Abstract
Motivation: In recent years, several biomedical event extraction (EE) systems have been developed. However, the nature of the annotated training corpora, as well as the training process itself, can limit the performance levels of the trained EE systems. In particular, most event-annotated corpora do not deal adequately with coreference. This impacts on the trained systems' ability to recognize biomedical entities, thus affecting their performance in extracting events accurately. Additionally, the fact that most EE systems are trained on a single annotated corpus further restricts their coverage.
Results: We have enhanced our existing EE system, EventMine, in two ways. First, we developed a new coreference resolution (CR) system and integrated it with EventMine. The standalone performance of our CR system in resolving anaphoric references to proteins is considerably higher than the best ranked system in the COREF subtask of the BioNLP'11 Shared Task. Secondly, the improved EventMine incorporates domain adaptation (DA) methods, which extend EE coverage by allowing several different annotated corpora to be used during training. Combined with a novel set of methods to increase the generality and efficiency of EventMine, the integration of both CR and DA have resulted in significant improvements in EE, ranging between 0.5% and 3.4% F-Score. The enhanced EventMine outperforms the highest ranked systems from the BioNLP'09 shared task, and from the GENIA and Infectious Diseases subtasks of the BioNLP'11 shared task.
Availability: The improved version of EventMine, incorporating the CR system and DA methods, is available at: http://www.nactem.ac.uk/EventMine/.
Contact: makoto.miwa@manchester.ac.uk
1 INTRODUCTION
The focus of text mining systems in biomedicine has recently shifted from extracting named entities of biological relevance to recognizing relations and events. Biomedical event extraction (EE) systems have already been integrated with a number of applications, such as semantic search, association mining for knowledge discovery, bioprocess extraction and pathway reconstruction (Ananiadou et al., 2010; Björne et al., 2010; Tsuruoka et al., 2011; Wang et al., 2011). The increasing requirement for high performance EE systems has motivated the development of several event annotated corpora to facilitate their training [e.g. GENIA (Kim et al., 2008), BioInfer (Pyysalo et al., 2007), GREC (Thompson et al., 2009), BioNLP Shared Tasks 2009 (ST09) (Kim et al., 2011a) and 2011 (ST11) (Kim et al., 2011b) corpora]. Events are structured descriptions of biological processes involving complex relationships (e.g. angiogenesis, metabolism and reactions) between biomedical entities, and are highly dependent on context. Events usually consist of triggers, which are often verbs (e.g. inhibit) or nominalizations (e.g. inhibition), and their typed arguments, which can be biomedical entities (e.g. gene) or other events (e.g. Regulation).
Typically, EE systems find both triggers and associated arguments, which can present a number of challenges. Triggers are expressed in diverse ways and their exact interpretation can depend upon context, e.g. ‘expression of [gene]’ is an event of type Gene Expression, but ‘expression of [mRNA]’ is of type Transcription. Furthermore, event arguments can be difficult to detect; since triggers are sublanguage dependent, their exact syntactic arguments are dictated by the domain. EE systems are thus highly dependent on the availability of training sets and external resources (e.g. dictionaries) that can expand the trigger candidates in the training set with semantically similar alternatives. Generally, machine learning methods are applied to syntactic parse results to disambiguate event types according to their surrounding contexts and to identify relations between the syntactic and semantic arguments of triggers.
There are two important issues that are scarcely dealt with by existing EE systems. First, the usual method of training on only a single annotated corpus can limit the coverage and scalability of the system. Secondly, coreference resolution (CR) is required for the correct interpretation of certain event arguments. This is illustrated in Figure 1, in which there are two coreferential links. The first link involves the mention this gene and its antecedent jun-B, whereas the second concerns the mention that and its antecedent expression. There are two gene expression events in the sentence, i.e. jun-B expression and jun-C expression, which can only be correctly recognized if these coreferential links are identified and resolved.
Fig. 1.

Coreference example. Two coreferential links are illustrated
This article reports on two enhancements that have been made to an existing EE system, EventMine (Miwa et al., 2010b), to address the issues outlined above. First, we constructed a new CR system, whose performance in resolving anaphoric references to genes and proteins considerably exceeds that of the best participating system in the Protein Coreference task (COREF) of ST11, which focussed on protein/gene name CR. Subsequent integration of our new CR system with EventMine clearly demonstrates an improvement in state-of-the-art EE performance on several event corpora. Secondly, our incorporation of domain adaptation (DA) methods into EventMine, which make it possible to use information from multiple annotated corpora when training the system, have been shown to further boost EE performance. The enhanced version of EventMine, incorporating both CR and DA, outperforms the highest ranked systems participating in ST09, and the GENIA and Infectious Diseases (ID) subtasks of ST11.
2 RELATED WORK
2.1 Biomedical EE
Driven by an increasing interest in biomedical EE, event corpora such as GENIA, BioInfer and GREC have been complemented by community shared tasks (STs), i.e. ST09 and ST11, which have provided standard evaluation benchmarks for EE. In ST09, the main task (GE09) focussed on event types relating to protein biology in the GENIA corpus. ST11 included several tasks: GE11, an extension of GE09, incorporated full text annotations in addition to abstracts, EPI [EPIgenetics and post-translational modifications (PTM)] concerned events relating to epigenetic change, and ID dealt with infectious diseases in full texts. Biomedical events in all ST tasks consist of: event types, e.g. Binding and Regulation; trigger expressions, e.g. bind and expression; arguments with entity types assigned, e.g. p53: Protein; argument roles (trigger-argument relations), e.g. Theme and Site; and finally information relating to event modification, e.g. negation and speculation. Figure 2 illustrates the event representation used in GE09.
