1200/JCO.2008.19.9331. [DOI] [PubMed] [Google Scholar]
- 156.Jassem J, Langer CJ, Karp DD, et al. Randomized, open label, phase III trial of figitumumab in combination with paclitaxel and carboplatin versus paclitaxel and carboplatin in patients with non-small cell lung cancer (NSCLC) J Clin Oncol. 2010;28(15 suppl):7500. doi: 10.1200/JCO.2013.54.4932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Gualberto A, Dolled-Filhart M, Gustavson M, et al. Molecular analysis of non-small cell lung cancer identifies subsets with different sensitivity to insulin-like growth factor I receptor inhibition. Clin Cancer Res. 2010;16:4654–4665. doi: 10.1158/1078-0432.CCR-10-0089. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 158.Vivanco I, Sawyers CL. The phosphatidylinositol 3-Kinase AKT pathway in human cancer. Nat Rev Cancer. 2002;2:489–501. doi: 10.1038/nrc839. [DOI] [PubMed] [Google Scholar]
- 159.Kawano O, Sasaki H, Endo K, et al. PIK3CA mutation status in Japanese lung cancer patients. Lung Cancer. 2006;54:209–215. doi: 10.1016/j.lungcan.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 160.Garnis C, Lockwood WW, Vucic E, et al. High resolution analysis of non-small cell lung cancer cell lines by whole genome tiling path array CGH. Int J Cancer. 2006;118:1556–1564. doi: 10.1002/ijc.21491. [DOI] [PubMed] [Google Scholar]
- 161.Yamamoto H, Shigematsu H, Nomura M, et al. PIK3CA mutations and copy number gains in human lung cancers. Cancer Res. 2008;68:6913–6921. doi: 10.1158/0008-5472.CAN-07-5084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.West KA, Linnoila IR, Belinsky SA, et al. Tobacco carcinogen-induced cellular transformation increases activation of the phosphatidylinositol 3′-kinase/Akt pathway in vitro and in vivo. Cancer Res. 2004;64:446–451. doi: 10.1158/0008-5472.can-03-3241. [DOI] [PubMed] [Google Scholar]
- 163.Soria JC, Lee HY, Lee JI, et al. Lack of PTEN expression in non-small cell lung cancer could be related to promoter methylation. Clin Cancer Res. 2002;8:1178–1184. [PubMed] [Google Scholar]
- 164.Marsit CJ, Zheng S, Aldape K, et al. PTEN expression in non-small-cell lung cancer: evaluating its relation to tumor characteristics, allelic loss, and epigenetic alteration. Hum Pathol. 2005;36:768–776. doi: 10.1016/j.humpath.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 165.Soria JC, Shepherd FA, Douillard JY, et al. Efficacy of everolimus (RAD001) in patients with advanced NSCLC previously treated with chemotherapy alone or with chemotherapy and EGFR inhibitors. Ann Oncol. 2009;20:1674–1681. doi: 10.1093/annonc/mdp060. [DOI] [PubMed] [Google Scholar]
- 166.Naing A, Kurzrock R, Burger A, et al. Phase I trial of cixutumumab combined with temsirolimus in patients with advanced cancer. Clin Cancer Res. 2011;17:6052–6060. doi: 10.1158/1078-0432.CCR-10-2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Maulik G, Madhiwala P, Brooks S, et al. Activated c-Met signals through PI3K with dramatic effects on cytoskeletal functions in small cell lung cancer. J Cell Mol Med. 2002;6:539–553. doi: 10.1111/j.1582-4934.2002.tb00453.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Brognard J, Clark AS, Ni Y, et al. Akt/protein kinase B is constitutively active in non-small cell lung cancer cells and promotes cellular survival and resistance to chemotherapy and radiation. Cancer Res. 2001;61:3986–3997. [PubMed] [Google Scholar]
- 169.Tsurutani J, West KA, Sayyah J, et al. Inhibition of the phosphatidy-linositol 3-kinase/Akt/mammalian target of rapamycin pathway but not the MEK/ERK pathway attenuates laminin-mediated small cell lung cancer cellular survival and resistance to imatinib mesylate or chemotherapy. Cancer Res. 2005;65:8423–8432. doi: 10.1158/0008-5472.CAN-05-0058. [DOI] [PubMed] [Google Scholar]
