Abstract
Objectives
Human milk oligosaccharides (HMO) are the third most abundant component of breast milk. Our laboratory has previously revealed gene clusters specifically linked to HMO metabolism in select bifidobacteria isolated from fecal samples of infants. Our objective was to test the hypothesis that growth of select bifidobacteria on HMO stimulates the intestinal epithelium.
Methods
Caco-2 and HT-29 cells were incubated with lactose (LAC) or HMO-grown Bifidobacterium longum subsp. infantis (B. infantis) or B. bifidum. Bacterial adhesion and translocation was measured by real-time quantitative PCR. Expression of pro- and anti-inflammatory cytokines and tight junction proteins was analyzed by real time reverse transcriptase. Distribution of tight junction proteins was measured using immunofluorescent microscopy.
Results
We showed that HMO-grown B. infantis had significantly higher rate of adhesion to HT-29 cells compared to B. bifidum. B. infantis also induced expression of a cell membrane glycoprotein, P-selectin glycoprotein ligand -1. Both B. infantis and B. bifidum grown on HMO caused less occludin relocalization and higher expression of anti-inflammatory cytokine, interleukin (IL)-10 compared to LAC-grown bacteria in Caco-2 cells. B. bifidum grown on HMO showed higher expression of junctional adhesion molecule and occludin in Caco-2 cell and HT-29 cells. There were no significant differences between LAC or HMO treatments in bacterial translocation.
Conclusions
This study provides evidence for the specific relationship between HMO-grown bifidobacteria and intestinal epithelial cells. To our knowledge, this is the first study describing HMO-induced changes in the bifidobacteria-intestinal cells interaction.
Keywords: milk oligosaccharides, bifidobacteria, intestinal epithelial cells, inflammation
INTRODUCTION
Breast feeding is associated with multiple benefits in infants [1]. There are well-documented differences in the intestinal microbiota between human milk-fed and formula-fed infants [2]. The predominance of beneficial bacteria, mainly bifidobacteria, in the gut microbiota of breast-fed infants is thought to result in part from the fermentation of oligosaccharides, non-digestible carbohydrates consisting of several types of linked monosaccharides [3]. Human milk oligosaccharides (HMOs) are the third most abundant component of human milk [4] and comprise more than 200 HMO structural species [5, 6]. Our research group has previously demonstrated that HMOs can selectively nourish a protective bifidobacterial microbiota isolated from fecal samples of breast-fed infants [7–10]. Using genomic analysis of Bifidobacterium longum subsp. infantis (B. infantis), our laboratory has revealed gene clusters specifically linked to oligosaccharide metabolism that are expressed only during growth on HMO but not during growth on lactose (LAC) or current commercial prebiotics (fructooligosaccharides or galactooligosaccharides) [11–14]. It was shown that other bifidobacteria (e.g. B. bifidum, B. longum subsp. longum and B. breve) also grow on HMO as the sole carbon source [10], however other mechanisms of HMO metabolism were proposed [12, 15].
The ability to adhere to the intestinal epithelium may play an important role in gut colonization, as it prevents the peristaltic elimination of bacteria [16, 17]. Adhesion promotes the modulation of the immune system [18, 19] and prevents pathogens from attaching to the gut mucosa [20]. Garrido and colleagues [13] characterized a family of solute binding proteins in B. infantis which have an affinity for mammalian glycans. Strain- and species-specific adhesion of bifidobacteria to epithelial cells is not a new phenomenon and has been described previously [21–23]. However, previous reports described minimal invasion of bifidobacteria, suggesting that bifidobacteria do not translocate [24].
Intestinal barrier function requires tight junctional (TJ) complexes, which are made up of complex lipoprotein structures [25]; disruption of TJs leads to compromised intestinal integrity[26–30]. Ewaschuk et al. [31] demonstrated that soluble factors produced by B. infantis increase and expression of occludin, a primary TJ protein in the human epithelial cell line, suggesting improved intestinal barrier function [31, 32]. There are numerous examples in the literature showing an evidence of a role for probiotic in TJ repair and maintenance [33–35]; these effects seem to be mediated by up-regulation of TJ proteins.
In addition, previous reports suggest that bifidobacteria showing higher adhesion to intestinal epithelial cells also had a higher anti-inflammatory capacity [36]. For example, B. infantis induced intestinal production of anti-inflammatory cytokine IL-10, while reducing production of pro-inflammatory cytokine TNF [37, 38]. Lipopolysaccharide (LPS), induces exaggerated activation of nearly all pro-inflammatory cytokines, chemokines, immune receptors and cell surface adhesion molecules [39]. Preising et al. [36] showed that bifidobacteria were able to inhibit LPS-induced secretion of IL-8, an early mediator of inflammatory responses, in Caco-2 cells. Those authors also reported variations between two epithelial cell lines, Caco-2 and HT-29 in this repose to bifidobacteria.
Stimuli such as cytokines have been reported to influence not only the expression of TJ proteins but also their association with cytoskeleton (rearrangement) or paracellular permeability [40]. Whether other genes (beside TJ) involved in bacteria-host interactions are up-regulated in epithelial cells upon contact with bifidobacteria remains to be elucidated. An example of a possible gene candidate is the alpha1-proteinase inhibitor (SERPING), which is synthesized by the intestinal epithelial cells, and is likely to act as an immunomodulatory factor [41]. Also, P-selectin glycoprotein ligand-1 (SELPLG) is a glycoprotein that binds to the cell adhesion molecule and play a key role in the inflammatory response [42]. Microarray studies in our laboratory have previously shown that incubation of HMO with B. infantis activated the transcriptional up-regulation of SELPLG [43].
This study focused on characterizing the interaction between HMO-grown bifidobacteria and cultured human intestinal epithelial cells. We hypothesize that growth on HMO will facilitate colonization and that will lead to the protected modulation of the host, including altered gene expression. The effects of HMO on the bifidobacterial-intestinal epithelium interaction are currently not known. Here, the analyzed interaction included adhesion, translocation, production of inflammatory cytokines, and expression and distribution of TJ proteins. To our knowledge, this is the first study describing HMO-induced changes in the bifidobacteria-intestinal epithelial cells interaction.
MATERIALS AND METHODS
Bacterial strains and culture conditions
The bifidobacterial strains used in this study were: Bifidobacterium bifidum ATCC 29521 (B. bifidum) and B. infantis ATCC 15697 (B. infantis). Cultures from stocks frozen at −80°C in glycerol were grown overnight anaerobically at 37°C in the semisynthetic de Man-Rogosa-Sharpe (MRS) broth (Becton Dickinson) supplemented with 1% (wt/vol) L-cysteine hydrochloride [44]. This medium was supplemented with 2% (wt/vol) lactose (LAC) (Sigma Aldrich) or HMO as the sole carbon source. Media was autoclaved separately from the carbohydrates, which was added to the cooled broth after sterile filtration (Millex-GV, 0.22 um, Millipore). All bacterial cultures used in the adhesion experiment were grown for 48h (exponential growth).