Fig. 2.
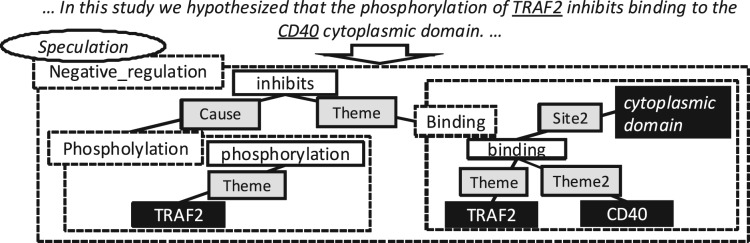
ST event representation. Events are represented as dotted rectangles with event types. Within events, solid rectangles represent participants, whereas trigger expressions are shown with a white background, argument roles with a grey background and arguments with a black background. The oval denotes an event modification, in this case speculation
State-of-the-art EE systems use a number of machine learning methods, including pipeline approaches (Björne and Salakoski, 2011; Miwa et al., 2010b), dual decomposition-based models (Riedel and McCallum, 2011), stacking-based multiple model integration approaches (Riedel et al., 2011) and search-based models (Vlachos and Craven, 2011).
2.2 Use of external resources in EE
Four machine learning-based and two rule-based systems were built to approach the COREF task. (Kim et al., 2011c) adapted a machine learning-based CR system originally developed for newswire text, by disabling several domain-specific features. This system achieved the best performance of all participating systems (i.e. 35% F-Score for protein/gene name CR). (Tuggener et al., 2011) developed a rule-based system that used a salience measure based on the output of a dependency parser. Since the general performance of all the CR systems was low, none of the ST11 participants made use of CR results to assist with the EE process.
Independently of the COREF task, (Yoshikawa et al., 2011) trained a CR system on the event-focussed coreference annotation in GENIA. They incorporated the CR results into their argument role detection system (based on Markov logic networks), which resulted in an improvement in the performance of role detection. However, the effect of CR on event recognition performance and the use of corpora other than GENIA are still open problems.
DA methods have rarely been applied in the context of EE. (Riedel and McCallum, 2011) used instance-based DA to tackle the ID task, resulting in the highest performance among the ST participants. They supplemented event instances from the ID corpus with instances from the GE11 corpus, after tuning the combination settings on the development set. (Vlachos and Craven, 2011) applied feature-based DA to the GE11 task, which improved the F-Score by 0.41% on the test set. They incorporated additional features specific to the target text types, i.e. abstracts and full texts, into their system. DA thus constitutes a promising means to improve EE results, although further research is required to investigate how best to transfer information from different corpora to the target corpus.
3 EVENT EXTRACTION
In Section 3.1, we provide an overview of our EE system, EventMine, whereas in Section 3.2, we explain several general modifications that have been made.
3.1 EventMine
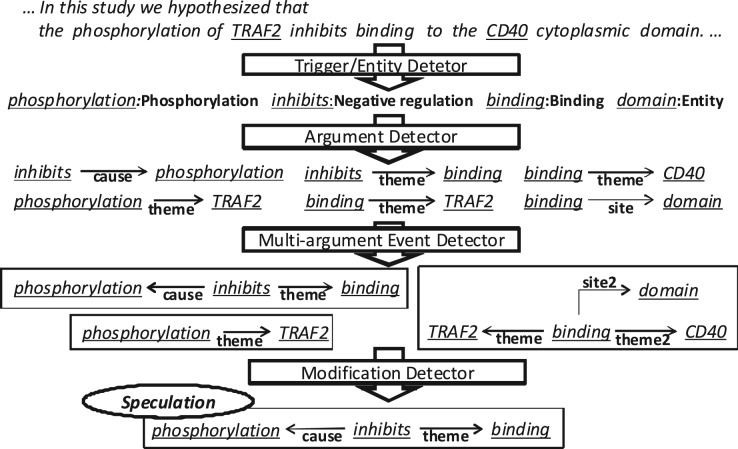
EventMine is a pipeline-based EE system, which extracts events from documents that already contain named entity annotations (e.g. Protein). Figure 3 illustrates the four different modules of the system, i.e. the trigger/entity detector, argument detector, multi-argument event detector and modification detector, together with sample outputs of each module. The trigger/entity detector identifies words (i.e. event triggers and entities) that are potential fragments of an event. Each word is assigned either an event type (e.g. Phosphorylation) if it is an event trigger, or otherwise an entity type (e.g. Entity). The categories assigned by this detector can be collapsed into more general types (e.g. Positive/Negative Regulation, Regulation → REGULATION) to increase the number of training instances and to facilitate the application of DA methods (see Section 4.3). The argument detector identifies possible trigger-argument pairs. Each argument can be either an entity or the trigger of another event, and is assigned a semantic role type (e.g. Theme and Cause). The multi-argument event detector combines multiple trigger-argument pairs found by the argument detector to create complete event structures, and assigns an event type to them. The modification detector assigns modification information (i.e. negation and speculation) to each event. Each module solves multi-class multi-label classification problems by applying a one-versus-rest SVM (Fan et al., 2008) to the output of the preceding module in the pipeline (except for the trigger/entity detector).
