Abstract
Summary: We introduce BRAT-BW, a fast, accurate and memory-efficient tool that maps bisulfite-treated short reads (BS-seq) to a reference genome using the FM-index (Burrows–Wheeler transform). BRAT-BW is significantly more memory efficient and faster on longer reads than current state-of-the-art tools for BS-seq data, without compromising on accuracy. BRAT-BW is a part of a software suite for genome-wide single base-resolution methylation data analysis that supports single and paired-end reads and includes a tool for estimation of methylation level at each cytosine.
Availability: The software is available in the public domain at http://compbio.cs.ucr.edu/brat/.
Contact: elenah@cs.ucr.edu
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
Bisulfite sequencing (BS-seq) combined with next-generation sequencing (NGS) instruments enables genome-wide methylation analysis at a single base-resolution. Bisulfite treatment of DNA followed by PCR converts unmethylated cytocines to thymines and leaves methylated cytocines unchanged (Frommer et al., 1992). Bisulfite-treated sequenced reads have to be aligned to the reference genome, but the treatment introduces the computational challenge of mapping both Cs and Ts in a read to Cs in the genome.
The most successful methods for mapping short reads either use hashing or data structures based on the Burrows–Wheeler transform (Burrows and Wheeler, 1994) where the latter approach is considered to yield more time efficient solutions than the former. Although several tools are available for BS-seq data, most of them still use hashing (including RMAP-bs, SOAP, MAQ and BRAT). The fastest tools for mapping BS-seq reads are Bismark (Krueger and Andrews, 2011) and BS-seeker (Chen et al., 2010). Both employ the mapping tool Bowtie (Langmead et al., 2009) that internally uses the FM-index (Ferragina and Manzini, 2000) based on the Burrows–Wheeler transform. As a consequence, both tools are required to post-process the output of Bowtie to remove ambiguous reads or reads with too many mismatches. Bismark synchronizes instances of FM-indexes run in parallel, which takes a toll on time-efficiency. BS-seeker outputs the results of distinct instances into separate files during mapping and then post-processes mapping results, which demands extra storage for intermediate results. Bismark and BS-seeker can therefore require large amount of primary memory to complete the processing. Both tools support two distinct types of bisulfite libraries: the first type yields sequenced reads that are bisulfite-converted versions of two original genomic strands (Lister et al., 2009); the second type produces reads that correspond to four possible strands, as a byproduct of PCR step (Cokus et al., 2008). To support the second type of libraries, Bismark and BS-seeker align reads to four distinct FM-indexes. Even though a type-1 bisulfite library would require only two FM-indexes, Bismark builds four FM-indexes in parallel requiring 16 GB of memory for human genome (Bowtie-2 with offrate 4). On the other hand, BS-seeker's memory footprint depends directly on the size of the input file: it may require up to 15 GB of memory for ∼30 M 32 bp-long reads (the typical number of reads/lane for the Illumina Genome Analyzer). Additionally, BS-seeker currently does not support paired-end reads and allows a limited number of mismatches per read, which makes it unsuitable for longer reads. Table 1 in the Supplementary Material summarizes the features of all the available tools for BS-seq data.
Table 1.
Comparing the efficiency of several BS-seq mapping tools
| Options | Time | RAM (GB) | Mapped reads (%) | ||
|---|---|---|---|---|---|
| 32 bp | Bismark | bowtie1, best, k=2, n=1, l=32, q | 94 m 26 s | 14.7 | 61.3 |
| BS-seeker | best, k=2, n=1 | 110 m 55 s | 15.0 | 64.2 | |
| BRAT | bs, m=1, S | 190 m 57 s | 2.9 | 61.2 | |
| BRAT-BW | S=16, C, F=1, m=1 | 99 m 23 s | 6.4 | 65.9 | |
| 62 bp | Bismark | bowtie1, best, k=2, l=32, n=1, e=150 | 158 m 22 s | 14.7 | 73.2 |
| BS-seeker | best, k=2, e=64, m=3 | 317 m 0 s | 14.0 | 72.4 | |
| BRAT | S, m=3, bs | 330 m 2 s | 2.9 | 68.7 | |
| BRAT-BW | S=16, m=3 | 104 m 54 s | 6.4 | 73.6 |
In this article we introduce BRAT-BW, a fast and accurate mapping tool that uses a very memory-efficient implementation of the FM-index. BRAT-BW is an evolution of BRAT (Harris et al., 2010), which uses about half as much memory compared with BS-seeker and Bismark. Additionally, its memory footprint does not depend on the size of the input sequenced reads, likely to continue to increase with future sequencing technologies advances. BRAT-BW supports both types of bisulfite libraries and handles single-end and paired-end reads. It has no limitation on the maximum length of the read or the number of allowed mismatches. BRAT-BW guarantees to find all matches as long as they have at most one mismatch in a prefix of length 32–64 bp (user defined) of the read.
There are several advantages of designing a tool for BS-seq data based on the FM-index from the ‘ground-up’ instead of relying on a general-purpose tool such as Bowtie. BRAT-BW processes both FM-indexes on a single processor, so no synchronization cost is required. In addition, the selection of correctly mapped unique reads is performed ‘on the fly’ during mapping, so no storage for intermediate results is necessary.
2 METHODS, RESULTS AND DISCUSSION
BRAT-BW uses the strategy proposed in (Lister et al., 2009) and employed by both Bismark and BS-seeker. Two FM-indexes are built on the positive strand of the reference genome: in the first, Cs are converted to Ts, and in the second, Gs are converted to As. Original reads with Cs converted to Ts are mapped to the first index, and reverse-complements of the reads with Gs changed to As are mapped to the second index. To achieve higher efficiency, BRAT-BW employs a multi-seed approach similar to Bowtie-2, by attempting to align a read starting from different locations within the read (details in the Supplementary Material).