- 170.Cassatella MA. On the production of TNF-related apoptosis-inducing ligand (TRAIL/Apo-2L) by human neutrophils. J Leukoc Biol. 2006;79:1140–1149. doi: 10.1189/jlb.1005558. [DOI] [PubMed] [Google Scholar]
- 171.Soria JC, Smit E, Khayat D, et al. Phase 1b study of dulanermin (recombinant human Apo2L/TRAIL) in combination with paclitaxel, carboplatin, and bevacizumab in patients with advanced non-squamous non-small-cell lung cancer. J Clin Oncol. 2010;28:1527–1533. doi: 10.1200/JCO.2009.25.4847. [DOI] [PubMed] [Google Scholar]
- 172.Greco FA, Bonomi P, Crawford J, et al. Phase 2 study of mapatumumab, a fully human agonistic monoclonal antibody which targets and activates the TRAIL receptor-1, in patients with advanced non-small cell lung cancer. Lung Cancer. 2008;61:82–90. doi: 10.1016/j.lungcan.2007.12.011. [DOI] [PubMed] [Google Scholar]
- 173.Von Pawel J, Harvey JH, Spigel DR, et al. A randomized phase II trial of mapatumumab, a TRAIL-R1 agonist monoclonal antibody, in combination with carboplatin and paclitaxel in patients with advanced NSCLC. J Clin Oncol. 2010;28(18 suppl):LBA7501. [Google Scholar]
- 174.LoRusso P, Hong D, Heath E, et al. First-in-human study of AMG 655, a pro-apoptotic TRAIL receptor-2 agonist, in adult patients with advanced solid tumors. J Clin Oncol. 2007;25(18 suppl):3534. [Google Scholar]
- 175.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 176.Ramalingam SS. Histone deacetylase, proteasome, and heat shock protein inhibitors for the treatment of lung cancer. J Thorac Oncol. 2010;5(12 suppl 6):S458–S460. doi: 10.1097/01.JTO.0000391365.56258.16. [DOI] [PubMed] [Google Scholar]
- 177.Mukhopadhyay NK, Weisberg E, Gilchrist D, et al. Effectiveness of trichostatin A as a potential candidate for anticancer therapy in non-small-cell lung cancer. Ann Thorac Surg. 2006;81:1034–1042. doi: 10.1016/j.athoracsur.2005.06.059. [DOI] [PubMed] [Google Scholar]
- 178.Ramalingam SS, Maitland ML, Frankel P, et al. Carboplatin and paclitaxel in combination with either vorinostat or placebo for first-line therapy of advanced non-small-cell lung cancer. J Clin Oncol. 2010;28:56–62. doi: 10.1200/JCO.2009.24.9094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Uehara Y. Natural product origins of Hsp90 inhibitors. Curr Cancer Drug Targets. 2003;3:325–330. doi: 10.2174/1568009033481796. [DOI] [PubMed] [Google Scholar]
- 180.Rajan A, Kelly RJ, Trepel JB, et al. A phase 1 study of PF-04929113 (SNX-5422), an orally bioavailable heat shock protein 90 inhibitor in patients with refractory solid tumor malignancies and lymphomas. Clin Cancer Res. 2011;17:6831–6839. doi: 10.1158/1078-0432.CCR-11-0821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Samuel TA, Sessa C, Britten C, et al. AUY922, a novel HSP90 inhibitor: final results of a first-in-human study in patients with advanced solid malignancies. J Clin Oncol. 2010;28(suppl 15):2528. [Google Scholar]
- 182.Sequist LV, Gettinger S, Senzer NN, et al. Activity of IPI-504, a novel heat-shock protein 90 inhibitor, in patients with molecularly defined non-small-cell lung cancer. J Clin Oncol. 2010;28:4953–4960. doi: 10.1200/JCO.2010.30.8338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Albanell J, Lonardo F, Rusch V, et al. High telomerase activity in primary lung cancers: association with increased cell proliferation rates and advanced pathologic stage. J Natl Cancer Inst. 1997;89:1609–1615. doi: 10.1093/jnci/89.21.1609. [DOI] [PubMed] [Google Scholar]
- 184.Hiyama K, Hiyama E, Ishioka S, et al. Telomerase activity in small-cell and non-small-cell lung cancers. J Natl Cancer Inst. 1995;87:895–902. doi: 10.1093/jnci/87.12.895. [DOI] [PubMed] [Google Scholar]
