Abstract
Abnormalities in the epigenetic regulation of chromatin structure and function can lead to aberrant gene expression and cancer development. Consequently, epigenetic therapies aim to restore normal chromatin modification patterns through the inhibition of various components of the epigenetic machinery. Histone deacetylase and DNA methyltransferase inhibitors represent the first putative epigenetic therapies, however these agents have pleiotropic effects and it remains unclear how they lead to therapeutic responses. More recently, drugs that inhibit histone methyltransferases were developed, perhaps representing more specific agents. Here we review emerging epigenetic targets in cancer and present recent models of promising epigenetic therapies.
Introduction
Cells in an organism, regardless of their function, contain identical genetic material, yet vary greatly in gene expression and phenotype. Gene transcription is controlled in part through the architecture of its chromatin, and through recruitment of transcription factors to specific regulatory elements. These mechanisms are regulated through covalent modifications of DNA and histone proteins that leave underlying DNA sequence unaltered. DNA methylation at the cytosine of the CpG dinucleotide is associated with gene silencing. CpG is underrepresented in the genome but clusters in “islands” often found at the 5’ end of a gene. Cancer genomes are globally hypomethylated, but by poorly understood mechanisms, CpG islands become hypermethylated in cancer, flagging in the recruitment of gene silencing complexes.
Post-translational modifications of histone proteins are mediated by enzymes that can add or subtract covalent attachments at specific residues. Histones can be methylated, acetylated, phosphorylated or ubiquitinated, and depending on the residue being modified, identical chemical modifications can have opposing consequences. In addition, certain histone modifications are dependent on each other, and can be found simultaneously on the same genomic loci under appropriate conditions, while others are mutually exclusive. Adding another layer of complexity, histone lysine residues can be mono-, di- or trimethylated while arginines can be monomethylated or symmetrically or asymmetrically dimethylated, with each modification having a specific biological effect. Collectively, the combination of covalent modifications (often referred to as a “Histone Code”) in cooperation with DNA methylation affect the structural state of chromatin and transcriptional status of a gene. The histone code is “read” by modules found within chromatin regulators including Bromo, Chromo and Tudor domains. Often, an enzyme that creates a specific histone modification contains a domain that recognizes that same mark. In this way additional molecules of the enzyme can be attracted to chromatin in a feed forward process allowing the modification to spread across a locus.
Increasing evidence links mutations, amplifications, deletions and rearrangements of genes encoding epigenetic regulators to cancer. Depending on the enzyme involved and the pathways affected, such alterations may lead to changes in gene expression and/or global changes in chromatin structure and function. Epigenetic effects can phenocopy loss of function gene mutation. Increased DNA methylation and repressive histone marks on a promoter silence gene transcription. Conversely, loss of DNA methylation and accumulation of activating marks can, similarly to chromosomal translocation or gene amplification, increase gene expression. Unlike genetic events, epigenetic changes can in theory be reversed by pharmacological intervention to block enzymes that add or remove modifications from histones (Writers and Erasers), prevent critical protein–protein interactions among transcription factors or block protein domains (Readers) from recognizing specific histone modification states.
Currently the only epigenetically directed therapies in clinical practice are inhibitors of DNMTs and histone deacetylases (HDACs). While these drugs yield global changes in DNA methylation and histone acetylation respectively, it remains uncertain whether the efficacy of these agents is linked to specific changes in gene expression. HDAC inhibitors have pleiotropic actions and can affect cytoplasmic as well as nuclear processes. Furthermore, both classes of agents elicit DNA damage responses and may be acting as low intensity cytotoxic agents. Here we review the development of a new generation of potentially more specific epigenetic therapies designed to reverse aberrant gene expression in cancer.
Histone Methyltransferases
Over the past decade, structurally distinct histone arginine and lysine methyltransferases (HMTs) were identified and linked to gene regulatory complexes. Other than DOT1L (KMT4), all lysine methyltransferases contain the conserved SET (Suppressor of variegation, Enhancer of zeste, and Trithorax) domain. More recently, the notion that demethylation occurs only upon synthesis of new histones was overturned with the discovery of enzymes that convert arginine to citruline, to remove arginine methylation, and lysine demethylases, including LSD1 (KDM1) and the Jumonji C family. Both HMTs and demethylases are altered in human cancer and represent putative therapeutic targets.
DOT1L (KMT4)
DOT1L is a unique HMT that has an AdoMet binding motif, similar to those of arginine methyltransferases (1). Additionally, while most histone modifications occur on the N-terminal tails, DOT1L methylates lysine 79 on histone H3 (H3K79), within the globular histone domain upon which DNA is wrapped. Furthermore, following monomethylation of lysine 79, DOT1L must dissociate and reassociate with histone for further methylation of the same residue. These unique properties and the fact that DOT1L is the only known H3K79 methyltransferase, make DOT1L an attractive target for specific malignancies. Dot1l is expressed throughout embryogenesis and in adult tissues. Its knockout in mice leads to defective angiogenesis in the yolk sac, and embryonic death (2). Loss of Dot1l leads to disruption of centromeres and telomeres suggesting that it is critical for the establishment or maintenance of heterochromatin structures (2). The majority of genes affected by the loss of Dot1l were activated, suggesting that H3K79 methylation by Dot1l played a role in transcriptional repression (3). This contrasts with genome-wide studies showing that actively transcribed genes are enriched for H3K79 methylation (4, 5). Furthermore, DOT1L is associated with the RNA polymerase II elongating complex, supporting the notion that DOT1L promotes gene expression (6).
DOT1L has an important role in aberrant gene regulation mediated by MLL (Mixed Lineage Leukemia/KTM2A) in leukemia. MLL translocations are found in >70% of infant leukemia and 10% of adult acute myeloid leukemia (AML), resulting in fusion of MLL to one of more than 70 different partners and deregulation of MLL target genes (7). MLL is a HMT specific for H3K4, a mark found at the promoters of active loci (5, 8). In leukemia, the gene-targeting domains of MLL are retained in the fusion protein while the C-terminal SET domain is lost, suggesting that aberrant gene activation is not linked to methylation by MLL. Many MLL fusion partners are part of a large multi-protein super elongation complex (SEC) whose function of promoting transcription possibly plays a role in leukemogenesis (9). Other MLL fusion partners, including the most common translocation partners AF4 and AF9, can recruit a smaller protein complex that includes DOT1L methyltransferase (6). MLL-AF fusion proteins bind to specific targets, such as HOX genes, resulting in the recruitment of additional DOT1L, enhanced H3K79 methylation and transcription. The importance of DOT1L for this process was highlighted by the fact that MLL-AF9 cannot immortalize bone marrow progenitors from Dotl1 null mice (10). Furthermore, elimination of Dot1l expression in MLL-AF9 transformed cells decreased expression of genes bound by MLL-AF9 but not other genes enriched for H3K79 methylation (11). These data suggested that targeting DOT1L might specifically affect leukemia-associated gene activation while sparing normal gene expression.