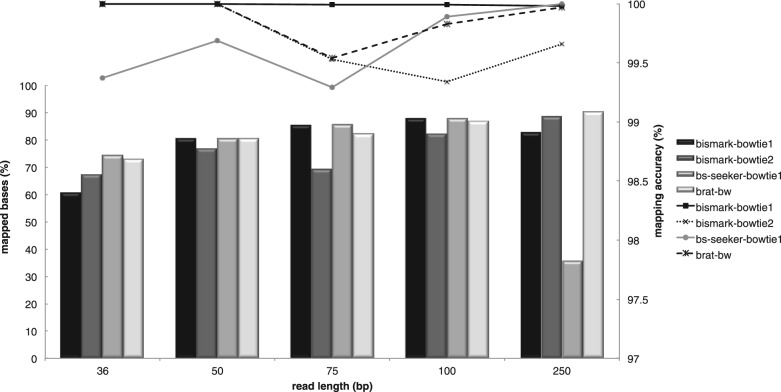
To assess the accuracy of our tool with that of Bismark and BS-seeker, we generated 1 M in silico reads of different lengths originated from the human genome (hg18), with ∼2% of errors introduced uniformly at random positions in each read. Our synthetic dataset consisted of a mix of 36 bp and 50 bp reads with one mismatch per read, 75 bp and 100 bp reads with two mismatches per read and 250 bp reads with five mismatches per read. Simulated reads and the parameters used to run the experiments are provided in the Supplementary Material. Bisulfite conversion rate was set to 98%. Figure 1 reports the total number of uniquely mapped reads and mapping accuracy estimated as the number of unique reads mapped to the original genomic positions divided by the sum of correctly and incorrectly uniquely mapped reads. A read is considered mapped incorrectly if it was mapped with a number of mismatches equal to a given threshold, but the reported location differed from the original genomic location. Bismark and BS-seeker handles differently the case when a C in a read has to be mapped to T in the genome: Bismark allows this mapping, whereas BS-seeker considers it a mismatch. We calculated the number of mismatches in the resulting mapped reads according to both policies. BRAT-BW allows a user to choose between the two policies. In all experiments, Bowtie's FM-index was built with an offrate 4. For BS-seeker, option p was disabled. For BS-seeker on 250 bp-long reads, we required the tool to map the first 150 bp with three mismatches (maximum allowed). Figure 1 shows that the performance of BRAT-BW in terms of mapped uniquely bases and mapping accuracy is comparable with the best results of the other tools. On longer reads, BRAT-BW shows slightly better mapping accuracy than Bismark with Bowtie-2. We carried out the same tests on BRAT (tool brat-large). Since brat-large does not allow mismatches in the first 24 bases of a read, the error model used to generate the simulated reads is severely affecting the performance of brat-large. Unlike real reads where the majority of sequencing errors tend to accumulate towards the 3′ end, a substantial portion of our simulated reads had mismatches in the first 24 bp. On 36, 50, 75, 100 and 250 bp reads, brat-large only mapped 27, 43, 40, 51 and 55% of reads, respectively, with mapping accuracy of 96.3, 98.8, 99.2, 99.7 and 99.96%, respectively.
Fig. 1.
Percentage of bases mapped uniquely (bars) and mapping accuracy (lines) on synthetic data, as a function of the read length
To evaluate time- and memory efficiency on real data, we used human reads (SRA #SRR020138, Lister et al., 2009) and prepared two datasets. The first one contains 32 bp-long reads obtained by selecting the high-quality prefix of that length. Each read was duplicated to obtain a realistic number of sequenced reads per lane (∼29.6 M in total). In the second dataset we trimmed reads by quality, selected the first 64 bases, then removed the first two bases, and duplicated each read (∼24.5 M in total). Table 1 shows that BRAT-BW used half as much memory as other tools. On short reads, the time and the total number of mapped reads was comparable among all tools considered here. On longer reads, BRAT-BW was 1.5, 2.7, 3 and 3 times faster than Bismark with Bowtie-1 and Bowtie-2, BS-seeker, and BRAT, respectively.
Supplementary Material
ACKNOWLEDGEMENTS
We thank F.Krueger for helpful comments and discussions.
Funding: NIH R01 AI85077-01A1 and NSF DBI-1062301 (in part).
Conflict of Interest: none declared.
REFERENCES
- Burrows M., Wheeler D. A block sorting lossless data compression algorithm. 1994. Technical Report #124. Digital Equipment Corporation. [Google Scholar]
- Chen P.Y., et al. BS Seeker: precise mapping for bisulfite sequencing. BMC. Bioinform. 2010;11:203. doi: 10.1186/1471-2105-11-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cokus S.J., et al. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature. 2008;452:215–219. doi: 10.1038/nature06745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferragina P., Manzini G. Proceedings of IEEE Foundation of Computer Science. Redondo Beach, CA: 2000. Opportunistic data structures with applications; pp. 390–398. [Google Scholar]
- Frommer M., et al. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. PNAS USA. 1992;89:1827–1831. doi: 10.1073/pnas.89.5.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris E.Y., et al. BRAT: bisulfite-treated reads analysis tool. Bioinformatics. 2010;26:572–573. doi: 10.1093/bioinformatics/btp706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger F., Andrews S. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics. 2011;27:1571–1572. doi: 10.1093/bioinformatics/btr167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., et al. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R., et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.