Fig. 3.
EventMine EE pipeline. Documents used as input to the system must be pre-annotated with entities. In this case, TRAF2 and CD40 have already been identified as Protein
EventMine uses five feature extraction functions to extract features representing a word or pair of words, together with their contexts. The word feature function extracts the surface representation of a word, including character types (e.g. number and symbol), n-grams (n=1; 2; 3; 4) of characters, base form and part-of-speech (POS). The neighbouring feature function extracts all 2-step dependency paths from a word, represented by the features extracted by the word feature function, plus word and dependency n-grams (n=2; 3; 4), word n-grams (n=2; 3) and dependency n-grams (n=2), where each word is represented by its base form. The word n-gram feature function extracts n-grams (n=1; 2; 3; 4) of words within a window of three words before or after the target word. Each word is represented by its base form, POS and its relative position (before or after the word). The pair n-gram feature function extracts n-grams (n=1; 2; 3; 4) of words within a window of three words before/after the first/last word in the pair. Each word is represented by its base form, POS and its relative position (before, between or after the pair). The shortest path feature function extracts the shortest paths between a pair of words, represented by the path length, word n-grams (n=2; 3; 4), dependency n-grams (n=2; 3; 4), consecutive word n-grams (n=1; 2; 3) representing governor-dependent relationships, and edge walks (word-dependency-word), vertex walks (dependency-word-dependency) and their sub-structures. Each word is represented by its base form.
These functions are combined in different ways to make features for each module, as shown in Table 1. Features are normalized using the L2-norm (Graf et al., 2003), both at the level of the feature extraction functions, and globally. EventMine accounts for the output of multiple syntactic parsers by extracting features from each parser's output separately. Following (Miwa et al., 2010b), we have used both the Enju parser (Miyao et al., 2009) and the GDep parser (Sagae and Tsujii, 2007).
Table 1.
Features for trigger/entity, argument, multi-argument event and modification detectors
| Detector | Type | Function |
|---|---|---|
| Trigger/entity | Target candidate | word |
| Words around candidate | word n-gram | |
| Path between candidate and all NEs | shortest path | |
| Arg. | Terminal nodes of candidate pair | word |
| Words around candidate pair | pair n-gram | |
| Path between candidate pair | shortest path | |
| Path between argument trigger and its closest NE | shortest path | |
| Confidences assigned to terminal nodes found by trigger/entity detector | – | |
| Multi-arg. | Included trigger-argument pairs | arg. detector |
| event | All pairs among arguments | arg. detector |
| All pairs sharing trigger outside of candidate event | arg. detector | |
| Confidences assigned to included pairs found by arg. detector | – | |
| Mod. | Trigger | neighbouring |
| Included trigger-argument pairs | pair n-gram |
3.2 Generalization and modification of EventMine
Prior to incorporating CR and DA into a EventMine, several modifications were made to its core functionality, both to enhance its performance and to increase its applicability to a wide variety of EE tasks.
First, the system was generalized, so that training can be carried out on different annotated corpora, which may contain a variety of event types and structures. Since each module is machine learning based, this modification only required a small amount of effort. Secondly, since the system uses several rich feature extraction functions, the memory usage is potentially high. Feature Hashing (FH) has been incorporated to map the features used by the system to a smaller dimension (limited to 220 features) by using a hash function (Shi et al., 2009), which results in a minimal decrease in performance (0.1% F-Score). By incorporating FH, the memory usage associated with constructing models for GE09 is reduced by three-quarters. Thirdly, cross validation (CV) is utilized to train each module. A pipeline model tends to overfit the training set by using it several times. To reduce this effect, each module is trained using the predictions made within the test folds of a 10-fold CV run on the previous module in the pipeline. The same partitioning of the data are used for CV in every module. CV reduces the amount of data available to train the subsequent modules, and is thus ineffective when there are only a small number of training instances. Fourthly, trigger filtering (TF) is used to help to reduce computation time, by selecting only those trigger/entity candidates whose base form matches that of the head word of an annotation in the training set. TF halves the time required to construct models for GE09. Fifthly, dictionary-based trigger expansion (DTE) increases the features that are shared among the training instances and alleviates the problem of unknown words. Using dictionaries, DTE expands trigger/entity candidates with related words (e.g. synonyms) and also expands the word feature function by adding words related to a candidate as additional features. Related words were found via the ‘hypernyms’ and ‘similar to’ relations in WordNet (Fellbaum, 1998), in addition to transcategorial operations (e.g. degrade→degradation) and synonyms in the UMLS Specialist Lexicon (Bodenreider, 2004). Sixthly and finally, a simplified version of the trigger/entity detector (STD) reduces computation time by omitting a feature extraction step from the original system (Miwa et al., 2010b), which was found to have no effect on overall performance.
4 INCORPORATING EXTERNAL RESOURCES INTO EE
In this section, we describe the two types of external resources that have been incorporated within EventMine. Our new CR system is described in Section 4.1, whereas its integration with EventMine is explained in Section 4.2. In Section 4.3, we outline the DA methods that have been used to incorporate information from external annotated corpora into EventMine.
