Summary
The mechanisms that dictate nuclear shape are largely unknown. Here we screened the budding yeast deletion collection for mutants with abnormal nuclear shape. A common phenotype was the appearance of a nuclear extension, particularly in mutants in DNA repair and chromosome segregation genes. Our data suggest that these mutations led to the abnormal nuclear morphology indirectly, by causing a checkpoint-induced cell cycle delay. Indeed, delaying cells in mitosis by other means also led to the appearance of nuclear extensions, while inactivating the DNA damage checkpoint pathway in a DNA repair mutant reduced the fraction of cells with nuclear extensions. Formation of a nuclear extension was specific to a mitotic delay, as cells arrested in S or G2 had round nuclei. Moreover, the nuclear extension always coincided with the nucleolus, while the morphology of DNA mass remained largely unchanged. Finally, we found that phospholipid synthesis continues unperturbed when cells delay in mitosis, and inhibiting phospholipid synthesis abolished the formation of nuclear extensions. Our data suggest a mechanism that promotes nuclear envelope expansion during mitosis. When mitotic progression is delayed, cells sequester the added membrane to the nuclear envelope associated with the nucleolus, possibly to avoid disruption of intra-nuclear organization.
Results and Discussion
Mutants in DNA repair and chromosome segregation genes exhibit abnormal nuclear morphology
The majority of eukaryotic cells undergo open mitosis, where the nuclear envelope (NE) disassembles at the beginning of mitosis and reassembles following chromosome segregation [1]. In contrast, many yeast undergo closed mitosis [2], where the nuclear envelope remains intact throughout the cell cycle, and chromosome segregation occurs within an elongating nucleus. In budding yeast, the nucleus is round throughout interphase, but adopts an hourglass shape during mitosis (Figure 1A, see arrow in left panel). Regulation of nuclear shape is a question of general importance, as changes in nuclear morphology are the hallmarks of aging and certain diseases, such as cancer [3–5]. Yet the mechanisms that govern NE shape changes throughout the cell cycle are poorly understood. The budding yeast Pah1p (homologous to the metazoan lipin) is a phosphatidic acid hydrolase that is activated by the Nem1p-Spo7p phosphatase complex and inactivated in mitosis by Cdk phosphorylation [6–8]. Pah1p is also phosphorylated by the cyclin-dependent kinase-cyclin complex Pho85p-Pho80p [9]. Cells lacking Pah1p, Nem1p, or Spo7p up-regulate phospholipid synthesis and exhibit nuclear extensions called “flares” [6, 10–12], suggesting a link between cell cycle regulated phospholipid synthesis and nuclear shape.
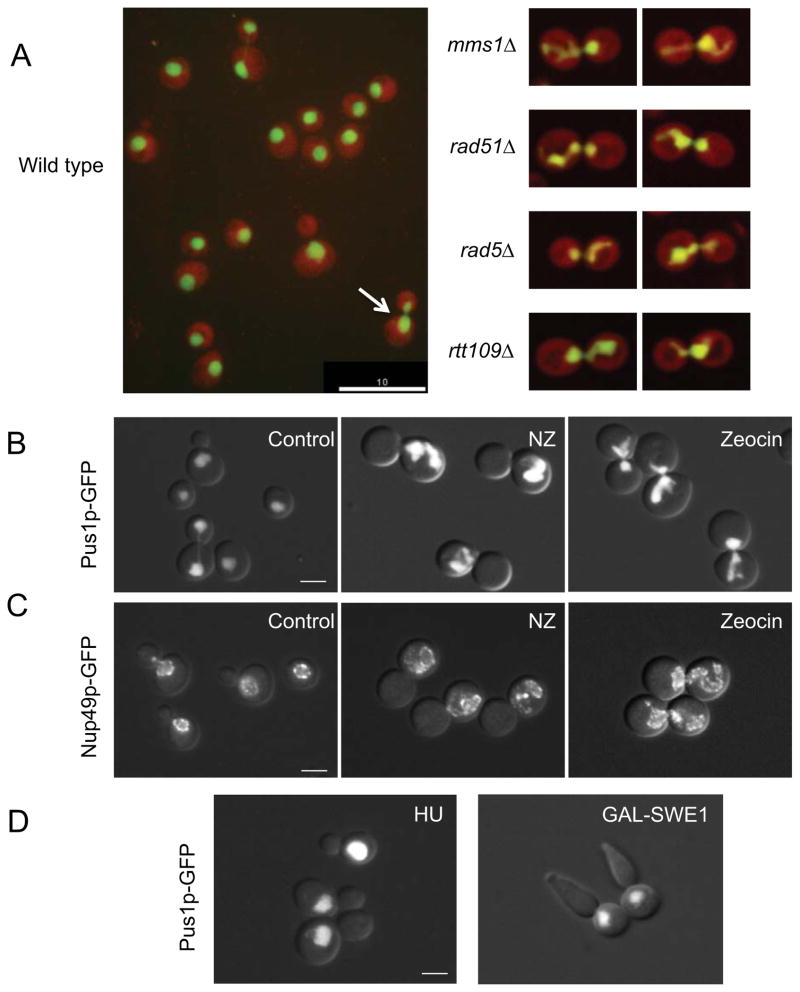
Figure 1. Mitotic delay leads to the formation of a nuclear extension.