HMO used in this study was kindly provided by the laboratories of Dr. Bruce German (Department of Food Science and Technology). The purification process of HMO was performed as described previously [45].
Epithelial cell lines
Enterocyte-like human colon adenocarcinoma (Caco-2) cells show marked characteristics of human epithelial cells, including the ability to differentiate as well as to polarize and form TJ complexes [46]. In the present study Caco-2 cells were obtained from the American Type Culture Collection (ATCC HTB-37, passages 31–37) and were maintained in our laboratory by culture in Dulbecco’s modified Eagle medium (DMEM) containing 10% heat-inactivated (30 min at 56°C) fetal calf serum, 1% nonessential amino acids, 50 IU/mL penicillin, and 50 μg/mL streptomycin. Cells were cultured routinely in 75-cm2 flasks at 37°C in a 5% CO2 constant-humidity environment with medium replaced every 2–3 days. Another human colon adenocarcinoma, HT-29 [47] cell culture (ATCC HTB-38, passages 39–44) was maintained in the same conditions except the McCoy’s 5A medium (Invitrogen) was used. Monolayers were subcultured at 80% confluence exposing the monolayers to 0.25% trypsin and 0.9mM EDTA (Invitrogen, CA) using split ratio of 1:10. Epithelial cell monolayers and Transwells (described below) were treated by lipopolysaccharide (LPS; Sigma, Saint Louis MO) at 100 ng/mL.
Adhesion assay
For adhesion assays, Caco-2 and HT-29 cells were seeded in 24-well plate (2 cm2/well; BD Falcon, Franklin Lakes, NJ). Adhesion experiments were performed 15d after confluence, a time when morphological and functional differentiation is complete [48, 49]. The viable cell number, counted in a Neubauer chamber, was about 6 × 105 cells per well. Cell monolayers were carefully washed twice with sterile phosphate-buffered saline (PBS; pH 7.3) before bacterial cells were added. B. infantis and B. bifidum from the exponentially grown 48h-old cultures were collected by centrifugation (4000g for 10 min), washed, and resuspended in DMEM for assays with Caco-2 cells and in McCoy’s medium for assays with HT-29 cells. For reference purposes (100% values), 1 ml aliquots of the original bacterial cell suspensions used in the adhesion assay were centrifuged, the cells resuspended in 200 μl trypsin/EDTA plus 200 μl PBS and then frozen and stored at −20°C until quantification of the bacteria. Approximately 1 × 108 cells of each strain were incubated with a monolayer of fully differentiated cells. All incubations were performed in biologically independent triplicates. Plates were then incubated at 37°C, 5% CO2 for 2h, after which all monolayers were washed gently three times with PBS to release unbound bacteria. The epithelial cells were detached from the plastic surface by incubation with 200 μl trypsin/EDTA per well (10 min, 37°C). To perform quantification of adherent bacteria, samples of bacteria plus epithelial cells were incubated at 37°C for 30 min in Gram-positive lysis buffer consisting of 20 mM Tris-Cl, 2 mM sodium EDTA, 1.2% Triton X-100 and lysosyme (final conc. 20 mg/ml). The adhesion capacity was determined using real-time quantitative PCR (qPCR; online-only appendix, http://links.lww.com/MPG/A104) and was expressed as the number of adherent bacteria divided by total number of bacteria added, multiplied by 100.
Bacterial translocation
For experiments testing translocation of HMO-grown bifidobacteria, Caco-2 or HT-29 cells were seeded on Transwell polycarbonate cell culture inserts (Corning Inc., NY; 24-mm diameter; 3.0-μm pore size) at 2–3 × 105 cells per square centimeter and grown for 14 days post-confluence to achieve fully differentiated monolayers. Translocation studies were conducted in Hanks’ balanced salt solution (HBSS), which reduces bacterial growth by approximately 1000-fold compared with DMEM, while maintaining bacterial viability [50]. This allows more accurate estimation of the numbers of bacteria crossing the epithelial monolayer. Complete differentiation was confirmed by measurement of transepithelial electrical resistance (TER) using a Millicell-ERS voltohmeter fitted with chopstick electrodes (Millipore Corp., Bedford MA) and cells were used when value was more than 1000 ohms/cm2. Culture medium was removed by washing monolayers twice with HBSS at 37°C. After equilibration for 30 minutes, any monolayers for which TER has not returned to within 10% of the value before removal of experimental media was discarded. Bifidobacteria were inoculated into the apical chamber of the Transwell to a final concentration of 108 bacteria/mL. After 2h incubation period (37°C; 5% CO2, 95% room air), during which translocation of bacteria was allowed to occur, the concentration of bacteria in the basolateral chamber was determined by qPCR (online-only appendix, http://links.lww.com/MPG/A104).
Statistical analysis
Data are expressed as mean ± standard deviation (SD) of the results of four independent trials conducted in triplicate. All statistical tests used a two-sided significance level of 0.05. A paired t-test was used to compare LAC and HMO experimental groups for each outcome measure. All reported significance levels represent two-tailed P-value. Expression of the cytokines and TJs was determined using ΔCt (online-only appendix, http://links.lww.com/MPG/A104). Translocation was expressed as 103 bacteria/cm2 of the monolayer.
RESULTS
Adhesion of HMO-grown bifidobacteria in Caco-2 and HT-29 cells
Numerous studies in our and other laboratories demonstrated the ability of two bifidobacterial strains, B. infantis and B. bifidum to grow on HMO as a sole carbon source. [10]. Based on those results, both B. infantis and B. bifidum were selected for experiments focused on the interaction between intestinal epithelial cells and bifidobacteria grown on HMO. Adhesion activity of B. infantis and B. bifidum grown on LAC or HMO was evaluated by a real-time PCR-based method (Fig. 1). In comparison with traditional techniques (e.g. viable counts) the analytical approach using real-time PCR is rapid, accurate and particularly useful for studying bacterial adhesion, especially when dealing with different phenotypes of in the genus Bifidobacterium [23]. Different adhesion could be observed between two bifidobacterial species tested. The levels of adhesion ranged from 8.1% to 20.4% for Caco-2 cells (Fig. 1a) and from 12.5% to 26.5% for HT-29 cells (Fig. 1b). B. bifidum grown on LAC was the most efficient strain in terms of adhesion to Caco-2 epithelial cells (20.4%), while HMO-grown B. infantis incubated with HT-29 cells adhered at the rate of 26.5%. There were numerical trends between HMO and LAC treatments, however the only significant difference was observed for B. infantis incubated with HT-29 cells (p < 0.05). The largest alteration in the binding among bacteria incubated with HT-29 cells was shown with B. infantis (8.5% for LAC and 26.5% for HMO) (Fig. 1b).