4.1 Rule-based coreference resolution system
Our novel rule-based CR system identifies coreferential links between genes and proteins, i.e. mentions and their antecedents. Using the COREF task training data, a set of rules was developed based on the output of the Enju parser, which consists of syntactic trees and predicate-argument structures. This is in contrast to the dependency parse results used by (Tuggener et al., 2011). Three rule-based detectors are used, i.e. a mention candidate detector, an antecedent candidate detector and a coreferential link detector.
The mention candidate detector identifies words and phrases that are potential mentions of genes and proteins. First, a full list of mention candidates is extracted, consisting of all noun phrases (NPs) with articles (e.g. these transcription factors), pronouns (e.g. they), possessive pronouns (e.g. its), relative pronouns (e.g. which and whose) and complementizers (e.g. that). Secondly, a filter is applied to remove mention candidates that are unlikely to refer to genes and proteins, i.e. NPs without the, this, that, these or those as their articles (e.g. our method), NPs whose head word is not one of the three most frequent head words in the training set (i.e. protein, gene and factor), NPs that contain words other than head words, articles and quantifiers (e.g. the three interacting proteins), pleonastic pronouns (e.g. it in ‘it is clear that …’) and personal pronouns except for it and they (e.g. we). This filtering step causes a large reduction in the number of false positive mentions found by the system, with only a minimal loss of true positive mentions. It also simplifies the detection of coreferential links.
The antecedent candidate detector selects all NPs apart from: full mention candidates found by the mention candidate detector, those containing sentence clauses and those sharing their head words with other larger NPs.
The coreferential link detector links each mention to its most suitable antecedent candidate. For complementizers and relative clauses, the parser output is used. For definite NPs and pronouns, a set of rules is applied to rank potential antecedents, and the top ranked antecedent in the list is linked to the mention. For each pair of possible antecedents, the following rules [motivated by (Raghunathan et al., 2010)] are applied, in the sequence indicated, until a particular rule distinguishes a more likely antecedent: Do both antecedents precede the mention? Is one of the antecedent-containing sentence clauses closer to clause containing the mention than the other? Do both of the antecedents share grammatical number with the mention? Are both antecedents not i-within-i (antecedents cannot be children of mentions, and vice versa)? Do both antecedents share head words with the mention? If both antecedents occur in the same sentence clause as the mention, which occurs closest to the mention? If both antecedents occur in a single clause, but not in the same clause as the mention, which is closest to the mention?
4.2 Incorporation of coreference resolution
The output of our CR system is used in two ways in the enhanced version of EventMine. First, parse result modification (PR) modifies the original parse results so that mentions and their antecedents share dependencies, i.e. dependencies from/to mentions are added to antecedents, and vice versa. Figure 4 illustrates how parse results are changed through the application of PR. In the modified parse tree, the two coreferring entities become closer, in terms of their dependencies. Secondly, feature extension (FE) extends the set of features used during EE, by making use of the CR results. Specifically, features of genes/proteins detected as events argument are extended by including features associated with all of their coreferential mentions. The latter modification helps the system to find explicit relations between participants.
Fig. 4.

Parse result modification (PR) using the CR output. In addition to the dependencies identified in the original parser output (upper solid arrows), shared dependencies are generated between the mention its and its antecedent SLP-76 (lower dotted arrows)
4.3 Domain adaptation with external corpora
The improved EventMine uses two DA methods to allow the information annotated in the main training corpus (target corpus) to be supplemented with information from other annotated corpora (source corpora).
The first DA method is instance weighting (IW). If there are annotated source (src) corpora whose event types and possible arguments match those in the target (tgt) corpus, event instances from the source corpora could be added to the training instances from the target corpus to increase the amount of training data. However, the EE results could become skewed if the distributions of positive and negative instances are different in the source and target corpora. To reduce the potential skewing effect, different weights are assigned to the positive (+) and negative (−) instances in the source and target corpora by modifying the objective function as follows.
| (1) |
Here, ℓ represents the loss function in SVM, l represents labels, c represents corpora and Cs represent the weights for the instances. The ratio of positive to negative instances in the source corpora is made consistent with the ratio in the target corpus, by setting Cs to satisfy C−srcn−srcC+srcn+src=C−tgtn−tgt: C+tgtn+tgt. The conditions C−tgt=1 and C+tgt=C+src are used for simplicity, and ensuring that C−src·n−src=C+src·n+src eases problems caused by an imbalanced distribution of positive and negative examples.
The second DA method concerns the use of a stacking model (SM). If a source corpus shares only a subset of its event types and arguments with the target corpus, then it is not appropriate to add training instances directly from the source corpus, because positive instances in the source corpora could correspond to negative instances in the target corpus. Instead, each module of EventMine can be trained separately on the source corpus, and its output can be incorporated as additional features within the same module trained on the target corpus. Also, since the multi-argument event detector shares features with the argument detector, as detailed in Table 1, the outputs of argument detectors trained on different source corpora can provide additional features to the multi-argument event detector trained on the target corpus.
5 EVALUATION
Our CR system has been evaluated on the COREF task data, whereas the enhanced EventMine has been evaluated on four different tasks, i.e. GE09, GE11, EPI and ID. The task and evaluation settings are described in Sections 5.1 and 5.2. We then provide the evaluation results in Section 5.3, compare the results with other systems in Section 5.4, and analyze errors in the results in Section 5.5.