Using the chemical structure of AdoMet, an obligatory HMT co-factor, as the starting point for the synthesis of candidates, Daigle and colleagues identified EPZ004777 as a potent DOT1L inhibitor (Figure 1A) (12). Despite its similarity to SAM, EPZ004777 shows remarkable selectivity for DOT1L in vitro and does not affect methylation by other HMTs, such as EZH2 (KMT6), SET7 (KMT7) and WHSC1 (NSD2/MMSET). In cell line models, EPZ004777 selectively decreased K79 methylation, but only inhibited proliferation and induced apoptosis of cells harboring MLL rearrangements, while blocking expression of MLL fusion target genes. Interestingly, it took nearly a week before EPZ004777 decreased H3K79 methylation and gene expression, suggesting that H3K79 demethylation occurred by the passive replacement of methylated histones by new histones over several cell divisions. DOT1L inhibitors, administered to mice harboring human leukemia cells expressing the MLL-AF9 fusion, were well tolerated and prolonged survival. This is critical given that genetic loss of Dot1l in mice leads to anemia, and depletion of hematopoietic stem cells (13). Together, these data indicate that many MLL-associated leukemias are addicted to the HMT activity of DOT1L and that this enzyme can be targeted. However, duo to poor pharmacokinetic properties of EPZ004777, better in vivo efficacy and toxicity assessment will require molecules that have superior in vivo properties.
Fig 1.
(A) Recruitment of DOT1L via MLL fusion proteins enhances gene expression, which can be inhibited by targeting DOT1L activity or MLL-menin interaction. (B) Different genetic alterations affect H3K27 methylation.
EZH2 (KMT6)
Enhancer of Zeste 2 (EZH2) is a HMT that catalyzes methylation of H3K27. Along with co-factors SUZ12, EED and RbAp46/48, EZH2 forms the Polycomb Repressive Complex 2 (PRC2) (14). EZH2 is overexpressed in a wide range of cancers, including advanced stage and high-grade prostate, breast and lung tumors (15). EZH2 and PRC2 are critical for the control of gene expression in embryonic stem (ES) cells, maintaining self-renewal while inhibiting differentiation (16), and these properties of EZH2 appear active when the gene is overexpressed in tumors. EZH2 overexpression induces cell migration and colony formation and induces genomic instability by repression of regulators of DNA repair (17). Conversely, EZH2 depletion suppresses proliferation, and attenuates tumor formation in vivo (18). A homologous polypeptide, EZH1, can also form a PRC2-like complex and may have overlapping or complementary roles to those of EZH2 (19). However, the exact interplay between EZH2 and EZH1 is insufficiently understood and it is unclear how this relationship may be affected in cancer.
Recently, somatic mutations and deletions of EZH2 were identified in hematological malignancies, leading to the gain or loss of EZH2 function. About 30% of diffuse large B-cell lymphomas (DLBCL) and 10% of follicular lymphomas (FL), contain a mutation at tyrosine 641 (Y641) within the SET-domain, predicted to alter the substrate recognition pocket within the enzyme (20). These mutations are always heterozygous, suggesting that they are either dominant to or cooperate with the wild-type EZH2 protein. Enzymatic studies showed that WT EZH2 converted unmethylated H3K27 to H3K27me1 and to a lesser extent the me2 and me3 states (21). By contrast EZH2Y641X failed to recognize unmethylated H3K27 but readily converted H3K27me1 (created by WT EZH2) to H3K27me2 or me3. Accordingly, DLBCL cells harboring EZH2Y641X display increased levels of H3K27me3 (Figure 1B). EZH2 is silenced in resting, mature B-cells and is transiently upregulated in germinal center B-cells, where along with BCL6 it blocks DNA-damage response pathways allowing cells to survive the somatic hypermutation of antibody maturation (22). By amplifying these functions and targeting additional pathways, EZH2 mutations may stimulate malignant transformation.
In myeloid neoplasia, EZH2 is most often affected by deletions and nonsense mutations that yield loss-of-function, and leukemia cell lines harboring EZH2 mutations show decreased H3K27 methylation (23). The presence of activating and inactivating EZH2 mutations in different cancers suggest a complex, context-dependent role of Polycomb proteins in oncogenesis. It is unclear whether EZH2 affects different sets of genes in different malignancies or whether global histone changes may interfere with other chromatin functions, such as replication and DNA repair. Nevertheless, the frequent occurrence of genetic lesions affecting H3K27 (Figure 1B), suggests that this mark is under tight control, which may present a challenge in the design of safe and effective EZH2 inhibitors. The S-adenosylhomocysteine (SAH) hydrolase inhibitor 3-Deazaneplanocin A (DZNeP) is the only EZH2 inhibitor described to date (24). DZNeP can inhibit HMTs by increasing SAH levels, inducing the degradation of EZH2 and leading to a global decrease of H3K27 methylation accompanied by apoptosis of cancer cells. However, in some cells, DZNeP decreases methylation of multiple other histone residues, perhaps due to the ability of SAH to compete with the AdoMet co-factor (25). Hence, more specific inhibitors of EZH2 are required to address various lesions in cancer. Recently introduced GSK126 molecule is the first-in-class inhibitor of EZH2, acting as a SAM competitor and affecting specifically the HMT activity of EZH2 (26). DLBCL cell lines expressing mutant EZH2 are highly sensitive to GSK126 suggesting that direct inhibition of EZH2 activity presents a promising avenue for treatment in the clinic.
NSD Proteins
The NSD (nuclear receptor SET domain-containing) family of histone methyltransferases, methylate H3K36, a modification implicate in transcription, DNA repair and alternative splicing. Actively transcribed genes are enriched for H3K36me3, where the mark precludes adventitious initiation of transcription from within the gene body (5, 27). The three members of this protein family (NSD1, 2 and 3) contain PHD and PWWP domains, important for chromatin recognition, as well as the SET domain. NSD1 (KMT3B) can methylate non-histone substrates such as NF-κB, a function likely preserved in NSD2 (WHSC1/MMSET) and NSD3 (WHSC1L1) (28), suggesting that chromatin-centric studies may be insufficient to decipher how HMTs drive disease.
NSD1 is mutated in Sotos syndrome, a disorder characterized by developmental overgrowth and cognitive disabilities (29). In a small number of cytogenetically normal cases of acute myeloid leukemia (AML), chromosomal translocation fuses the N-terminus of NUP98 to the C-terminal portion of NSD1, including the SET domain (30). NUP98-NSD1 protein can immortalize progenitor bone marrow cells, activate Hox gene expression and induce AML in mice (31), in a manner dependent on H3K36 methylation. NSD3 is also translocated with NUP98 locus in rare cases of AML (32) and increased NSD3 expression occurs in about 15% of breast cancers (33). Furthermore, NSD3 readily transformed breast cell lines and cells overexpressing NSD3 cease proliferation upon its depletion (34). Deletions of the WHSC1 gene lead to Wolf Hirschhorn syndrome (WHS) characterized by cognitive and developmental defects (35). WHSC1 was linked to malignancy due to its rearrangement with the immunoglobulin locus and overexpression in ~20% of cases of multiple myeloma. Knockdown of WHSC1 in myeloma cells slows cell growth, alters cellular adhesion and induces apoptosis (36). WHSC1 levels are also frequently elevated in advanced stage solid tumors (37). WHSC1 was recently shown to be a critical factor for repair of double stranded DNA damage, but how this function correlates with its oncogenic activity is uncertain (38).