(A) Images of strains from the automated microscopy screen. Cells are expressing a nuclear marker, Pus1p-GFP (green), and a cytoplasmic marker, TdTomato (red). Left: a field of wild type cells. The arrow points to a typical large budded cell at early stages of mitosis, with an elongated nucleus. Right: typical examples of cells of the indicated genetic backgrounds with nuclei exhibiting nuclear extensions. Scale bar =10μm. Additional images are in Supplemental Figure S1. (B and C) Wild type cells (KW913) expressing Pus1p-GFP (pane B) or Nup49p-GFP (pane C) were either untreated or treated with nocodazole (NZ) or zeocin for 3 hours. The nucleus in nocodazole treated cells does not traverse the bud neck due to the absence of microtubules. Scale bar =3μm. (D) Left panel: Wild type cells (KW926) were treated with hydroxyurea (HU) to induce an S phase arrest. Right panel: Cells over expressing SWE1 (MWY1152) and expressing Pus1p-GFP were arrested in G2 arrest by adding galactose. In both cases the arrest was achieved after three hours. Typical examples are shown. Scale bar =3μm. Quantification of the nuclear morphology of these strains and the control cells is presented in Table 1.
To identify additional pathways that affect nuclear shape, we carried out a high-content, genome-wide screen of roughly 4300 mutant strains from the S. cerevisiae deletion collection. The deletion strains expressed the nuclear Pus1p-GFP, which fills the entire nucleoplasm [12, 13], and a cytoplasmic tdTomato marker (Figure 1A). Deletion strains were grown individually, imaged and analyzed for nuclear shape computationally and by visual inspection. Strains identified as having abnormal nuclei by either approach were retested.
A striking phenotype seen in several mutant strains was the presence of a nuclear extension (Figure 1A and Supplemental Figure S1). A similar nuclear phenotype was previously noted by Thrower et al in cells delayed in mitosis due to dicentric chromosome breakage [14]. Our list of gene deletions resulting in this phenotype included DNA recombination and DNA repair genes, and genes that control spindle function/chromosome segregation (Supplemental Table S1). Interestingly, fission yeast defective in spindle assembly also exhibit abnormal nuclear shape [15]. Also represented were genes involved in lipid and fatty acid synthesis, including NEM1 (the strain collection tested did not include spo7Δ or pah1Δ strains). Nuclear extensions in DNA recombination/repair mutants and spindle function/chromosome segregation mutants were more common in mitotic (large budded) cells, a trend not seen in lipid/fatty synthesis mutants (Supplemental Table S1 and see below).
To further confirm the observations from the high throughput screen, we first focused on the DNA recombination and repair mutations. Ten deletion strains were recreated and analyzed for the presence of abnormal nuclei. These strains were compared to an isogenic nem1Δ strain, which exhibits a nuclear extension called a flare [6, 11, 12]. Of the ten DNA recombination/repair mutants retested, nine exhibited a statistically significant increase over wild type in the fraction of cells with a nuclear extension (Supplemental Figure S1B). Unlike nem1Δ cells, which were equally likely to exhibit a nuclear flare in interphase or mitosis, the incidence of abnormal nuclei in DNA recombination and repair mutant strains was 2–5 fold higher in mitotic cells than in interphase cells (Supplemental Figure S1C). The preponderance of abnormal nuclei in mitotic cells was also observed in cft4Δ and cin8Δ mutants, which are defective in spindle-related functions (data not shown). Interestingly, while few wild type cells exhibited abnormal nuclei, these nuclei, too, were primarily in mitotic cells (Supplemental Figure S1C).