Figure 1.
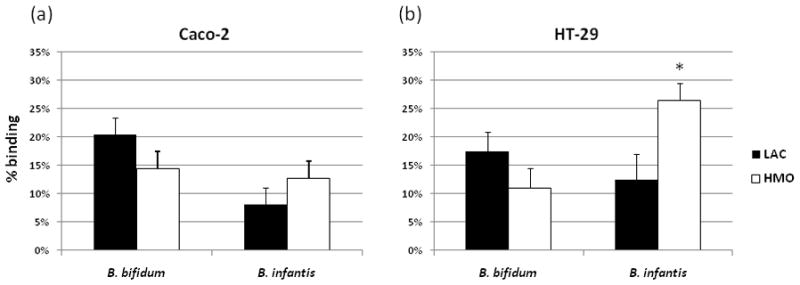
Adhesion of B. bifidum and B. infantis grown on LAC or HMO to the Caco-2 (a) or HT-29 (b) cells. Bifidobacteria were incubated with fully differentiated epithelial cells for 2h at 37°C in anaerobic conditions. Quantification of the bacterial genomes was based on the presence of 16s rRNA. The results are presented as the percentage of bacteria recovered vs. applied on monolayers. Values are the mean ± SD and * indicates statistical significance (p < 0.05).
HMO-grown bifidobacteria alter tight junction protein expression
Caco-2 and HT-29 cells were incubated with LAC- or HMO-grown bifidobacteria and expression of occludin, ZO-1 and junctional adhesion molecule (JAM-A) was measured. These particular proteins were selected as they have been implicated in TJ barrier function and are known to be expressed in intestinal cells [51]. Caco-2 incubated with B. bifidum cultured on HMO expressed higher expression of occludin (3.6 fold induction) while incubation with both HMO-grown bifidobacterial species resulted in enhanced protein expression of JAM-A compared to LAC-grown bacteria - these changes were from 5.7 fold induction for B. bifidum to 1.7 for B. infantis (Fig. 2a). There were no differences in ZO-1 expression between the LAC and HMO treatments in Caco-2 cells. In HT-29 cells B. infantis grown on HMO had 2-fold increase of ZO-1 expression compared to LAC. Occludin expression was not altered in HT-29 cells incubated with HMO-grown bifidobacteria; however JAM-A was 4 fold higher for HMO-grown B. bifidum.
Figure 2.
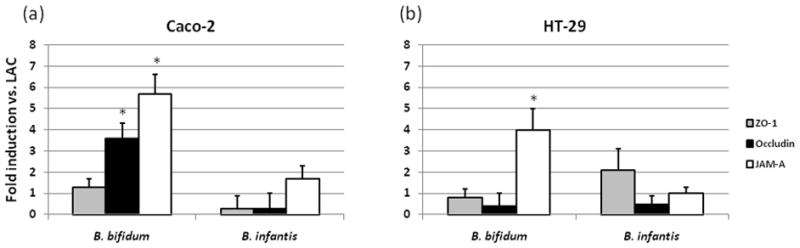
Analysis of the effects of HMO- grown bifidobacteria on tight junction protein mRNA expression in Caco-2 (a) or HT-29 (b) cells. Cells were incubated for 2h, then the expression was measured by real-time PCR, normalized to β-actin and presented as fold induction relative to LAC-grown bifidobacteria. The data are the means ± SD where * indicates statistical significance between LAC and HMO (p < 0.05).
B. bifidum and B. infantis grown on HMO prevent occludin relocation into the cytoplasm
We used immunofluorescence assay to evaluate the localization of occludin in epithelial cells incubated with bifidobacteria grown on LAC or HMO. Under normal conditions (no bacterial treatment), occludin was localized in a pattern consistent with their distribution in tight junctions in Caco-2 cells (Supplemental Fig. 1, http://links.lww.com/MPG/A104), while cells incubated with HMO-grown bifidobacteria caused mild occludin relocalization to the cytoplasm. LAC treatment caused considerable relocalization of occludin and it was characterized by discontinuities in membrane staining and submembranous internalization of these proteins. Thus both B. infantis and B. bifidum grown on HMO prevented the significant intracellular redistribution of occludin. HT-29 cells showed the similar pattern of occludin distribution (data not shown).
Immunomodulatory and inflammatory markers in the epithelial cells are affected by HMO-grown bifidobacteria
To determine whether growth of bifidobacteria on HMO induces the immunomodulation in epithelial cells we analyzed the expression of two factors, SELPLG and SERPING. B. infantis grown on HMO significantly up-regulated the expression of SELPLG (7 fold induction vs. LAC) compared to B. bifidum (Fig. 3). No significant differences were observed in the expression of SERPING in B. infantis or B. bifidum. Expression of those two immunomodulatory markers was not affected by HMO treatment in HT-29 cells (data not shown). We further assessed the expression of pro- and anti-inflammatory cytokines in Caco-2 and HT-29 cells incubated with bifidobacteria grown on HMO and LAC. The results for expression of the genes for IL-8, IL-10 and TNF are shown in Figure 4. Caco-2 cells incubated with HMO-grown B. bifidum and B. infantis had significantly increased expression of IL-10 (fold induction vs. LAC = 4.6 and 2.5, respectively). HMO-grown B. infantis showed increased expression of IL-8 while B. bifidum did not show induction of IL-8 expression in Caco-2 cells compared with cells incubated with LAC-grown bacteria (Fig. 4a). B. infantis grown on HMO lowered the expression of pro-inflammatory TNF (0.6 fold vs. LAC). HMO-grown bifidobacteria had no effect on secretion of measured cytokines in HT-29 cells with values comparable to that of LAC-grown bifidobacteria (Fig. 4b). LPS of Gram negative bacteria triggers an inflammatory response and the subsequent overexpression of proinflammatory cytokines, e.g. IL-8 [52]. LPS treatment increased expression of all inflammatory cytokines, including 5-fold induction for IL-8 for both Caco-2 and HT-29 cells, however no significant differences were noted between LAC and HMO treatments (data not shown).
Figure 3.