Overexpression of WHSC1 in myeloma leads to a genome-wide increase of H3K36me2 and concomitant decrease of H3K27me3 (Figure 1B) (36). The NUP98-NSD1 fusion has a similar but more localized effect on chromatin of HOX genes, activating their expression (31). These data are consistent with previous reports indicating that H3K36 methylation inhibits PRC2-mediated H3K27 methylation (39). Which genes and pathways are deregulated by this epigenetic switch in myeloma remains to be fully defined. Nevertheless, this is another example of altered H3K27 methylation as a possible driver of tumorigenesis, suggesting that inhibitors of NSD proteins may be useful for specific tumors.
H3K9 Methyltransferases
The first enzymatically active SET domain-containing HMT described was SUV39H1 (KMT1), belonging to the family of H3K9 specific methyltransferases (40). While H3K9 methylation is generally associated with transcriptional repression related to heterochromatin formation and DNA methylation, there is also evidence linking K9 methylation to transcriptionally active loci (41). H3K9 HMTs have been found upregulated in various human tumors. Consistent with this finding, downregulation of G9a (KMT1C), another H3K9-specific HMT, suppresses tumor cell growth and invasion in mice (42). Two small molecules, Chaetocin and BIX01294, specifically inhibit H3K9 HMTs through AdoMet and histone tail competitive inhibition, respectively (43–45). Given the role of H3K9 methylation in proviral silencing, it is unclear whether the drugs would have undesirable effects and what their effect may be in normal cells.
Arginine Methyltransferases (RMTs)
There have been nine different arginine methyltransferases identified to date (PRMT1-9) and methylation of arginines is associated with both positive and negative regulation of transcription. Consequently, recent evidence points to the role of arginine methylation in oncogenesis, including hematologic and solid tumors (46). Similar to lysine methyltransferases, RMTs can modify non-histone targets, including p53 and affecting its target gene specificity (47). Moreover, RMTs function can be affected by oncogenic pathways. For example, constitutively active JAK2V617F mutant phosphorylates PRMT5 function, impairing its methyltransferase activity and promoting myeloproliferation (48). While initial arginine methyltransferase inhibitors (AMIs) have been tested in vitro for their efficacy and selectivity, data regarding their usefulness in cellular and animal models has been sparse. Better understanding of the roles RMTs may play in disease will enhance the opportunity for testing these and other newer classes of AMIs.
Histone Demethylases
Histone methylation is counteracted by histone demethylases (HDMs), including LSD1 (KDM1), a flavin adenine dinucleotide (FAD)-dependent amine oxidase, and Jumonji C (JmjC) lysine and arginine demethylases, which use α-ketoglutarate and Fe++ ions as co-factors. While LSD1 can only remove methyl groups from mono- or di-methylated H3K4 or H3K9, JmjC enzymes can catalyze removal of trimethyl marks. Like HMTs, HDMs function can be altered in cancer.
UTX (KDM6A)
H3K27, the site of frequent epigenetic anomalies, is demethylated by UTX/UTY and JMJD3, all belonging to the JmjC subgroup. UTX, found on a pseudoautosomal region of Xp11.2 and UTY, encoded on the Y chromosome, share high degree of homology. JMJD3 (KDM6B) is a related protein, but is missing a tetratrico peptide repeat interaction domain found in UTX/UTY. The roles of UTX/UTY and JMJD3 in development are context dependent, tissue specific and non-overlapping (49, 50).
JMJD3 displays occasional mutation in cancer of uncertain significance (51). By contrast, somatic loss-of-function mutations of UTX are fond in multiple myeloma (10%), esophageal squamous cell (8%), renal carcinomas (1%), and occasionally in breast, AML, glioblastoma, colorectal and bladder cancer (52). UTX mutant tumors tend to suffer deletion of UTY, suggesting a drive towards complete loss-of-function of these proteins. Genes affected by UTX mutation displayed increased levels of H3K27me3 but global levels of H3K27 methylation are not altered, suggesting that UTX is primarily involved in chromatin demethylation at specific regulatory sequences (Figure 1B). Indeed, UTX forms a complex with MLL2, an H3K4 HMT (53), suggesting a mechanism where UTX removes repressive H3K27 methylation while MLL2 places activating methylation. Loss of UTX function may lead to inappropriate gene silencing by Polycomb complexes and as a result such tumors might also be amenable to therapy with EZH2 inhibitors. By contrast, tumors with loss of EZH2 function might be amenable to UTX/JMJD3 inhibitors, with an aim of restoring levels of H3K27me3.
IDH1/2-Metabolic Mutations Affecting Epigenetic Regulation
Genome sequencing of glioblastoma and AML identified recurrent, heterozygous missense mutations in IDH1 and IDH2, encoding isocitrate dehydrogenases that catalyze decarboxylation of isocitrate to α-ketoglutarate (α-KG) in the cytoplasm and mitochondria respectively (54, 55). IDH1/2 lesions occur in ~70% of low-grade brain tumors and 20% of AML and can be found in other myeloid neoplasms and chondrosarcoma (56, 57). Instead of oxidizing isocitrate to α-KG, mutant IDH reduces α-KG to 2-hydroxyglutarate (2-HG) (58). As a result, α-KG levels drop and 2-HG levels, normally barely detectable, are dramatically elevated. This has profound effects on the epigenetic state of the cell.
2-HG is a competitive inhibitor of α-KG-dependent enzymes. In leukemia, IDH mutations disrupt the function of TET2, which encodes an enzyme that converts 5-methylcytosine to 5-hydroxymethylcytosine, a step on the way to DNA demethylation (59). In AML, mutations in IDH1/2 and TET2 are mutually exclusive, suggesting that they have a common function. Indeed, increase in 2-HG inactivates TET proteins and leads to DNA hypermethylation. The 2-HG/α-KG imbalances also can inhibit prolyl hydroxylases which downregulate HIF-1α, a transcription factor involved in response to hypoxia and implicated in malignancy (60). However, recent data also suggest that (R)-but not (S)-enantiomer of 2HG can enhance activity of prolyl hydroxylase EGLN, leading to diminished HIF-1α levels (61). Furthermore, 2HG also inhibits JmjC histone demethylases (60) and IDH1 mutations can induce hypermethylation of histone H3 lysine residues 4, 9, 27 and 79 (Figure 1B). In addition, α-KG-dependent arginine demethylases, like JMJD6, are also likely to be affected by elevated 2HG (62). Why IDH1/2 mutations are found in such a limited set of tumors remains uncertain but might be related to the dependence of brain, blood, gallbladder and cartilage on specific sets of α-KG-dependent epigenetic regulators to control cell growth. Lastly, reduction of α-KG to 2HG is predicted to deplete NADPH, a stoichiometric co-factor for sirtuins, histone deacetylases that affect chromatin and non-histone proteins. Drugs targeting mutant IDH proteins are in development. Enforced expression of mutant IDH1 or IDH2 in erythroleukemia cells stimulated growth factor independent myeloid growth and down regulation of GATA1- an effect reversed by an agent that inhibits the mutant IDH enzyme (63). Further Identification of the key enzymes inhibited by 2HG and biomarkers of on-target effects will be required to interpret the action of such drugs.