5.1 Corpora and task settings
GE09/11 both deal with nine event types involving proteins, consisting of five simple event types (e.g. Gene Expression and Phosphorylation), one Binding type (Binding) and three Regulation types (Positive/Negative Regulation and Regulation). Simple event types require a single core Theme argument, Binding events require an arbitrary number of core Theme arguments and Regulation events can include recursive event structures. EPI defines 15 event types relating to Protein, consisting of seven simple event types (e.g. Acetylation, Phosphorylation and Methylation), their inverse event types (e.g. Deacetylation is the inverse of Acetylation) and one Regulation event type (Catalysis, a subtype of Positive Regulation). The event types in EPI are largely disjoint from those in the other corpora. ID covers events relating to five named entities (e.g. Protein Chemical and Organism) and defines the same event types as GE09/11, with the addition of Process, which requires no arguments. All the tasks cover secondary arguments (i.e. Site/Location arguments) for certain event types and also modifications (i.e. negation and speculation). Table 2 provides some statistics regarding the annotated corpora used in each task.
Table 2.
Statistics for training and development sets
| Corpus | Abstracts | Full texts | Sentences | Events/coref. links |
|---|---|---|---|---|
| COREF | 950 | 0 | 7982 | 2786 |
| GE09 | 950 | 0 | 7982 | 10 410 |
| GE11 | 950 | 10 | 10 761 | 13 560 |
| EPI | 800 | 0 | 7827 | 2452 |
| ID | 0 | 20 | 3412 | 2679 |
In the last column, the number of coreferential links is shown for COREF, and the number of events is shown for the other corpora.
GE09/11 defined three tasks: Task 1 required the identification of core arguments (Theme and Cause), Task 2 was concerned with finding secondary arguments, and Task 3 required the detection of modifications on Task 1 events. Our evaluation concerns Task 1, since it was undertaken by all participating systems. EPI and ID defined both a core task (the same as Task 1 in GE09/11), and a full task, which additionally required modifications and secondary arguments to be found. We focus on the full task, since its results were considered as the primary evaluation metric for EPI and ID.
5.2 Evaluation settings
Each corpus was split into sentences by the GENIA sentence splitter (Sætre et al., 2007), and was subsequently parsed by both the Enju 2.4.1 parser with the GENIA model, and the GDep beta2 parser. The CR system used the Enju parse results, whereas EventMine used the results of both parsers. Liblinear-java (Fan et al., 2008; http://www.bwaldvogel.de/liblinear-java/) was used for classification, with the bias term set.
Certain semantic types were generalized following trigger/entity detection, to increase both the number of training instances and the number of event representations that were shared among corpora, as explained in Section 3.1. In terms of triggers, all the Regulation event types were collapsed into a single type, REGULATION, whereas Phosphorylation in GE09/11 and ID, and simple event types in EPI, were generalized as PTM. In terms of arguments, all event and entity types were generalized as EVENT and ENTITY, respectively.
Of the newly introduced methods described in Section 3.2, FH, TF and STD were utilized for all tasks. CV and DTE were used for all tasks except EPI. CV considerably reduced the recall for this task, due to the small size of the EPI corpus, and the incorporation of DTE did not improve the performance. The version of EventMine that incorporates these methods is referred as to ‘the base system’.
Unless otherwise stated, the evaluation metrics follow those originally defined for each task. Each task provided training, development and test sets. Following (Björne and Salakoski, 2011), EventMine was first trained on the training set for each task, and its performance was evaluated on the development set, to determine the optimal settings (e.g. CV and DA). Subsequently, the system was trained on both the training and development sets using the optimal settings, and then evaluated on the test set using the evaluation systems provided by the ST organizers.
5.3 Evaluation of EventMine with external resources
Table 3 shows the performance of our CR system on the COREF task data. The results of the best performing system in the the original COREF task evaluation (Kim et al., 2011c) are also shown, demonstrating that our system performs significantly better. The results obtained on the development set, using both exact matching and head word matching (the latter are shown in parentheses) revealed that our mention detector found 81.4% (81.4%) of gold standard annotated mentions, our antecedent detector found 57.1 (99.6%) of gold standard antecedents, and the coreferential link detector found 70.2% (79.7%) of gold standard links between mentions and antecedents. In comparison, the recall of (Kim et al., 2011c) is much lower than ours in detecting exact antecedents (41.2%), but marginally higher in detecting mentions (85.1%).
Table 3.
Performance of rule-based CR systems on the development and test sets of the COREF task
| Recall | Precision | F-Score | |
|---|---|---|---|
| Development | 53.5 | 69.8 | 60.5 |
| Test | 50.4 | 62.7 | 55.9 |
| Test (Kim et al., 2011c) | 22.2 | 73.3 | 34.1 |
Recall, Precision and F-Score were evaluated according to the protein evaluation criteria of the COREF task. The best performing system participating in the original COREF evaluation is shown for reference.