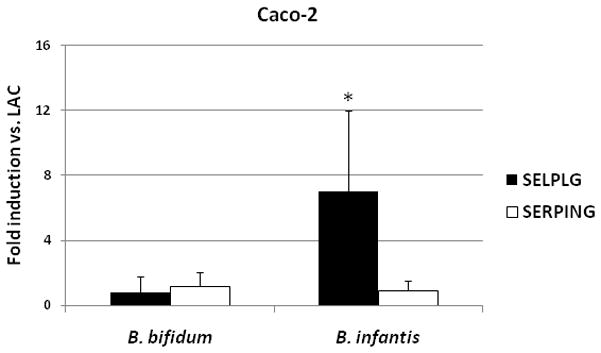
Effects of HMO-grown B. bifidum and B. infantis on mRNA expression of SELPLG and SERPING in Caco-2 cells was measured by real-time PCR, normalized to β-actin and presented as fold induction relative to LAC-grown bifidobacteria. The data are the means ± SD where * indicates statistical significance between LAC and HMO (p < 0.05).
Figure 4.
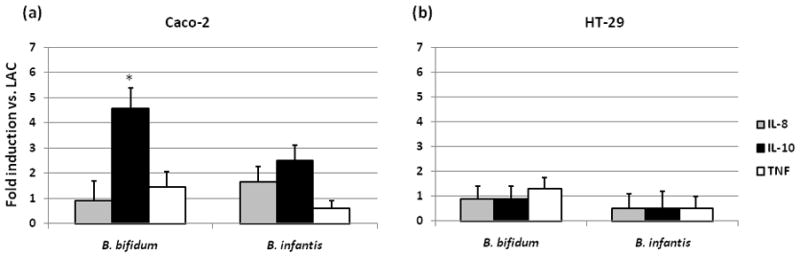
Cytokine mRNA expression in Caco-2 (a) and HT-29 (b) cells incubated with bifidobacterial strains grown on HMO was measured by real-time PCR, normalized to β-actin and presented as fold induction relative to LAC-grown bifidobacteria. * indicates statistical significance between LAC and HMO (p < 0.05).
Translocation of HMO-grown bifidobacteria in Caco-2 and HT-29 cells
To examine whether the observed changes in the expression and localization of TJ, as well as immune activation, are associated with alterations in the epithelial barrier we tested translocation of bifidobacteria. We observed minimal translocation of LAC and HMO-grown bifidobacteria in Caco-2 and HT-29 cells (Fig. 5a and 5b). No significant difference in translocation was observed between bacteria grown on LAC or HMO in monolayers not treated with LPS. Translocation in HT-29 cells was at the lower rate (0.3 – 0.45 × 103/cm2 of the monolayer) compared to Caco-2 cells. No correlation between adhesion and translocation was observed for two bifidobacterial species incubated with Caco-2 or HT-29 cells. LPS treatment dramatically increased translocation in both Caco-2 and HT-29 cells, however there were no significant differences between the bacteria or the sugar source treatments (Fig. 5a and 5b).
Figure 5.
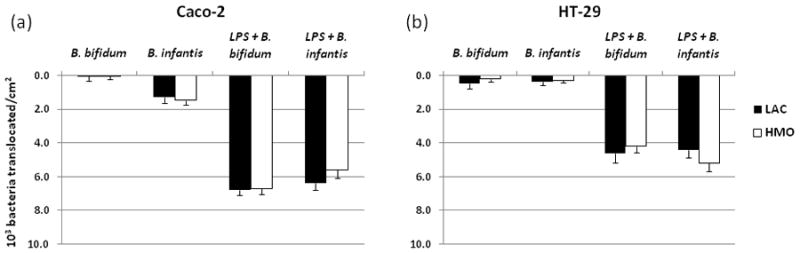
Translocation of bifidobacteria in Caco-2 (a) and HT-29 (b) monolayers. Cells were incubated with bacteria apically for 2h, and then basolateral translocation was measured. Data are means ± SD. Significance was at p < 0.05.
DISCUSSION
This study characterizes for the first time the interactions between HMO-grown bifidobacteria and intestinal epithelial cells. Bacterial adhesion is critical to these interactions since the ability to adhere also represents a significant prerequisite for the transient intestinal colonization [17]. HMO are naturally evolved substrates that provide a structure-specific growth advantage to co-evolved bifidobacteria and result in beneficial interactions with the host [10]. Unlike lactose, HMO pass mainly unabsorbed and undigested through the small intestine into the colon, where they are fermented to short-chain fatty acids creating an acidic environment[53]. We used lactose as a standard comparative in this study to exemplify different phenotypes of bifidobacteria consuming those sugars, since both lactose and HMO are milk’s constituents delivered during lactation. In addition, the core structures of HMOs consist of lactose [5].
The ability of select bifidobacteria to consume complex oligosaccharides from human milk likely enables this genus to be one of the most abundant colonizers of the breastfed infant gut [7, 8]. Previous studies have shown that among three gut commensals (B. infantis, Lactobacillus gasseri and Escherichia coli) only B. infantis was able to ferment HMO as a sole carbon source and achieve high cell densities [9, 54]. Although growth of B. bifidum on HMO was weaker than B. infantis, direct consumption of HMO was observed previously [10]. Those two species were shown to metabolize HMO via different mechanisms [7, 8, 10, 55]. In the present study both B. infantis and B. bifidum were selected for analysis of the interaction with the intestinal epithelial cells.
We used two epithelial cell lines to test the abilities of HMO-grown B. bifidum and B. infantis to adhere to the intestinal cells. The first adhesion model is based on the human colon adenocarcinoma cell line Caco-2, whose characteristics simulate structural and functional distinctiveness of mature enterocytes in vitro [46]. The second cell line used in this study, HT-29, maintains a constant proliferation rate with practically no further differentiation [24, 56]. We used incubation period of 2 h, since it was previously shown sufficient to enable epithelial cells to adapt to the presence of bacteria without acidification of the medium [57].
Although the adhesive phenotype is frequently present in the members of the Bifidobacterium species [52, 58, 59], of importance for the present study are the specific changes in adhesion induced by HMO growth. Previous reports demonstrated a strongly adhesive phenotype of B. bifidum MIMBb75 [60] which was able to adhere to both Caco-2 and HT-29 cells [23]. Our observations support these data. However, unique for the present study, HMO had a significant impact on the binding abilities of tested bifidobacteria. As seen in Figure 1, high percentage of B. infantis grown on HMO adhered to HT-29 epithelial cell lines. The proliferation rate of HT-29 cells is constant and is higher than that in Caco-2 cells [56], thus the interaction between bifidobacteria and HT-29 cells is expected to be greater than the interaction with Caco-2 cells. Interestingly, HMO-grown B. infantis induced expression of SELPLG (annotated as a cell membrane glycoprotein) in Caco-2 cells but not in HT-29 cells. These results suggest a synergistic effect on the gut ability to sense and modulate genes playing a role in the binding and signaling to bacterial gut commensals. Here, we show that HMO-grown B. infantis (but not B. bifidum) facilitates colonization and leads to a protective modulation of the host’s intestinal epithelium.