Blockade of Epigenetic Readers
The epigenetic enzymes are an obvious choice for targeted therapy, but equivalently important are modules that interpret the epigenetic code. Directly or through recruitment of other transcriptional regulators, these proteins play essential roles in the control of gene expression.
BRD4
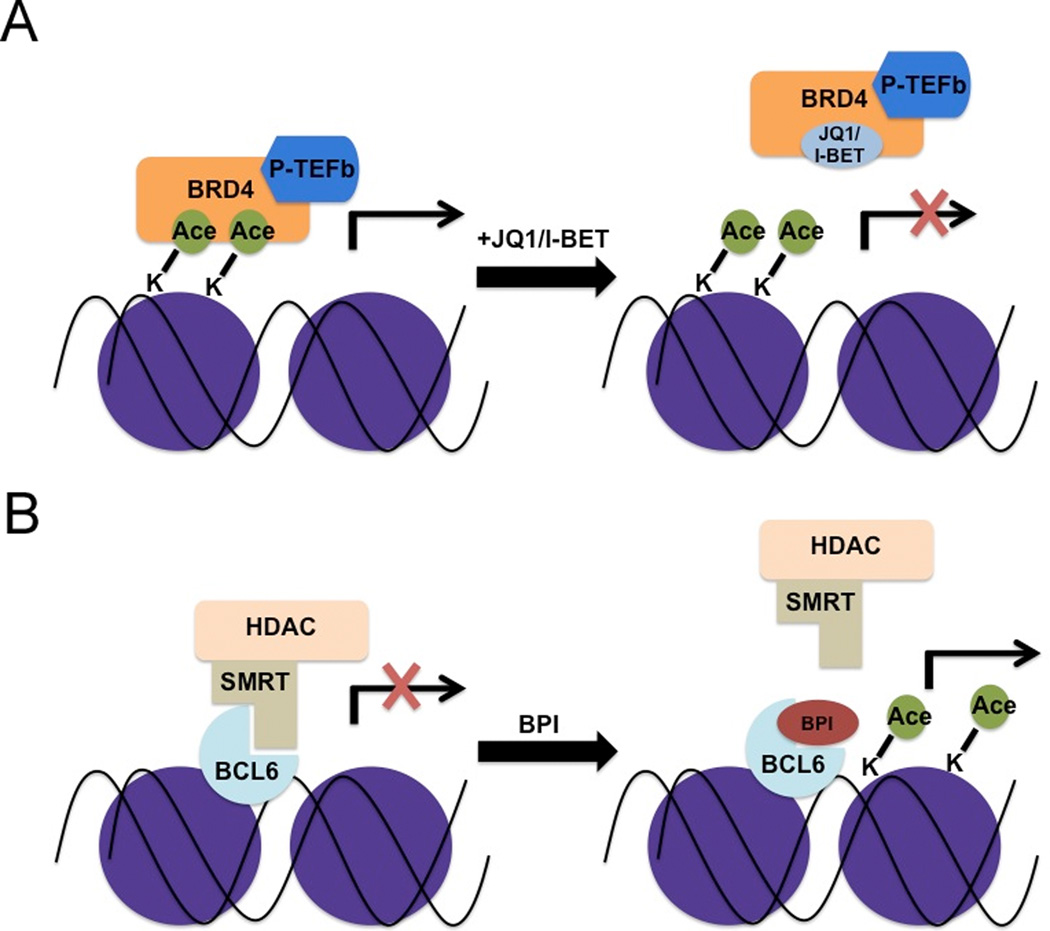
Bromodomain-containing proteins affect gene regulation through their ability to bind acetylated histone lysine residues. BRD4, a BET (Bromodomain and Extra-Terminal) protein, stimulates transcriptional elongation by recruitment of the P-TEFb complex, which phosphorylates and increases the processivity of RNA polymerase II, leading to expression of growth-promoting genes (64). In rare, t(15;19)-associated lethal midline carcinoma, the resulting BRD4-NUT fusion contains two BRD4 bromodomains that aberrantly bind and sequester acetylated histones, leading to a global hypoacetylation of chromatin, a block of differentiation and stimulation of proliferation (65). Treatment of BRD4-NUT positive cell lines with HDAC inhibitors is a true form of epigenetic differentiation therapy that leads to chromatin acetylation, the induction of sheets of differentiated, squamous epithelium and terminal cell division (66). An alternative way to inhibit BRD4 is to block its ability to bind to acetylated lysine (Figure 2). A synthetic molecule, I-BET, binds to the acetylated lysine-binding pocket of BET proteins preventing their recruitment to target genes (67). This limits the recruitment of P-TEFb and Pol II, decreases histone acetylation and transcription. Filippakopoulus et al. identified JQ1, a molecule that blocks BET/Acetyl histone interaction, with greatest specificity for the BRD3 and BRD4 (68). JQ1 displaces BRD4 from chromatin, induces differentiation and growth arrest of a midline carcinoma cell line in vitro and in mouse xenografts, phenocopying changes seen by siRNA-induced knockdown of the BRD4-NUT (Figure 2A).
Fig 2.
(A) BRD4 interaction with acetylated histones activates gene expression. Small molecule inhibitors (JQ1 or I-BET) that bind BRD4, preventing this interaction, lead to repression of BRD4 transcriptional targets. (B) Inhibition of BCL6-SMRT complex interaction by BPI (BCL6 Peptide Inhibitor) decreases HDAC recruitment, increasing histone acetylation and activating gene expression.
Bromodomain inhibitors may have broader uses as well. JQ1 was applied to a panel of multiple myeloma cells and led to decreased cell growth (69, 70). Gene expression profiling showed that this was accompanied with the decline of c-MYC target genes, explained by a dramatic decrease in c-MYC transcription. Notably, JQ1 inhibited growth of cell lines resistant to dexamethasone and melphalan, two standard agents for myeloma. While many chemotherapeutic drugs become ineffective when cells are cultured on stromal cells, growth inhibition by JQ1 was unaffected. Furthermore, JQ1 decreasing disease burden and extending survival in a c-MYC driven mouse model of myeloma. The utility of BRD4 blockade was further extended to leukemia, as an shRNA screen in a mouse AML model identified Brd4 as a critical gene for leukemia cell survival (71). Accordingly, JQ1 blocked the ability of Brd4 to bind to c-myc regulatory elements of these cells and inhibited their growth.