- 185.Frias C, Garcia-Aranda C, De Juan C, et al. Telomere shortening is associated with poor prognosis and telomerase activity correlates with DNA repair impairment in non-small cell lung cancer. Lung Cancer. 2008;60:416–425. doi: 10.1016/j.lungcan.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 186.Shay JW, Wright WE. Telomerase activity in human cancer. Curr Opin Oncol. 1996;8:66–71. doi: 10.1097/00001622-199601000-00012. [DOI] [PubMed] [Google Scholar]
- 187.Ouellette MM, Wright WE, Shay JW. Targeting telomerase-expressing cancer cells. J Cell Mol Med. 2011;15:1433–1442. doi: 10.1111/j.1582-4934.2011.01279.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Reya T, Morrison SJ, Clarke MF, et al. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 189.Wang JC, Dick JE. Cancer stem cells: lessons from leukemia. Trends Cell Biol. 2005;15:494–501. doi: 10.1016/j.tcb.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 190.Clarke MF, Dick JE, Dirks PB, et al. Cancer stem cells—perspectives on current status and future directions: AACR workshop on cancer stem cells. Cancer Res. 2006;66:9339–9344. doi: 10.1158/0008-5472.CAN-06-3126. [DOI] [PubMed] [Google Scholar]
- 191.Lapidot T, Sirard C, Vormoor J, et al. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature. 1994;367:645–648. doi: 10.1038/367645a0. [DOI] [PubMed] [Google Scholar]
- 192.Chen YC, Hsu HS, Chen YW, et al. Oct-4 expression maintained cancer stem-like properties in lung cancer-derived CD133-positive cells. PLoS One. 2008;3:e2637. doi: 10.1371/journal.pone.0002637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Jiang F, Qiu Q, Khanna A, et al. Aldehyde dehydrogenase 1 is a tumor stem cell-associated marker in lung cancer. Mol Cancer Res. 2009;7:330–338. doi: 10.1158/1541-7786.MCR-08-0393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Kitamura H, Okudela K, Yazawa T, et al. Cancer stem cell: implications in cancer biology and therapy with special reference to lung cancer. Lung Cancer. 2009;66:275–281. doi: 10.1016/j.lungcan.2009.07.019. [DOI] [PubMed] [Google Scholar]
- 195.Ho MM, Ng AV, Lam S, et al. Side population in human lung cancer cell lines and tumors is enriched with stem-like cancer cells. Cancer Res. 2007;67:4827–4833. doi: 10.1158/0008-5472.CAN-06-3557. [DOI] [PubMed] [Google Scholar]
- 196.Eramo A, Lotti F, Sette G, et al. Identification and expansion of the tumorigenic lung cancer stem cell population. Cell Death Differ. 2008;15:504–514. doi: 10.1038/sj.cdd.4402283. [DOI] [PubMed] [Google Scholar]
- 197.Wicha MS, Liu S, Dontu G. Cancer stem cells: an old idea—a paradigm shift. Cancer Res. 2006;66:1883–1890. doi: 10.1158/0008-5472.CAN-05-3153. discussion 1886-1895. [DOI] [PubMed] [Google Scholar]
- 198.Watkins DN, Berman DM, Burkholder SG, et al. Hedgehog signalling within airway epithelial progenitors and in small-cell lung cancer. Nature. 2003;422:313–317. doi: 10.1038/nature01493. [DOI] [PubMed] [Google Scholar]
- 199.Yuan Z, Goetz JA, Singh S, et al. Frequent requirement of hedgehog signaling in non-small cell lung carcinoma. Oncogene. 2007;26:1046–1055. doi: 10.1038/sj.onc.1209860. [DOI] [PubMed] [Google Scholar]
- 200.He B, You L, Uematsu K, et al. A monoclonal antibody against Wnt-1 induces apoptosis in human cancer cells. Neoplasia. 2004;6:7–14. doi: 10.1016/s1476-5586(04)80048-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.You L, He B, Xu Z, et al. Inhibition of Wnt-2-mediated signaling induces programmed cell death in non-small-cell lung cancer cells. Oncogene. 2004;23:6170–6174. doi: 10.1038/sj.onc.1207844. [DOI] [PubMed] [Google Scholar]