Ewaschuk et al. [31] recently showed that B. infantis-conditioned medium lowered the mucosal permeability, changed expression of TJ proteins and prevented occludin redistribution into the cytoplasm. Our goal was to test whether growth on HMO affects the permeability of epithelium and expression and localization of TJ proteins which are positioned around the apical end of the lateral cell membrane [31]. The redistribution of TJ from the intercellular junctions into the intracellular compartment would be an implication of a defective barrier function. We analyzed the distribution of occludin using immunofluorescence method (Supplemental Fig. 1, http://links.lww.com/MPG/A104). Under baseline conditions (no bacteria) occludin was not found in the cytoplasm of the epithelial cells. When B. infantis or B. bifidum were incubated with epithelial monolayers, relocalization of the occludin to the cytoplasm occurred. However, this relocalization of occludin was less apparent when bacteria were grown on HMO compared to LAC. Incubation with LAC-grown bifidobacteria was characterized by discontinuities in membrane staining and submembranous internalization of these proteins.
Previous studies strongly suggest that alterations in TJ composition and protein localization may have a role in the pathogenesis of chronic inflammatory conditions [25]. Defining the immunomodulatory capacity of HMO-grown bifidobacteria seems relevant in order to understand their contribution to the establishment of the mucosal tolerance and immune responses in the early stages of life. Several studies have evaluated the effects of different bifidobacteria in the production of cytokines by intestinal epithelial cells [36, 61, 62]. however reports in the literature are often inconclusive [36]. For example, researchers noticed minimal impact of probiotic strains B. animalis subsp. lactis Bb12 and, to a lesser extent, B. longum NCC2705 [60] on the cytokine expression induction. In another study, 8 out of 19 bifidobacteria, including two B. bifidum strains, were demonstrated to stimulate the production of IL-8 [63].. Our findings show that B. infantis attenuates baseline IL-8 secretion in HT-29 cells [64]. As expected, the LPS treatment increased expression of IL-8 in Caco-2 cells [65, 66]; however, the expression of IL-8was not affected by HMO treatment compared to LAC in the present study.
Previous studies have demonstrated that TNF increase permeability by inducing redistribution of various TJ proteins by internalization [40, 67]. Here, the expression of TNF by either Caco-2 or HT-29 cells was not significantly altered in the HMO group, however it is possible that numerical trends toward higher expression of TNF by HMO-grown B. bifidum could turn the mucosal immune system on stand-by and prevent the release of severe inflammation. Other cytokines such as IL-10 have been shown to decrease permeability. We observed higher expression of IL-10 in Caco-2 incubated with HMO-grown B. bifidum and B. infantis, suggesting potential anti-inflammatory properties of HMO growth.. Overall, the response of Caco-2 and HT-29 cells to LPS treatment was consistent with the previously published research [65, 66]; however HMO did not have any significant effects on the levels of cytokine expression in LPS-treated cells.
Both bifidobacterial species tested in the present study, regardless whether grown on HMO or LAC, translocated at the minimal rate through the intestinal epithelial cells, confirming previous reports that the genus Bifidobacterium is generally non-invasive [24]. Despite the observed changes in TJ expression and localization, growth on HMO did not seem to affect the bacterial translocation regardless of the LPS presence.
The concept of pre- and probiotics has attracted increasing attention in recent years. A number of publications show anti-inflammatory effects of probiotics in vivo and in vitro [68, 69] and live probiotics and commensals have been shown to affect monolayer barrier function in cultured human epithelial cells [20]. Our results support the concept of synbiotics as a synergistic combination of probiotics and prebiotics [70]. According to our data, the growth on HMO as a sole carbon source enhances epithelial binding and can potentially induce anti-inflammatory response in the intestinal epithelial cells; however it did not have any effects in the presence of the massive inflammatory stimulation by LPS. Is it possible that B. infantis, as well as other bifidobacteria, exert the beneficial effect on human physiology in a prophylactic fashion, serving to enhance barrier function? Further studies are needed to establish the role of HMO-grown bifidobacteria in the infant’s developing gut.
Supplementary Material
Acknowledgments
Authors thank Man Ki Tsui for technical assistance. The study described was supported by Award Number F32AT006642 from the National Center For Complementary & Alternative Medicine. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Center For Complementary & Alternative Medicine or the National Institutes of Health. This publication was made possible in part by grant support from the University of California Discovery Grant Program, the California Dairy Research Foundation, Dairy Management Inc., the Gates Foundation, USDA NRI-CSREES Award 2008-35200-18776, NIEHS Superfund P42 ES02710, and by NIH awards R01HD059127, 1R01HD061923 and R21AT006180.
Footnotes
Supplemental digital content is available for this article. Direct URL citations appear in the printed text, and links to the digital files are provided in the HTML text of this article on the journal’s Web site (www.jpgn.org).
The authors report no conflicts of interest.