These encouraging studies should motivate the targeting of other histone reading modules, such as PHD fingers, chromodomains, Tudor domains and PWWP domains. The crystal structures of many of these domains bound to their cognate histone residues are solved and may facilitate the design of such molecules. However, because many of these domains are widely distributed in many epigenetic regulators and may show high degree of structural and functional similarity, the specificity of histone binding domain inhibitors may be crucial for their clinical utility.
Protein Interaction Blockade
While epigenetic enzymes and chromatin binding domains represent more traditional drug targets, recent evidence indicated that indeed transcription factors and their interplay with co-factors could be targeted.
BCL6
BCL6, a transcription factor critical for the development of germinal center B cells, represses the activity of p53 in a developmental stage-specific manner to prevent apoptosis in response to DNA damage generated during somatic mutagenesis used to produce antibody diversity (72). Aberrant overexpression of BCL6 in diffuse large B cell lymphoma stimulates malignancy by continuous inhibition of such pathways. BCL6 contains a BTB repression domain that is unique in its ability to tightly bind to the co-repressor NCOR and related SMRT protein through a specific groove (73). A BCL6 peptide inhibitor (BPI), based upon the portion of SMRT bound to BCL6, blocks BCL6-mediated repression, activates BCL6 target genes and selectively inhibits growth of DLBCL cell lines overexpressing BCL6 (Figure 2B) (74). The utility of this approach may extend beyond lymphoma. BCL6 is upregulated in chronic myelogenous leukemia in response to the tyrosine kinase inhibitor imatinib, possibly representing a general response to cellular stress. Treatment of mice with anti-BCL6 peptide and imantinib eradicated BCR-ABL positive leukemia cells while imantinib alone did not (75). BCL6 represses the histone acetyl transferase p300 suggesting that the combination of BCL6 blockade and histone deacetylase inhibitors could be a useful combination for some forms of lymphoma (76).
MLL-Menin Inhibitors
Recruitment of the wild type MLL and MLL fusion proteins to target sequences require direct interaction with menin, a tumor suppressor gene that plays a role in transcriptional activation (77). Menin is required for transcription of MLL target genes and for induction of leukemia by MLL-fusion proteins. Recently, Grembecka et al. identified a small molecule (MI-2) that specifically block MLL-menin interaction (Figure 1A) (78). Consequently, MI-2 induced growth arrest and differentiation, and blocked colony formation of mouse progenitor cells transduced with MLL-AF9 or MLL-ENL. Treatment of human leukemia prevented recruitment of MLL fusion proteins to their gene targets and suppressed cell growth, findings that phenocopy the effects of menin downregulation. However, because wild type MLL also requires menin, it remains to be determined whether these agents will adversely affect MLL-dependent normal hematopoiesis.
Conclusions
The past decade led to the identification and characterization of new enzymes and protein-protein interactions required for the epigenetic regulation of gene expression, a subset of which are dysregulated in cancer. Broadly acting therapies such as HDAC inhibitors have now been succeeded by molecules that target specific epigenetic regulators and specific transcription factors. Novel inhibitors affecting H3K27 are clearly needed, as deregulation of this modification seems to be a unifying component in a number of different malignancies. Inhibitors of H3K36 methylation, agents that block aberrant activity of mutant IDH and molecules that interfere with the “reading” of the histone code all represent agents that should be prioritized for development.
Summary.
The use of DNA methyltransferase (DNMT) and histone deacetylase (HDAC) inhibitors in patients has validated the use of drugs targeted to epigenetic enzymes and strengthened the need for development of additional therapies. In this review, we summarize recently discovered epigenetic abnormalities, their implications for cancer, and the approaches taken for discovering small molecule inhibitors targeting various properties of the epigenetic machinery.
Rationale for Epigenetic Therapies.
-
-
Growing evidence points to deregulation of the epigenetic machinery as a critical lesion in cancer.
-
-
Epigenetic modifications are reversible, making enzymes that modify histone and DNA attractive targets for pharmacological interventions.
-
-
DNA methyltransferase and HDAC inhibitors have been approved for clinical use but have many non-specific effects
-
-
Preclinical evidence suggest that targeting the writers, readers and erasers of the histone code may prove beneficial for many types of cancer.
-
-
Inhibition of H3K79 methyltransferase DOT1L may be useful in 11q23-related leukemias, associated with MLL rearrangement.
-
-
Inhibition of the chromatin reader BRD4 may prove beneficial for many tumors as BRD4 appears critical for MYC transcriptional pathways
-
-
Alteration of H3K27 methylation is a common occurrence in cancer and may arise from anomalies of histone methylation or demethylation.
Acknowledgments
Grant Support
Research in the Licht laboratory is supported by the Multiple Myeloma Research Foundation Fellowship (R.P), RO1CA123204, a Leukemia and Lymphoma Society Specialized Center of Research Award a Physical Sciences Oncology Center grant U54CA143869 (J.D.L).
Footnotes
Conflict of interest disclosure: J.D. Licht receives research support from Epizyme and is a consultant for Glaxo Smith Kline.