Nuclear extensions form as a consequence of a mitotic delay
In budding yeast, both the DNA damage checkpoint pathway and the spindle checkpoint pathway delay the cell cycle in mitosis, prior to anaphase [16–19]. Thus, the abnormal nuclear morphology seen in both DNA repair/ recombination mutants and spindle function/chromosome segregation mutants could have been an indirect consequence of a cell cycle delay caused by checkpoint activation. If that were the case, then other conditions that delay cells in mitosis should also cause nuclear extensions. To test this, we quantified the fraction of cells with a nuclear extension following treatment with nocodazole (which depolymerizes microtubules and leads to a mid-mitosis delay through spindle checkpoint activation), or bleomycin or zeocin (which cause DNA double strand breaks and lead to a mid-mitosis delay due to DNA damage checkpoint activation). We also examined nuclear morphology in a cdc16 mutant grown at 34°C, a condition that blocks anaphase initiation by inactivating the anaphase promoting complex, but without activating a checkpoint [20, 21]. In untreated cells, the majority of cells exhibited round or hourglass shaped nuclei, as expected (Table 1 and Figures 1B, 1C). In contrast, the majority of cells treated with nocodazole, bleomycin or zeocin had nuclear extensions (Table 1 and Figures 1B, 1C), as did cdc16 mutants (Table 1). Nuclear extensions were also seen in cells expressing non-degradable Pds1p, which leads to a pre-anaphase delay by blocking sister chromatid separation [22] (data not shown). Finally, if nuclear extensions in DNA recombination/repair mutants formed as an indirect consequence of a mitotic delay, rather than a direct consequence of loss of DNA repair/recombination activity, then inactivating the DNA damage checkpoint pathway should reduce the fraction of DNA repair deficient cells that exhibit nuclear extensions. Indeed, deletion of the RAD9 gene, a component of the DNA damage checkpoint pathway, in rad51Δ cells led to both fewer cell cycle-delayed cells and a lower fraction of cells with nuclear extensions compared to rad51Δ alone (Table 1). Taken together, our data show that delaying cell cycle progression prior to anaphase initiation, by a variety of independent means, results in the formation of a nuclear extension.
Table 1.
Cell cycle dependence of nuclear extensions
| Condition | Percent large budded cells with a single nucleusa | Percent cells with nuclear extensionsb | |
|---|---|---|---|
| 1 | Wild type, untreated | 32.17 ± 4.62 | 21.7 ± 0.56 |
| 2 | Nocodazole | 83.09 ± 1.29 | 87.5 ± 1.96* |
| 3 | Bleomycin | 80.81 ± 7.95 | 55.58 ± 4.77* |
| 4 | Hydroxyurea | 70.24 ± 3.68 | 26.75 ± 3.47 |
| 5 | Wild type at 34°C | 8.5 ± 0.71 | 12.5 ± 3.54 |
| 6 | cdc16-123 at 34°C | 76.5 ± 7.78 | 84.0 ±1.41* |
| 7 | GAL-SWE1c | 74.87 ± 7.91 | 29.38 ± 5.78* |
| 8 | Wild type + nocodazolec | 73.19 ± 8.58 | 86.87 ± 4.73 |
| 9 | Wild type at 34°C | 7.0 ± 0 | 5.0 ± 2.83 |
| 10 | cdc6-1 at 34°C | 73.5 ± 3.54 | 10.5 ± 6.36 |
| 11 | rad9Δ | 29.94 ± 8.18 | 26.46 ± 3.47* |
| 12 | rad51Δ | 43.97 ± 2.67 | 46.15 ± 9.73 |
| 13 | rad51Δrad9Δ | 27.9 ± 5.17 | 32.94 ± 7.64* |
| 14 | GAL-SPO7+NEM1 in nocodazolec | 92.97 ± 3.32 | 23.37 ± 2.78* |
| 15 | Vectors only in nocodazolec,d | 93.04 ± 0.33 | 92.88 ± 3.41 |
| 16 | GAL-OPI1 in nocodazolec | 92.23 ± 2.02 | 61.91 ± 4.13* |
| 17 | Vector only in nocodazolec,d | 90.94 ± 4.25 | 96.85 ± 1.52 |
For different treatments, a large budded with a single nucleus state reflects a different cell cycle stage. For example, nocodazole treatment and growth of cdc16-123 cells at 34°C arrests cells in mid-mitosis, hydroxyurea and growth of cdc6-1 cells at 34°C arrests cells at early stages of DNA replication, and over expression of SWE1 arrests cells in G2. For untreated cells, large budded cells with a single nucleus are typically in G2 or early mitosis (before anaphase)
Numbers are average ± standard deviation, based on at least three biological replicates. Statistical analyses were done using students T-test. In each section, the reference control is shown in bold and the values that are statistically significantly different from the control (p< 0.05) are indicated with an asterisk (*).
Cells were grown in the presence of galactose.
Vectors only refers to the same vector backbones used in the construction of the GAL-SPO7 and GAL-NEM1, or GAL-OPI1
Stone et al [23] reported that arresting cells in G1 with mating pheromone resulted in nuclear extensions. To examine whether any cell cycle arrest results in nuclear extensions, cells were arrested in S phase with hydroxyurea, which limits deoxyribonuclotide availability [24], or by growing cdc6-1 cells at 34°C, a condition that blocks the initiation of DNA replication [25]. Cells were also arrested in G2 by over-expression of SWE1, an inhibitor of the mitotic cyclin-dependent kinase (Cdk1) [26]. In both S phase and G2 arrested cells, the fraction of cells with abnormal nuclei was significantly lower than in mitotic arrested cells, and similar to that of cycling wild type cells (Figure 1D and Table 1). Thus, the formation of a nuclear extension is not a general consequence of a cell cycle arrest, but happens specifically during a mitotic delay or following a G1 arrest by pheromone treatment [23].