References
- 1.James DC, Lessen R. Position of the American Dietetic Association. promoting and supporting breastfeeding. J Am Diet Assoc. 2009;109(11):1926–1942. doi: 10.1016/j.jada.2009.09.018. [DOI] [PubMed] [Google Scholar]
- 2.Nakamura N, Gaskins HR, Collier CT, et al. Molecular Ecological Analysis of Fecal Bacterial Populations from Term Infants Fed Formula Supplemented with Selected Blends of Prebiotics. Appl Environ Microbiol. 2009;75:1121–1128. doi: 10.1128/AEM.02359-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.German JB, Freeman SL, Lebrilla CB, et al. Human milk oligosaccharides: evolution, structures and bioselectivity as substrates for intestinal bacteria. Nestle Nutr Workshop Ser Pediatr Program. 2008;62:205–218. doi: 10.1159/000146322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kunz C, Rudloff S, Baier W, et al. Oligosaccharides in human milk: structural, functional, and metabolic aspects. Annual Review of Nutrition. 2000;20:699–722. doi: 10.1146/annurev.nutr.20.1.699. [DOI] [PubMed] [Google Scholar]
- 5.Chichlowski M, German JB, Lebrilla CB, et al. The Influence of Milk Oligosaccharides on Microbiota of Infants: Opportunities for Formulas. Annu Rev Food Sci Technol. 2011;2:331–351. doi: 10.1146/annurev-food-022510-133743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Niñonuevo MR, Lebrilla CB. Mass spectrometric methods for analysis of oligosaccharides in human milk. Nutrition Reviews. 2009;67:S216–S226. doi: 10.1111/j.1753-4887.2009.00243.x. [DOI] [PubMed] [Google Scholar]
- 7.LoCascio RG, Ninonuevo M, Kronewitter S, et al. A versatile and scalable strategy for glycoprofiling bifidobacterial consumption of human milk oligosaccharides. Microbial Biotechnology. 2009;2:333–342. doi: 10.1111/j.1751-7915.2008.00072.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.LoCascio RG, Ninonuevo MR, Freeman SL, et al. Glycoprofiling of bifidobacterial consumption of human milk oligosaccharides demonstrates strain specific, preferential consumption of small chain glycans secreted in early human lactation. J Agric Food Chem. 2007;55:8914–8919. doi: 10.1021/jf0710480. [DOI] [PubMed] [Google Scholar]
- 9.Ward RE, Ninonuevo M, Mills DA, et al. In Vitro Fermentation of Breast Milk Oligosaccharides by Bifidobacterium infantis and Lactobacillus gasseri. Appl Environ Microbiol. 2006;72:4497–4499. doi: 10.1128/AEM.02515-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ward RE, Niñonuevo M, Mills DA, et al. In vitro fermentability of human milk oligosaccharides by several strains of bifidobacteria. Molecular Nutrition & Food Research. 2007;51:1398–1405. doi: 10.1002/mnfr.200700150. [DOI] [PubMed] [Google Scholar]
- 11.Sela DA, Chapman J, Adeuya A, et al. The genome sequence of Bifidobacterium longum subsp. infantis reveals adaptations for milk utilization within the infant microbiome. Proceedings of the National Academy of Sciences. 2008;105:18964–18969. doi: 10.1073/pnas.0809584105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sela DA, Li Y, Lerno L, et al. An Infant-associated Bacterial Commensal Utilizes Breast Milk Sialyloligosaccharides. Journal of Biological Chemistry. 2011;286:11909–11918. doi: 10.1074/jbc.M110.193359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garrido D, Kim JH, German JB, et al. Oligosaccharide Binding Proteins from Bifidobacterium longum subsp.infantis Reveal a Preference for Host Glycans. PLoS ONE. 2011;6:e17315. doi: 10.1371/journal.pone.0017315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.LoCascio RG, Desai P, Sela DA, et al. Broad Conservation of Milk Utilization Genes in Bifidobacterium longum subsp. infantis as Revealed by Comparative Genomic Hybridization. Appl Environ Microbiol. 2010;76:7373–7381. doi: 10.1128/AEM.00675-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zivkovic AM, German JB, Lebrilla CB, et al. Human milk glycobiome and its impact on the infant gastrointestinal microbiota. Proceedings of the National Academy of Sciences. 2010 doi: 10.1073/pnas.1000083107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Erickson AK, Willgohs JA, McFarland SY, et al. Identification of two porcine brush border glycoproteins that bind the K88ac adhesin of Escherichia coli and correlation of these glycoproteins with the adhesive phenotype. Infect Immun. 1992;60(3):983–988. doi: 10.1128/iai.60.3.983-988.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tuomola E, Crittenden R, Playne M, et al. Quality assurance criteria for probiotic bacteria. The American Journal of Clinical Nutrition. 2001;73:393S–398S. doi: 10.1093/ajcn/73.2.393s. [DOI] [PubMed] [Google Scholar]
- 18.Schiffrin E, Brassart D, Servin A, et al. Immune modulation of blood leukocytes in humans by lactic acid bacteria: criteria for strain selection. Am J Clin Nutr. 1997;66:515S–520. doi: 10.1093/ajcn/66.2.515S. [DOI] [PubMed] [Google Scholar]
- 19.He F, Ouwehand AC, Isolauri E, et al. Comparison of mucosal adhesion and species identification of bifidobacteria isolated from healthy and allergic infants. FEMS Immunology & Medical Microbiology. 2001;30:43–47. doi: 10.1111/j.1574-695X.2001.tb01548.x. [DOI] [PubMed] [Google Scholar]
- 20.Resta-Lenert S, Barrett KE. Live probiotics protect intestinal epithelial cells from the effects of infection with enteroinvasive Escherichia coli (EIEC) Gut. 2003;52(7):988–997. doi: 10.1136/gut.52.7.988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bernet MF, Brassart D, Neeser JR, et al. Adhesion of human bifidobacterial strains to cultured human intestinal epithelial cells and inhibition of enteropathogen-cell interactions. Appl Environ Microbiol. 1993;59(12):4121–4128. doi: 10.1128/aem.59.12.4121-4128.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Crociani J, Grill JP, Huppert M, et al. Adhesion of different bifidobacteria strains to human enterocyte-like Caco-2 cells and comparison with in vivo study. Lett Appl Microbiol. 1995;21(3):146–148. doi: 10.1111/j.1472-765x.1995.tb01027.x. [DOI] [PubMed] [Google Scholar]
- 23.Candela M, Seibold G, Vitali B, et al. Real-time PCR quantification of bacterial adhesion to Caco-2 cells: Competition between bifidobacteria and enteropathogens. Research in Microbiology. 2005;156:887–895. doi: 10.1016/j.resmic.2005.04.006. [DOI] [PubMed] [Google Scholar]