References
- 1.Min J, Feng Q, Li Z, Zhang Y, Xu RM. Structure of the catalytic domain of human DOT1L, a non-SET domain nucleosomal histone methyltransferase. Cell. 2003;112:711–723. doi: 10.1016/s0092-8674(03)00114-4. [DOI] [PubMed] [Google Scholar]
- 2.Jones B, Su H, Bhat A, Lei H, Bajko J, Hevi S, et al. The histone H3K79 methyltransferase Dot1L is essential for mammalian development and heterochromatin structure. PLoS genetics. 2008;4:e1000190. doi: 10.1371/journal.pgen.1000190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barry ER, Krueger W, Jakuba CM, Veilleux E, Ambrosi DJ, Nelson CE, et al. ES cell cycle progression and differentiation require the action of the histone methyltransferase Dot1L. Stem Cells. 2009;27:1538–1547. doi: 10.1002/stem.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Steger DJ, Lefterova MI, Ying L, Stonestrom AJ, Schupp M, Zhuo D, et al. DOT1L/KMT4 recruitment and H3K79 methylation are ubiquitously coupled with gene transcription in mammalian cells. Molecular and cellular biology. 2008;28:2825–2839. doi: 10.1128/MCB.02076-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 6.Bitoun E, Oliver PL, Davies KE. The mixed-lineage leukemia fusion partner AF4 stimulates RNA polymerase II transcriptional elongation and mediates coordinated chromatin remodeling. Human molecular genetics. 2007;16:92–106. doi: 10.1093/hmg/ddl444. [DOI] [PubMed] [Google Scholar]
- 7.Popovic R, Zeleznik-Le NJ. MLL: how complex does it get? Journal of cellular biochemistry. 2005;95:234–242. doi: 10.1002/jcb.20430. [DOI] [PubMed] [Google Scholar]
- 8.Nakamura T, Mori T, Tada S, Krajewski W, Rozovskaia T, Wassell R, et al. ALL-1 is a histone methyltransferase that assembles a supercomplex of proteins involved in transcriptional regulation. Molecular cell. 2002;10:1119–1128. doi: 10.1016/s1097-2765(02)00740-2. [DOI] [PubMed] [Google Scholar]
- 9.Lin C, Smith ER, Takahashi H, Lai KC, Martin-Brown S, Florens L, et al. AFF4, a component of the ELL/P-TEFb elongation complex and a shared subunit of MLL chimeras, can link transcription elongation to leukemia. Molecular cell. 2010;37:429–437. doi: 10.1016/j.molcel.2010.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang MJ, Wu H, Achille NJ, Reisenauer MR, Chou CW, Zeleznik-Le NJ, et al. Histone H3 lysine 79 methyltransferase Dot1 is required for immortalization by MLL oncogenes. Cancer research. 2010;70:10234–10242. doi: 10.1158/0008-5472.CAN-10-3294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bernt KM, Zhu N, Sinha AU, Vempati S, Faber J, Krivtsov AV, et al. MLL-rearranged leukemia is dependent on aberrant H3K79 methylation by DOT1L. Cancer cell. 2011;20:66–78. doi: 10.1016/j.ccr.2011.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Daigle SR, Olhava EJ, Therkelsen CA, Majer CR, Sneeringer CJ, Song J, et al. Selective killing of mixed lineage leukemia cells by a potent small-molecule DOT1L inhibitor. Cancer cell. 2011;20:53–65. doi: 10.1016/j.ccr.2011.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jo SY, Granowicz EM, Maillard I, Thomas D, Hess JL. Requirement for Dot1l in murine postnatal hematopoiesis and leukemogenesis by MLL translocation. Blood. 2011;117:4759–4768. doi: 10.1182/blood-2010-12-327668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morey L, Helin K. Polycomb group protein-mediated repression of transcription. Trends in biochemical sciences. 2010;35:323–332. doi: 10.1016/j.tibs.2010.02.009. [DOI] [PubMed] [Google Scholar]
- 15.Albert M, Helin K. Histone methyltransferases in cancer. Seminars in cell & developmental biology. 2010;21:209–220. doi: 10.1016/j.semcdb.2009.10.007. [DOI] [PubMed] [Google Scholar]
- 16.Bracken AP, Dietrich N, Pasini D, Hansen KH, Helin K. Genome-wide mapping of Polycomb target genes unravels their roles in cell fate transitions. Genes & development. 2006;20:1123–1136. doi: 10.1101/gad.381706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kleer CG, Cao Q, Varambally S, Shen R, Ota I, Tomlins SA, et al. EZH2 is a marker of aggressive breast cancer and promotes neoplastic transformation of breast epithelial cells. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:11606–11611. doi: 10.1073/pnas.1933744100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gonzalez ME, Li X, Toy K, DuPrie M, Ventura AC, Banerjee M, et al. Downregulation of EZH2 decreases growth of estrogen receptor-negative invasive breast carcinoma and requires BRCA1. Oncogene. 2009;28:843–853. doi: 10.1038/onc.2008.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shen X, Liu Y, Hsu YJ, Fujiwara Y, Kim J, Mao X, et al. EZH1 mediates methylation on histone H3 lysine 27 and complements EZH2 in maintaining stem cell identity and executing pluripotency. Molecular cell. 2008;32:491–502. doi: 10.1016/j.molcel.2008.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morin RD, Johnson NA, Severson TM, Mungall AJ, An J, Goya R, et al. Somatic mutations altering EZH2 (Tyr641) in follicular and diffuse large B-cell lymphomas of germinal-center origin. Nature genetics. 2010;42:181–185. doi: 10.1038/ng.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sneeringer CJ, Scott MP, Kuntz KW, Knutson SK, Pollock RM, Richon VM, et al. Coordinated activities of wild-type plus mutant EZH2 drive tumor-associated hypertrimethylation of lysine 27 on histone H3 (H3K27) in human B-cell lymphomas. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:20980–20985. doi: 10.1073/pnas.1012525107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Velichutina I, Shaknovich R, Geng H, Johnson NA, Gascoyne RD, Melnick AM, et al. EZH2-mediated epigenetic silencing in germinal center B cells contributes to proliferation and lymphomagenesis. Blood. 2010;116:5247–5255. doi: 10.1182/blood-2010-04-280149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ernst T, Chase AJ, Score J, Hidalgo-Curtis CE, Bryant C, Jones AV, et al. Inactivating mutations of the histone methyltransferase gene EZH2 in myeloid disorders. Nature genetics. 2010;42:722–726. doi: 10.1038/ng.621. [DOI] [PubMed] [Google Scholar]
- 24.Tan J, Yang X, Zhuang L, Jiang X, Chen W, Lee PL, et al. Pharmacologic disruption of Polycomb-repressive complex 2-mediated gene repression selectively induces apoptosis in cancer cells. Genes & development. 2007;21:1050–1063. doi: 10.1101/gad.1524107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miranda TB, Cortez CC, Yoo CB, Liang G, Abe M, Kelly TK, et al. DZNep is a global histone methylation inhibitor that reactivates developmental genes not silenced by DNA methylation. Mol Cancer Ther. 2009;8:1579–1588. doi: 10.1158/1535-7163.MCT-09-0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Creasy CL, McCabe MT, Korenchuk S, Diaz E, Ott H, Thompson CS, et al. A novel selective EZH2 inhibitor exhibits anti-tumor activity in lymphoma with EZH2 activating mutations [abstract]. Proceedings of the 103rd Annual Meeting of the American Association for Cancer Research; AACR; 2012 Mar 31-Apr 4; Chicago, Illinois. Philadelphia (PA). 2012. p. 492. Abstract nr 4700. [Google Scholar]