The positive effects of incorporating CR into the EE pipeline when EventMine is trained on the GE09 data are summarized in Table 4. Both CR integration methods introduced in Section 4.2, i.e. PR and FE, improved event recognition performance for all event types. The best results are achieved when the two methods are combined. Following (Kim et al., 2011a), we performed a statistical significance test on the development set, using the approximate randomization method. The difference between the best results obtained using the CR integration methods (+PR+FE; 58.81%) and the results obtained using the original version of EventMine (EM10) (Miwa et al., 2010a), 57.22%, is statistically significant ( p<0.05). Although we cannot run a significance test on the test set, since gold standard annotation was not provided, the report in (Kim et al., 2011a) strongly suggests that the 1.99% difference in F-Score between the results of +PR+FE (58.27%) and EM10 (56.28%) when applied to the test set is statistically significant.
Table 4.
EE performance on the development (dev) and test sets of GE09, incorporating the CR results
| SVT | BIND | REG | TOT | ||||
|---|---|---|---|---|---|---|---|
| F | F | F | R | P | F | ||
| Dev | Base | 79.41 | 49.18 | 46.78 | 54.28 | 62.62 | 58.15 |
| +PR | 78.60 | 50.92 | 47.32 | 55.00 | 62.10 | 58.34 | |
| +FE | 80.31 | 48.69 | 47.31 | 54.11 | 63.70 | 58.51 | |
| +PR+FE | 80.16 | 50.52 | 47.48 | 55.00 | 63.17 | 58.81 | |
| Test | +PR+FE | 73.55 | 59.91 | 45.99 | 52.67 | 65.19 | 58.27 |
| UMass | 72.6 | 52.6 | 46.9 | – | – | 57.4 | |
F-Scores are shown for Simple (SVT), Binding (BIND) and Regulation (REG) events, together with overall recall, precision and F-Scores for all events (TOT). The results of the best reported system for GE09, UMass (Riedel and McCallum, 2011), are shown for reference.
Table 5 summarizes the results obtained when both DA and CR are incorporated into the EE process, for each of the four event corpora introduced above. The DA methods introduced in Section 4.3 were only used when training EventMine on the EPI and ID corpora, since these corpora contain many fewer event instances than GE09/11 (Table 2). We did not use other event corpora (e.g. BioInfer and GREC) as source corpora, since they are much smaller than GE09/11, with different event types and representations. For the ID task, IW was used, with the GE11 training and development sets as the source corpus, since ID and GE11 share all event types except for Process. For the EPI task, SM was adopted, using the ID model trained with GE11 as the source model, since EPI shares only the Phosphorylation and Catalysis event types with the other corpora. The use of these DA methods resulted in improvements over the base system of 1.3% F-Score for EPI, and 1.8% for ID. These results are promising, given that EPI shares few event types with the source model and that ID contains entities with different semantic classes to those in GE11. For comparative purposes, simple addition of instances from GE11, in combination with the +PR+FE setting, achieved an F-Score of 50.58% for the ID task, which is 0.66% lower than when IW is used (51.24%). This reveals that the distributions of GE11 and ID are similar, but not identical. A further experiment involved applying IW to GE11, with ID as the source corpus. However, this caused a 1.1% drop in F-Score, as GE11 does not contain the event type Process or named entities other than Protein. Table 5 also shows that by supplementing DA methods with CR results, EE performance can be further improved. A larger improvement can be observed for GE11 and ID (which both include full texts) than for GE09 and EPI. This seems reasonable, given that event descriptions in full texts are less restricted by space than in abstracts, resulting in more frequent occurrences of coreference.
Table 5.
F-Scores achieved through application of EventMine to the development sets of all corpora
| Corpus | Base | +DA | +PR +FE +DA |
|---|---|---|---|
| GE09 | 58.15 | – | 58.81 (+0.66) |
| GE11 | 55.67 | – | 56.73 (+1.06) |
| EPI (+ID (+GE11)) | 50.96 | 52.26 (+1.30) | 52.39 (+0.13) |
| ID (+GE11) | 47.88 | 49.64 (+1.76) | 51.24 (+1.60) |
The performance of the base system is compared with versions of the system incorporating DA and additionally CR (+PR+FE).
5.4 Comparison with other systems
At the bottom of Table 4, we compare the performance of EventMine (with the +PR+FE configuration) on the GE09 task with the highest performing system on the task, i.e. UMass (Riedel and McCallum, 2011), which uses a dual-decomposition-based joint model for EE. Tables 6 and 7 provide a comparison of EventMine with the two top performing systems in ST11, i.e. FAUST (Riedel et al., 2011) and UTurku (Björne and Salakoski, 2011), on the GE11, EPI and ID tasks. FAUST is an extension of UMass, which uses the output of a dependency-parsing-based EE system, whereas UTurku is a pipeline-based system, whose modules are similar to those of EM10, but they use different set of features. For all three ST11 tasks, EventMine uses the +PR+FE+DA configuration, which achieved the best results on the development sets, as shown in Table 5.
Table 6.
Overall recall/precision/F-Scores achieved for EE on the ST11 test sets
| System | GE11 Task 1 | EPI | ID |
|---|---|---|---|
| EventMine | 53.35/63.48/57.98 | 49.06/55.39/52.03 | 60.55/54.97/57.63 |
| FAUST | 49.41/64.75/56.04 | 28.88/44.51/35.03 | 48.03/65.97/55.59 |
| UTurku | 49.56/57.65/53.30 | 52.69/53.98/53.33 | 37.85/48.62/42.57 |
Primary evaluation criteria are employed. Results for two top systems participating in the original evaluation, FAUST (Riedel et al., 2011) and UTurku (Björne and Salakoski, 2011) are shown for reference. The highest scores are shown in bold.