- 202.Winn RA, Van Scoyk M, Hammond M, et al. Antitumorigenic effect of Wnt 7a and Fzd 9 in non-small cell lung cancer cells is mediated through ERK-5-dependent activation of peroxisome proliferator-activated receptor gamma. J Biol Chem. 2006;281:26943–26950. doi: 10.1074/jbc.M604145200. [DOI] [PubMed] [Google Scholar]
- 203.Fukui T, Kondo M, Ito G, et al. Transcriptional silencing of secreted frizzled related protein 1 (SFRP 1) by promoter hypermethylation in non-small-cell lung cancer. Oncogene. 2005;24:6323–6327. doi: 10.1038/sj.onc.1208777. [DOI] [PubMed] [Google Scholar]
- 204.Mazieres J, He B, You L, et al. Wnt inhibitory factor-1 is silenced by promoter hypermethylation in human lung cancer. Cancer Res. 2004;64:4717–4720. doi: 10.1158/0008-5472.CAN-04-1389. [DOI] [PubMed] [Google Scholar]
- 205.Uematsu K, He B, You L, et al. Activation of the Wnt pathway in non small cell lung cancer: evidence of dishevelled overexpression. Oncogene. 2003;22:7218–7221. doi: 10.1038/sj.onc.1206817. [DOI] [PubMed] [Google Scholar]
- 206.Wissmann C, Wild PJ, Kaiser S, et al. WIF1, a component of the Wnt pathway, is down-regulated in prostate, breast, lung, and bladder cancer. J Pathol. 2003;201:204–212. doi: 10.1002/path.1449. [DOI] [PubMed] [Google Scholar]
- 207.Dang TP, Eichenberger S, Gonzalez A, et al. Constitutive activation of Notch3 inhibits terminal epithelial differentiation in lungs of transgenic mice. Oncogene. 2003;22:1988–1997. doi: 10.1038/sj.onc.1206230. [DOI] [PubMed] [Google Scholar]
- 208.Politi K, Feirt N, Kitajewski J. Notch in mammary gland development and breast cancer. Semin Cancer Biol. 2004;14:341–347. doi: 10.1016/j.semcancer.2004.04.013. [DOI] [PubMed] [Google Scholar]
- 209.Parr C, Watkins G, Jiang WG. The possible correlation of Notch-1 and Notch-2 with clinical outcome and tumour clinicopathological parameters in human breast cancer. Int J Mol Med. 2004;14:779–786. doi: 10.3892/ijmm.14.5.779. [DOI] [PubMed] [Google Scholar]
- 210.Hainaud P, Contreres JO, Villemain A, et al. The role of the vascular endothelial growth factor-Delta-like 4 ligand/Notch4-ephrin B2 cascade in tumor vessel remodeling and endothelial cell functions. Cancer Res. 2006;66:8501–8510. doi: 10.1158/0008-5472.CAN-05-4226. [DOI] [PubMed] [Google Scholar]
- 211.Von Hoff DD, LoRusso PM, Rudin CM, et al. Inhibition of the hedge hog pathway in advanced basal-cell carcinoma. N Engl J Med. 2009;361:1164–1172. doi: 10.1056/NEJMoa0905360. [DOI] [PubMed] [Google Scholar]
- 212.Luistro L, He W, Smith M, et al. Preclinical profile of a potent gamma-secretase inhibitor targeting notch signaling with in vivo efficacy and pharmacodynamic properties. Cancer Res. 2009;69:7672–7680. doi: 10.1158/0008-5472.CAN-09-1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Wei P, Walls M, Qiu M, et al. Evaluation of selective gamma-secretase inhibitor PF-03084014 for its antitumor efficacy and gastrointestinal safety to guide optimal clinical trial design. Mol Cancer Ther. 2010;9:1618–1628. doi: 10.1158/1535-7163.MCT-10-0034. [DOI] [PubMed] [Google Scholar]
- 214.Campbell PJ, Stephens PJ, Pleasance ED, et al. Identification of somatically acquired rearrangements in cancer using genome-wide massively parallel paired-end sequencing. Nat Genet. 2008;40:722–729. doi: 10.1038/ng.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Pleasance ED, Stephens PJ, O’Meara S, et al. A small-cell lung cancer genome with complex signatures of tobacco exposure. Nature. 2010;463:184–190. doi: 10.1038/nature08629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216. [Accessed October 22, 2011];The Cancer Genome Atlas. Available at: http://cancergenome.nih.gov/
- 217. [Accessed October 22, 2011];Lung Cancer Mutation Consortium. Available at: http://www.golcmc.com/
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.