- 27.Carrozza MJ, Li B, Florens L, Suganuma T, Swanson SK, Lee KK, et al. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell. 2005;123:581–592. doi: 10.1016/j.cell.2005.10.023. [DOI] [PubMed] [Google Scholar]
- 28.Lu T, Jackson MW, Wang B, Yang M, Chance MR, Miyagi M, et al. Regulation of NF-kappaB by NSD1/FBXL11-dependent reversible lysine methylation of p65. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:46–51. doi: 10.1073/pnas.0912493107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Faravelli F. NSD1 mutations in Sotos syndrome. American journal of medical genetics Part C, Seminars in medical genetics. 2005;137C:24–31. doi: 10.1002/ajmg.c.30061. [DOI] [PubMed] [Google Scholar]
- 30.Hollink IH, van den Heuvel-Eibrink MM, Arentsen-Peters ST, Pratcorona M, Abbas S, Kuipers JE, et al. NUP98/NSD1 characterizes a novel poor prognostic group in acute myeloid leukemia with a distinct HOX gene expression pattern. Blood. 2011;118:3645–3656. doi: 10.1182/blood-2011-04-346643. [DOI] [PubMed] [Google Scholar]
- 31.Wang GG, Cai L, Pasillas MP, Kamps MP. NUP98-NSD1 links H3K36 methylation to Hox-A gene activation and leukaemogenesis. Nature cell biology. 2007;9:804–812. doi: 10.1038/ncb1608. [DOI] [PubMed] [Google Scholar]
- 32.Rosati R, La Starza R, Veronese A, Aventin A, Schwienbacher C, Vallespi T, et al. NUP98 is fused to the NSD3 gene in acute myeloid leukemia associated with t(8;11)(p11.2;p15) Blood. 2002;99:3857–3860. doi: 10.1182/blood.v99.10.3857. [DOI] [PubMed] [Google Scholar]
- 33.Angrand PO, Apiou F, Stewart AF, Dutrillaux B, Losson R, Chambon P. NSD3, a new SET domain-containing gene, maps to 8p12 and is amplified in human breast cancer cell lines. Genomics. 2001;74:79–88. doi: 10.1006/geno.2001.6524. [DOI] [PubMed] [Google Scholar]
- 34.Zhou Z, Thomsen R, Kahns S, Nielsen AL. The NSD3L histone methyltransferase regulates cell cycle and cell invasion in breast cancer cells. Biochemical and biophysical research communications. 2010;398:565–570. doi: 10.1016/j.bbrc.2010.06.119. [DOI] [PubMed] [Google Scholar]
- 35.Bergemann AD, Cole F, Hirschhorn K. The etiology of Wolf-Hirschhorn syndrome. Trends in genetics : TIG. 2005;21:188–195. doi: 10.1016/j.tig.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 36.Martinez-Garcia E, Popovic R, Min DJ, Sweet SM, Thomas PM, Zamdborg L, et al. The MMSET histone methyl transferase switches global histone methylation and alters gene expression in t(4;14) multiple myeloma cells. Blood. 2011;117:211–220. doi: 10.1182/blood-2010-07-298349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hudlebusch HR, Santoni-Rugiu E, Simon R, Ralfkiaer E, Rossing HH, Johansen JV, et al. The histone methyltransferase and putative oncoprotein MMSET is overexpressed in a large variety of human tumors. Clinical cancer research : an official journal of the American Association for Cancer Research. 2011;17:2919–2933. doi: 10.1158/1078-0432.CCR-10-1302. [DOI] [PubMed] [Google Scholar]
- 38.Pei H, Zhang L, Luo K, Qin Y, Chesi M, Fei F, et al. MMSET regulates histone H4K20 methylation and 53BP1 accumulation at DNA damage sites. Nature. 2011;470:124–128. doi: 10.1038/nature09658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yuan W, Xu M, Huang C, Liu N, Chen S, Zhu B. H3K36 methylation antagonizes PRC2-mediated H3K27 methylation. The Journal of biological chemistry. 2011;286:7983–7989. doi: 10.1074/jbc.M110.194027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rea S, Eisenhaber F, O'Carroll D, Strahl BD, Sun ZW, Schmid M, et al. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. 2000;406:593–599. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- 41.Vakoc CR, Mandat SA, Olenchock BA, Blobel GA. Histone H3 lysine 9 methylation and HP1gamma are associated with transcription elongation through mammalian chromatin. Molecular cell. 2005;19:381–391. doi: 10.1016/j.molcel.2005.06.011. [DOI] [PubMed] [Google Scholar]
- 42.Chen MW, Hua KT, Kao HJ, Chi CC, Wei LH, Johansson G, et al. H3K9 histone methyltransferase G9a promotes lung cancer invasion and metastasis by silencing the cell adhesion molecule Ep-CAM. Cancer research. 2010;70:7830–7840. doi: 10.1158/0008-5472.CAN-10-0833. [DOI] [PubMed] [Google Scholar]
- 43.Greiner D, Bonaldi T, Eskeland R, Roemer E, Imhof A. Identification of a specific inhibitor of the histone methyltransferase SU(VAR)3-9. Nature chemical biology. 2005;1:143–145. doi: 10.1038/nchembio721. [DOI] [PubMed] [Google Scholar]
- 44.Kubicek S, O'Sullivan RJ, August EM, Hickey ER, Zhang Q, Teodoro ML, et al. Reversal of H3K9me2 by a small-molecule inhibitor for the G9a histone methyltransferase. Molecular cell. 2007;25:473–481. doi: 10.1016/j.molcel.2007.01.017. [DOI] [PubMed] [Google Scholar]
- 45.Chang Y, Zhang X, Horton JR, Upadhyay AK, Spannhoff A, Liu J, et al. Structural basis for G9a-like protein lysine methyltransferase inhibition by BIX-01294. Nature structural & molecular biology. 2009;16:312–317. doi: 10.1038/nsmb.1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Copeland RA, Solomon ME, Richon VM. Protein methyltransferases as a target class for drug discovery. Nature reviews Drug discovery. 2009;8:724–732. doi: 10.1038/nrd2974. [DOI] [PubMed] [Google Scholar]
- 47.Jansson M, Durant ST, Cho EC, Sheahan S, Edelmann M, Kessler B, et al. Arginine methylation regulates the p53 response. Nature cell biology. 2008;10:1431–1439. doi: 10.1038/ncb1802. [DOI] [PubMed] [Google Scholar]
- 48.Liu F, Zhao X, Perna F, Wang L, Koppikar P, Abdel-Wahab O, et al. JAK2V617F-mediated phosphorylation of PRMT5 downregulates its methyltransferase activity and promotes myeloproliferation. Cancer cell. 2011;19:283–294. doi: 10.1016/j.ccr.2010.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hubner MR, Spector DL. Role of H3K27 demethylases Jmjd3 and UTX in transcriptional regulation. Cold Spring Harbor symposia on quantitative biology. 2010;75:43–49. doi: 10.1101/sqb.2010.75.020. [DOI] [PubMed] [Google Scholar]
- 50.Agger K, Cloos PA, Christensen J, Pasini D, Rose S, Rappsilber J, et al. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature. 2007;449:731–734. doi: 10.1038/nature06145. [DOI] [PubMed] [Google Scholar]
- 51.Catalogue of Somatic Mutations in Cancer [Internet] London (UK): Wellcome Trust Sanger Institute; [cited 2012 Apr 8]. c2012 Available from: http://www.sanger.ac.uk/genetics/CGP/cosmic/ [Google Scholar]