Table 7.
Detailed EE F-Scores achieved on the ST11 test sets
| EventMine | FAUST | UTurku | |
|---|---|---|---|
| GE11 simple | 76.01 | 73.90 | 72.11 |
| GE11 binding | 56.64 | 48.49 | 43.28 |
| GE11 regulation | 45.46 | 44.94 | 42.72 |
| GE11 full texts | 58.13 | 52.67 | 50.72 |
| GE11 abstracts | 57.92 | 57.46 | 54.37 |
| GE11 Task 1 (core arguments) | 57.98 | 56.04 | 53.30 |
| GE11 site | 56.35 | 44.92 | 49.72 |
| GE11 location | 47.42 | 48.98 | – |
| GE11 Task 2 (secondary arguments) | 54.47 | 45.86 | 37.96 |
| GE11 Task 3 (modification) | 26.24 | – | 26.86 |
| EPI catalysis | 28.76 | 6.58 | 7.06 |
| EPI full task | 52.03 | 35.03 | 53.33 |
| EPI core task | 67.52 | 68.59 | 68.86 |
| EPI modification | 30.61 | – | 28.07 |
| ID simple | 61.12 | 68.47 | 62.67 |
| ID binding | 31.50 | 31.30 | 22.22 |
| ID process | 70.57 | 65.69 | 41.57 |
| ID regulation | 47.28 | 47.07 | 39.49 |
| ID full task | 57.63 | 55.59 | 42.57 |
| ID core task | 59.15 | 57.57 | 43.93 |
| ID modification | 17.48 | – | 26.89 |
The highest scores are shown in bold.
The tables show that EventMine is the only system that can achieve state-of-the-art performance on all corpora and with different types of events. The relatively small differences between the results for GE09 (Table 5) and GE11 abstracts (Table 7) demonstrate that the general performance of EventMine remains constant, even when the training data includes full texts as well as abstracts. The high level of performance on full texts (GE11 full texts and ID, see Table 7) was partly due to the integration of CR, which has been shown to be effective on full texts in the development sets (Table 5). EventMine also performed particularly well on events with multiple arguments, such as Binding and Regulation, since the multi-argument event detector is specifically tailored to detecting of events with multiple arguments, as described in Section 3.1. EventMine slightly underperformed other systems in the detection of modifications. The performance was affected both by the use of CV, which reduced the upper bound of the recall, and the lower number of training examples than for other modules. For instance, although the use of CV with the base system resulted in an improvement in F-Score of 1.4% for EE, it also reduced the performance of modification detection by 5.8 F-Score for the GE09 task. Modification detection in the ID task was also affected by DA, since the frequency of modifications in the GE11 corpus is three times larger than those found in the ID corpus.
5.5 Error analysis
We analysed 100 errors (56 missing events and 44 incorrect events) produced by the system (+PR+FE) when it was applied to the development set for Task 1 in GE11. Missing events are cases where the system was unable to find gold standard event structures, causing false negatives (FNs). The trigger/entity, argument and multi-argument event detectors missed 22, 28 and 6 events, respectively. Incorrectly recognized events correspond to events that either only partially match manually annotated gold standard events, or else are spuriously recognized. These caused both false positives (FPs) and FNs. Of the 44 incorrect events, 11 were assigned the wrong event type, 19 contained incorrect arguments, whereas the remaining 14 were completely spurious, which caused FPs.
We found two major problems, which require novel solutions. The first problem involves inference. As an example, consider the expression affect in ‘A affected B’, which could correspond to any of the three Regulation event types, if information that can be inferred from surrounding events or facts is not taken into account. The second problem is concerned with missing semantic types of NPs. For example, the trigger expression express is annotated as Gene Expression in ‘the fetal genes are expressed’ but as Positive Regulation in ‘expressing the fetal developmental program’. Both cases have fetal as an event argument, which is annotated as Protein. However, genes and program are not annotated with their semantic types. Assigning semantic types to these NPs could help with the disambiguation of the event types.
6 CONCLUSION
This article has reported on a number of enhancements to the EventMine EE system. First, several methods were incorporated to increase both the generality and efficiency of the system. Secondly, a newly constructed, rule-based CR system was integrated into EventMine, and DA methods were applied to utilize information from multiple annotated corpora. The CR system considerably outperformed systems that participated in the GE11 COREF task. The incorporation of CR and DA into the EE process significantly improved the performance of EventMine on four different event-annotated corpora, demonstrating the adaptable nature of the system. The results surpass those of systems that participated in the GE09 task of ST09 and the GE11 and ID tasks of ST11.
As future work, we will explore further novel ways of integrating external resources, by reducing the effect of differences among corpora. We will also investigate how EventMine can be embedded into other applications, e.g. semantic search engines and pathway curation and reconstruction systems.
Funding: Biotechnology and Biological Sciences Research Council (BBSRC; BB/G013160/1).
Conflict of Interest: none declared.