- 24.Moroni O, Kheadr E, Boutin Y, et al. Inactivation of Adhesion and Invasion of Food-Borne Listeria monocytogenes by Bacteriocin-Producing Bifidobacterium Strains of Human Origin. Appl Environ Microbiol. 2006;72:6894–6901. doi: 10.1128/AEM.00928-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chichlowski M, Hale LP. Bacterial-mucosal interactions in inflammatory bowel disease--an alliance gone bad. Am J Physiol Gastrointest Liver Physiol. 2008;295:G1139–1149. doi: 10.1152/ajpgi.90516.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ciccocioppo R, Finamore A, Ara C, et al. Altered Expression, Localization, and Phosphorylation of Epithelial Junctional Proteins in Celiac Disease. American Journal of Clinical Pathology. 2006;125:502–511. doi: 10.1309/DTYR-A91G-8R0K-TM8M. [DOI] [PubMed] [Google Scholar]
- 27.Coeffier M, Gloro R, Boukhettala N, et al. Increased Proteasome-Mediated Degradation of Occludin in Irritable Bowel Syndrome. Am J Gastroenterol. 2010;105:1181–1188. doi: 10.1038/ajg.2009.700. [DOI] [PubMed] [Google Scholar]
- 28.Gassler N, Rohr C, Schneider A, et al. Inflammatory bowel disease is associated with changes of enterocytic junctions. American Journal of Physiology - Gastrointestinal and Liver Physiology. 2001;281:G216–G228. doi: 10.1152/ajpgi.2001.281.1.G216. [DOI] [PubMed] [Google Scholar]
- 29.Zeissig S, Bürgel N, Günzel D, et al. Changes in expression and distribution of claudin 2, 5 and 8 lead to discontinuous tight junctions and barrier dysfunction in active Crohn’s disease. Gut. 2007;56:61–72. doi: 10.1136/gut.2006.094375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Khailova L, Dvorak K, Arganbright KM, et al. Bifidobacterium bifidum improves intestinal integrity in a rat model of necrotizing enterocolitis. Am J Physiol Gastrointest Liver Physiol. 2009;297:G940–949. doi: 10.1152/ajpgi.00141.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ewaschuk JB, Diaz H, Meddings L, et al. Secreted bioactive factors from Bifidobacterium infantis enhance epithelial cell barrier function. AJP - Gastrointestinal and Liver Physiology. 2008;295:G1025–1034. doi: 10.1152/ajpgi.90227.2008. [DOI] [PubMed] [Google Scholar]
- 32.Turner JR. Molecular Basis of Epithelial Barrier Regulation: From Basic Mechanisms to Clinical Application. Am J Pathol. 2006;169:1901–1909. doi: 10.2353/ajpath.2006.060681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zyrek AA, Cichon C, Helms S, et al. Molecular mechanisms underlying the probiotic effects of <i>Escherichia coli</i> Nissle 1917 involve ZO-2 and PKCζ redistribution resulting in tight junction and epithelial barrier repair. Cellular Microbiology. 2007;9:804–816. doi: 10.1111/j.1462-5822.2006.00836.x. [DOI] [PubMed] [Google Scholar]
- 34.Seth A, Yan F, Polk DB, et al. Probiotics ameliorate the hydrogen peroxide-induced epithelial barrier disruption by a PKC- and MAP kinase-dependent mechanism. Am J Physiol Gastrointest Liver Physiol. 2008;294:G1060–1069. doi: 10.1152/ajpgi.00202.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Resta-Lenert S, Barrett KE. Probiotics and Commensals Reverse TNF-[alpha]- and IFN-[gamma]-Induced Dysfunction in Human Intestinal Epithelial Cells. Gastroenterology. 2006;130:731–746. doi: 10.1053/j.gastro.2005.12.015. [DOI] [PubMed] [Google Scholar]
- 36.Preising J, Philippe D, Gleinser M, et al. Selection of Bifidobacteria Based on Adhesion and Anti-Inflammatory Capacity In Vitro for Amelioration of Murine Colitis. Appl Environ Microbiol. 2010;76:3048–3051. doi: 10.1128/AEM.03127-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tanabe S, Kinuta Y, Saito Y. Bifidobacterium infantis suppresses proinflammatory interleukin-17 production in murine splenocytes and dextran sodium sulfate-induced intestinal inflammation. Int J Mol Med. 2008;22(2):181–185. [PubMed] [Google Scholar]
- 38.Sheil B, MacSharry J, O’Callaghan L, et al. Role of interleukin (IL-10) in probiotic-mediated immune modulation: an assessment in wild-type and IL-10 knock-out mice. Clinical & Experimental Immunology. 2006;144:273–280. doi: 10.1111/j.1365-2249.2006.03051.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Aderem A, Ulevitch RJ. Toll-like receptors in the induction of the innate immune response. Nature. 2000;406:782–787. doi: 10.1038/35021228. [DOI] [PubMed] [Google Scholar]
- 40.Bruewer M, Luegering A, Kucharzik T, et al. Proinflammatory Cytokines Disrupt Epithelial Barrier Function by Apoptosis-Independent Mechanisms. The Journal of Immunology. 2003;171:6164–6172. doi: 10.4049/jimmunol.171.11.6164. [DOI] [PubMed] [Google Scholar]
- 41.Faust D, Hormann S, Friedrich-Sander M, et al. Butyrate and the cytokine-induced α1-proteinase inhibitor release in intestinal epithelial cells. European Journal of Clinical Investigation. 2001;31:1060–1063. doi: 10.1046/j.1365-2362.2001.00927.x. [DOI] [PubMed] [Google Scholar]
- 42.Bode L, Rudloff S, Kunz C, et al. Human milk oligosaccharides reduce platelet-neutrophil complex formation leading to a decrease in neutrophil β 2 integrin expression. Journal of Leukocyte Biology. 2004;76:820–826. doi: 10.1189/jlb.0304198. [DOI] [PubMed] [Google Scholar]
- 43.LoCascio RG. Microbiology. Davis, University of California; Davis: 2009. Glycomic and genetic characterization of the metabolism of human milk oligosaccharides by Bifidobacterium species; p. 241. [Google Scholar]
- 44.Barrangou R, Altermann E, Hutkins R, et al. Functional and comparative genomic analyses of an operon involved in fructooligosaccharide utilization by Lactobacillus acidophilus. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:8957–8962. doi: 10.1073/pnas.1332765100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gnoth MJ, Rudloff S, Kunz C, et al. Investigations of the in Vitro Transport of Human Milk Oligosaccharides by a Caco-2 Monolayer Using a Novel High Performance Liquid Chromatography-Mass Spectrometry Technique. Journal of Biological Chemistry. 2001;276:34363–34370. doi: 10.1074/jbc.M104805200. [DOI] [PubMed] [Google Scholar]
- 46.Pinto M, Robine-Leon S, Appay MD, et al. Enterocyte-like differentiation and polarization of the human colon carcinoma cell line Caco-2 in culture. Biol Cell. 1983;47:323–330. [Google Scholar]