- 52.van Haaften G, Dalgliesh GL, Davies H, Chen L, Bignell G, Greenman C, et al. Somatic mutations of the histone H3K27 demethylase gene UTX in human cancer. Nature genetics. 2009;41:521–523. doi: 10.1038/ng.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lee MG, Villa R, Trojer P, Norman J, Yan KP, Reinberg D, et al. Demethylation of H3K27 regulates polycomb recruitment and H2A ubiquitination. Science. 2007;318:447–450. doi: 10.1126/science.1149042. [DOI] [PubMed] [Google Scholar]
- 54.Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W, et al. IDH1 and IDH2 mutations in gliomas. The New England journal of medicine. 2009;360:765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mardis ER, Ding L, Dooling DJ, Larson DE, McLellan MD, Chen K, et al. Recurring mutations found by sequencing an acute myeloid leukemia genome. The New England journal of medicine. 2009;361:1058–1066. doi: 10.1056/NEJMoa0903840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tefferi A, Lasho TL, Abdel-Wahab O, Guglielmelli P, Patel J, Caramazza D, et al. IDH1 and IDH2 mutation studies in 1473 patients with chronic-, fibrotic- or blast-phase essential thrombocythemia, polycythemia vera or myelofibrosis. Leukemia : official journal of the Leukemia Society of America, Leukemia Research Fund, UK. 2010;24:1302–1309. doi: 10.1038/leu.2010.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Amary MF, Bacsi K, Maggiani F, Damato S, Halai D, Berisha F, et al. IDH1 and IDH2 mutations are frequent events in central chondrosarcoma and central and periosteal chondromas but not in other mesenchymal tumours. The Journal of pathology. 2011;224:334–343. doi: 10.1002/path.2913. [DOI] [PubMed] [Google Scholar]
- 58.Ward PS, Patel J, Wise DR, Abdel-Wahab O, Bennett BD, Coller HA, et al. The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting alpha-ketoglutarate to 2-hydroxyglutarate. Cancer cell. 2010;17:225–234. doi: 10.1016/j.ccr.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Figueroa ME, Abdel-Wahab O, Lu C, Ward PS, Patel J, Shih A, et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer cell. 2010;18:553–567. doi: 10.1016/j.ccr.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xu W, Yang H, Liu Y, Yang Y, Wang P, Kim SH, et al. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of alpha-ketoglutarate-dependent dioxygenases. Cancer cell. 2011;19:17–30. doi: 10.1016/j.ccr.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Koivunen P, Lee S, Duncan CG, Lopez G, Lu G, Ramkissoon S, et al. Transformation by the (R)-enantiomer of 2-hydroxyglutarate linked to EGLN activation. Nature. 2012;483:484–488. doi: 10.1038/nature10898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chang B, Chen Y, Zhao Y, Bruick RK. JMJD6 is a histone arginine demethylase. Science. 2007;318:444–447. doi: 10.1126/science.1145801. [DOI] [PubMed] [Google Scholar]
- 63.Yen K, Wnag F, Choe S, Schalm S, Hansen E, Straley K, et al. IDH mutations and tumorgenicity [abstract]. Proceedings of the 103rd Annual Meeting of the American Association for Cancer Research; AACR; 2012 Mar 31-Apr 4; Chicago, Illinois. Philadelphia (PA). 2012. p. 363. Abstract nr LB-164. [Google Scholar]
- 64.Yang Z, He N, Zhou Q. Brd4 recruits P-TEFb to chromosomes at late mitosis to promote G1 gene expression and cell cycle progression. Molecular and cellular biology. 2008;28:967–976. doi: 10.1128/MCB.01020-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.French CA, Ramirez CL, Kolmakova J, Hickman TT, Cameron MJ, Thyne ME, et al. BRD-NUT oncoproteins: a family of closely related nuclear proteins that block epithelial differentiation and maintain the growth of carcinoma cells. Oncogene. 2008;27:2237–2242. doi: 10.1038/sj.onc.1210852. [DOI] [PubMed] [Google Scholar]
- 66.Schwartz BE, Hofer MD, Lemieux ME, Bauer DE, Cameron MJ, West NH, et al. Differentiation of NUT midline carcinoma by epigenomic reprogramming. Cancer research. 2011;71:2686–2696. doi: 10.1158/0008-5472.CAN-10-3513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nicodeme E, Jeffrey KL, Schaefer U, Beinke S, Dewell S, Chung CW, et al. Suppression of inflammation by a synthetic histone mimic. Nature. 2010;468:1119–1123. doi: 10.1038/nature09589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, et al. Selective inhibition of BET bromodomains. Nature. 2010;468:1067–1073. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mertz JA, Conery AR, Bryant BM, Sandy P, Balasubramanian S, Mele DA, et al. Targeting MYC dependence in cancer by inhibiting BET bromodomains. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:16669–16674. doi: 10.1073/pnas.1108190108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146:904–917. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zuber J, Shi J, Wang E, Rappaport AR, Herrmann H, Sison EA, et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature. 2011;478:524–528. doi: 10.1038/nature10334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Phan RT, Dalla-Favera R. The BCL6 proto-oncogene suppresses p53 expression in germinal-centre B cells. Nature. 2004;432:635–639. doi: 10.1038/nature03147. [DOI] [PubMed] [Google Scholar]
- 73.Melnick A, Carlile G, Ahmad KF, Kiang CL, Corcoran C, Bardwell V, et al. Critical residues within the BTB domain of PLZF and Bcl-6 modulate interaction with corepressors. Molecular and cellular biology. 2002;22:1804–1818. doi: 10.1128/MCB.22.6.1804-1818.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Polo JM, Dell'Oso T, Ranuncolo SM, Cerchietti L, Beck D, Da Silva GF, et al. Specific peptide interference reveals BCL6 transcriptional and oncogenic mechanisms in B-cell lymphoma cells. Nat Med. 2004;10:1329–1335. doi: 10.1038/nm1134. [DOI] [PubMed] [Google Scholar]
- 75.Hurtz C, Hatzi K, Cerchietti L, Braig M, Park E, Kim YM, et al. BCL6-mediated repression of p53 is critical for leukemia stem cell survival in chronic myeloid leukemia. J Exp Med. 2011;208:2163–2174. doi: 10.1084/jem.20110304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cerchietti LC, Hatzi K, Caldas-Lopes E, Yang SN, Figueroa ME, Morin RD, et al. BCL6 repression of EP300 in human diffuse large B cell lymphoma cells provides a basis for rational combinatorial therapy. J Clin Invest. 2010 doi: 10.1172/JCI42869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yokoyama A, Somervaille TC, Smith KS, Rozenblatt-Rosen O, Meyerson M, Cleary ML. The menin tumor suppressor protein is an essential oncogenic cofactor for MLL-associated leukemogenesis. Cell. 2005;123:207–218. doi: 10.1016/j.cell.2005.09.025. [DOI] [PubMed] [Google Scholar]
- 78.Grembecka J, He S, Shi A, Purohit T, Muntean AG, Sorenson RJ, et al. Menin-MLL inhibitors reverse oncogenic activity of MLL fusion proteins in leukemia. Nature chemical biology. 2012 doi: 10.1038/nchembio.773. [DOI] [PMC free article] [PubMed] [Google Scholar]