REFERENCES
- Ananiadou S., et al. Event extraction for systems biology by text mining the literature. Trends Biotechnol. 2010;28:381–390. doi: 10.1016/j.tibtech.2010.04.005. [DOI] [PubMed] [Google Scholar]
- Björne J., Salakoski T. Proceedings of BioNLP Shared Task 2011 Workshop. Portland, OR: ACL; 2011. Generalizing biomedical event extraction; pp. 183–191. [Google Scholar]
- Björne J., et al. Complex event extraction at PubMed scale. Bioinformatics. 2010;26:i382–i390. doi: 10.1093/bioinformatics/btq180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodenreider O. The Unified Medical Language System (UMLS): integrating biomedical terminology. Nucleic Acids Res. 2004;32:D267–D270. doi: 10.1093/nar/gkh061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan R.-E., et al. LIBLINEAR: a library for large linear classification. J. Mach. Learn. Res. 2008;9:1871–1874. [Google Scholar]
- Fellbaum C. WordNet: An Electronic Lexical Database. Cambridge, MA: The MIT Press; 1998. [Google Scholar]
- Graf A., et al. Classification in a normalized feature space using support vector machines. IEEE Trans. Neural Netw. 2003;14:597–605. doi: 10.1109/TNN.2003.811708. [DOI] [PubMed] [Google Scholar]
- Kim J.-D., et al. Corpus annotation for mining biomedical events from literature. BMC Bioinform. 2008;9:10. doi: 10.1186/1471-2105-9-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.-D., et al. Extracting bio-molecular events from literature – the bionlp' 09 shared task. Comput. Intell. 2011a;27:513–540. [Google Scholar]
- Kim J.-D., et al. Proceedings of BioNLP Shared Task 2011 Workshop. Portland, OR: ACL; 2011b. Overview of BioNLP Shared Task 2011; pp. 1–6. [Google Scholar]
- Kim Y., et al. Proceedings of BioNLP Shared Task 2011 Workshop. Portland, OR: ACL; 2011c. The taming of reconcile as a biomedical coreference resolver; pp. 89–93. [Google Scholar]
- Miwa M., et al. Proceedings of COLING 2010. Beijing, China: Coling 2010 Organizing Committee; 2010a. Evaluating dependency representations for event extraction; pp. 779–787. [Google Scholar]
- Miwa M., et al. Event extraction with complex event classification using rich features. J. Bioinform. Comput. Biol. 2010b;8:131–146. doi: 10.1142/s0219720010004586. [DOI] [PubMed] [Google Scholar]
- Miyao Y., et al. Evaluating contributions of natural language parsers to protein-protein interaction extraction. Bioinformatics. 2009;25:394–400. doi: 10.1093/bioinformatics/btn631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyysalo S., et al. BioInfer: a corpus for information extraction in the biomedical domain. BMC Bioinform. 2007;8:50. doi: 10.1186/1471-2105-8-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghunathan K., et al. Proceedings of EMNLP 2010. Cambridge, MA: ACL; 2010. A multi-pass sieve for coreference resolution; pp. 492–501. [Google Scholar]
- Riedel S., McCallum A. Proceedings of EMNLP 2011. Edinburgh, Scotland, UK: ACL; 2011. Fast and robust joint models for biomedical event extraction; pp. 1–12. [Google Scholar]
- Riedel S., et al. Proceedings of BioNLP Shared Task 2011 Workshop. Portland, OR: ACL; 2011. Model combination for event extraction in BioNLP 2011; pp. 51–55. [Google Scholar]
- Sætre R., et al. Proceedings of the Second BioCreative Challenge Evaluation Workshop. Madrid, Spain: CNIO; 2007. AKANE System: protein-protein interaction pairs in BioCreAtIvE2 Challenge, PPI-IPS subtask; pp. 209–212. [Google Scholar]
- Sagae K., Tsujii J. Proceedings of the CoNLL Shared Task Session of EMNLP-CoNLL 2007. Prague, Czech Republic: ACL; 2007. Dependency parsing and domain adaptation with LR models and parser ensembles; pp. 1044–1050. [Google Scholar]
- Shi Q., et al. Hash kernels for structured data. J. Mach. Learn. Res. 2009;10:2615–2637. [Google Scholar]
- Thompson P., et al. Construction of an annotated corpus to support biomedical information extraction. BMC Bioinform. 2009;10:349. doi: 10.1186/1471-2105-10-349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuruoka Y., et al. Discovering and visualizing indirect associations between biomedical concepts. Bioinformatics. 2011;27:i111–i119. doi: 10.1093/bioinformatics/btr214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuggener D., et al. Proceedings of BioNLP Shared Task 2011 Workshop. Portland, OR: ACL; 2011. An incremental model for the coreference resolution task of bionlp 2011; pp. 151–152. [Google Scholar]
- Vlachos A., Craven M. Proceedings of BioNLP Shared Task 2011 Workshop. Portland, OR: ACL; 2011. Biomedical event extraction from abstracts and full papers using search-based structured prediction; pp. 36–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., et al. Automatic extraction of angiogenesis bioprocess from text. Bioinformatics. 2011;27:2730–2737. doi: 10.1093/bioinformatics/btr460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa K., et al. Coreference based event-argument relation extraction on biomedical text. J. Biomed. Semant. 2011;2(Suppl. 5):S6. doi: 10.1186/2041-1480-2-S5-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]