- 47.Cencic A, Langerholc T. Functional cell models of the gut and their applications in food microbiology -- A review. International Journal of Food Microbiology. 2010;141:S4–S14. doi: 10.1016/j.ijfoodmicro.2010.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jovani M, Barbera R, Farre R, et al. Calcium, Iron, and Zinc Uptake from Digests of Infant Formulas by Caco-2 Cells. Journal of Agricultural and Food Chemistry. 2001;49:3480–3485. doi: 10.1021/jf010106t. [DOI] [PubMed] [Google Scholar]
- 49.Gretchen JM, Michael LS, Raymond PG. Characterization of Caco-2 and HT29-MTX cocultures in an in vitro digestion/cell culture model used to predict iron bioavailability. The Journal of nutritional biochemistry. 2009;20:494–502. doi: 10.1016/j.jnutbio.2008.05.006. [DOI] [PubMed] [Google Scholar]
- 50.Clark E, Hoare C, Tanianis-Hughes J, et al. Interferon gamma induces translocation of commensal Escherichia coli across gut epithelial cells via a lipid raft-mediated process. Gastroenterology. 2005;128:1258–1267. doi: 10.1053/j.gastro.2005.01.046. [DOI] [PubMed] [Google Scholar]
- 51.Anderson JM, Van Itallie CM. Physiology and function of the tight junction. Cold Spring Harb Perspect Biol. 2009;1(2):a002584. doi: 10.1101/cshperspect.a002584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Riedel CU, Foata F, Goldstein DR, et al. Interaction of bifidobacteria with Caco-2 cells--adhesion and impact on expression profiles. International Journal of Food Microbiology. 2006;110:62–68. doi: 10.1016/j.ijfoodmicro.2006.01.040. [DOI] [PubMed] [Google Scholar]
- 53.Ogawa K, Ben RA, Pons S, et al. Volatile fatty acids, lactic acid, and pH in the stools of breast-fed and bottle-fed infants. J Pediatr Gastroenterol Nutr. 1992;15:248–252. doi: 10.1097/00005176-199210000-00004. [DOI] [PubMed] [Google Scholar]
- 54.Ninonuevo MR, Ward RE, LoCascio RG, et al. Methods for the quantitation of human milk oligosaccharides in bacterial fermentation by mass spectrometry. Anal Biochem. 2007;361:15–23. doi: 10.1016/j.ab.2006.11.010. [DOI] [PubMed] [Google Scholar]
- 55.Katayama T, Sakuma A, Kimura T, et al. Molecular Cloning and Characterization of Bifidobacterium bifidum 1,2-{alpha}-L-Fucosidase (AfcA), a Novel Inverting Glycosidase (Glycoside Hydrolase Family 95) J Bacteriol. 2004;186:4885–4893. doi: 10.1128/JB.186.15.4885-4893.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zweibaum A, Pinto M, Chevalier G, et al. Enterocytic differentiation of a subpopulation of the human colon tumor cell line HT-29 selected for growth in sugar-free medium and its inhibition by glucose. J Cell Physiol. 1985;122(1):21–29. doi: 10.1002/jcp.1041220105. [DOI] [PubMed] [Google Scholar]
- 57.Boesten R, Schuren F, Willemsen L, et al. Bifidobacterium breve– HT-29 cell line interaction: modulation of TNF-α induced gene expression. Beneficial Microbes. 2011;2:115–128. doi: 10.3920/BM2011.0005. [DOI] [PubMed] [Google Scholar]
- 58.Gagnon M, Kheadr EE, Le Blay G, et al. In vitro inhibition of Escherichia coli O157:H7 by bifidobacterial strains of human origin. International Journal of Food Microbiology. 2004;92:69–78. doi: 10.1016/j.ijfoodmicro.2003.07.010. [DOI] [PubMed] [Google Scholar]
- 59.Perez PF, Minnaard Y, Disalvo EA, et al. Surface Properties of Bifidobacterial Strains of Human Origin. Appl Environ Microbiol. 1998;64:21–26. doi: 10.1128/aem.64.1.21-26.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Guglielmetti S, Tamagnini I, Mora D, et al. Implication of an Outer Surface Lipoprotein in Adhesion of Bifidobacterium bifidum to Caco-2 Cells. Appl Environ Microbiol. 2008;74:4695–4702. doi: 10.1128/AEM.00124-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Haller D, Bode C, Hammes WP, et al. Non-pathogenic bacteria elicit a differential cytokine response by intestinal epithelial cell/leucocyte co-cultures. Gut. 2000;47:79–87. doi: 10.1136/gut.47.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Parlesak A, Haller D, Brinz S, et al. Modulation of Cytokine Release by Differentiated CACO-2 Cells in a Compartmentalized Coculture Model with Mononuclear Leucocytes and Nonpathogenic Bacteria. Scandinavian Journal of Immunology. 2004;60:477–485. doi: 10.1111/j.0300-9475.2004.01495.x. [DOI] [PubMed] [Google Scholar]
- 63.Morita H, Fuse FHT, et al. Adhesion of lactic acid bacteria to caco-2 cells and their effect on cytokine secretion. Microbiol Immunol. 2002;46(4):293–297. doi: 10.1111/j.1348-0421.2002.tb02698.x. [DOI] [PubMed] [Google Scholar]
- 64.O’Hara AM, O’Regan P, Fanning A, et al. Functional modulation of human intestinal epithelial cell responses by Bifidobacterium infantis and Lactobacillus salivarius. Immunology. 2006;118(2):202–215. doi: 10.1111/j.1365-2567.2006.02358.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pozo-Rubio T, Mujico JR, Marcos A, et al. Immunostimulatory effect of faecal Bifidobacterium species of breast-fed and formula-fed infants in a peripheral blood mononuclear cell/Caco-2 co-culture system. Br J Nutr. 2011;106(8):1216–1223. doi: 10.1017/S0007114511001656. [DOI] [PubMed] [Google Scholar]
- 66.Candela M, Perna F, Carnevali P, et al. Interaction of probiotic Lactobacillus and Bifidobacterium strains with human intestinal epithelial cells: Adhesion properties, competition against enteropathogens and modulation of IL-8 production. International Journal of Food Microbiology. 2008;125:286–292. doi: 10.1016/j.ijfoodmicro.2008.04.012. [DOI] [PubMed] [Google Scholar]
- 67.Li Q, Zhang Q, Wang M, et al. Interferon-[gamma] and tumor necrosis factor-[alpha] disrupt epithelial barrier function by altering lipid composition in membrane microdomains of tight junction. Clinical Immunology. 2008;126:67–80. doi: 10.1016/j.clim.2007.08.017. [DOI] [PubMed] [Google Scholar]
- 68.Quigley EMM. Therapies Aimed at the Gut Microbiota and Inflammation: Antibiotics, Prebiotics, Probiotics, Synbiotics, Anti-inflammatory Therapies. Gastroenterology Clinics of North America. 2011;40:207–222. doi: 10.1016/j.gtc.2010.12.009. [DOI] [PubMed] [Google Scholar]
- 69.Grimoud J, Durand H, de Souza S, et al. In vitro screening of probiotics and synbiotics according to anti-inflammatory and anti-proliferative effects. International Journal of Food Microbiology. 2010;144:42–50. doi: 10.1016/j.ijfoodmicro.2010.09.007. [DOI] [PubMed] [Google Scholar]
- 70.Roberfroid MB. Prebiotics and synbiotics: concepts and nutritional properties. Br J Nutr. 1998;80(4):S197–202. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