Nuclear extensions form at the nuclear envelope adjacent to the nucleolus
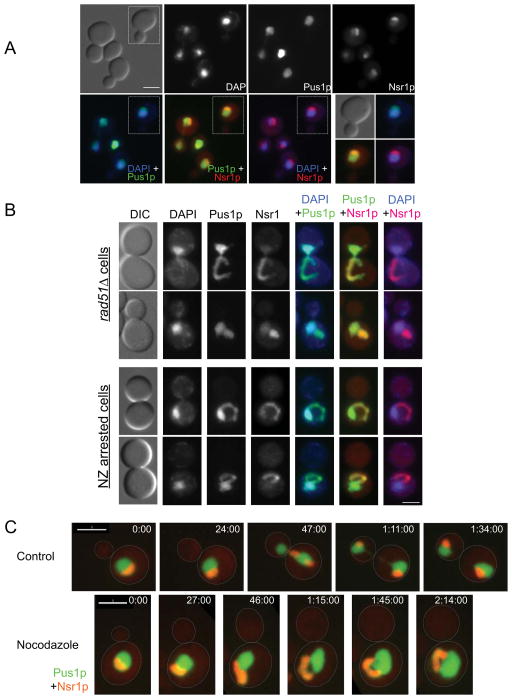
In budding yeast, the nucleolus forms a crescent-shaped structure that caps the DNA mass (Figure 2A). In cells disrupted for the PAH1 pathway (e.g. spo7Δ mutant), a nuclear flare forms at the NE that is adjacent to the nucleolus, without altering the overall structure of the DAPI-staining DNA mass [6, 11, 12]. To examine if a similar relationship exists between the nucleolus and the nuclear extension of mitotically delayed cells, we expressed both Pus1p-GFP and a nucleolar marker fused to mCherry (Nsr1p-CR). Two conditions are shown: rad51Δ cells and nocodazole arrested cells. When examined by DAPI staining, the DNA mass appeared overall normal (Figure 2B), although occasionally a thin thread of DNA could be seen extending away from the bulk of the DNA (Supplemental Figure S2A). Pus1p-GFP revealed the existence of a nuclear extension, and the nucleolar marker Nsr1p-CR showed that the nuclear extension associated with the nucleolus (Figure 2B), similar to spo7Δ flares. In nocodazole treated cells, the nucleolus co-localized perfectly with the nuclear extension in 88.9% of cells (n=81). In the remaining 11.1% the nucleolus still filled the main nuclear extension, but there were additional extensions, adjacent to the nucleolus, that did not conclusively contain nucleolar material (for example, see Supplemental Movie S2 described below). The association of the nuclear extension with the nucleolus was seen in under all tested conditions that generated a mitotic delay, including DNA recombination/repair and spindle/chromosome segregation mutants, cells arrested in mid-mitosis with non-degradable Pds1p, and cells treated with zeocin (data not shown).
Figure 2. The mitotic nuclear extension coincides with the nucleolus.
(A) The nucleolus in wild type cells. Wild type cells (KW926) were fixed, stained with DAPI and imaged for Pus1p-GFP (green in the overlays), Nsr1p-mCherry (Nsr1p-CR, red in the overlays) and DAPI (blue in the overlays). The green/red overlap causes Nsr1p-CR to appear orange. Scale bar =3μm. (B) Nuclear morphology in rad51Δ cells (opt two rows) and wild type cells treated with nocodazole (NZ, bottom two rows). rad51Δ and nocodazole treated wild type cells (KW926) were fixed and imaged as described in (A). Shown are typical examples of cells with nuclear extensions. Note that the DNA in these cells is similar to the controls (panel A), while the nuclear extension (i.e. the part of the nucleus containing Pus1p-GFP but no DNA) coincides with the nucleolar marker Nsr1p-CR. Scale bar =3μm. (C) Wild type cells (KW926), either without (control) or with nocodazole treatment were imaged at the indicated time points. Pus1p-GFP is in green and Nsr1p-CR is in red (appears orange due to the overlay). For nocodazole treated cells, panels with only Pus1p-GFP or Nsr1p-CR are shown in Supplemental Figure S2B. Scale bar = 3 μm.
To determine whether the nuclear extension forms at the NE associated with the nucleolus, or whether the nucleolus moves into the extension subsequently, we followed Pus1p-GFP and Nsr1p-CR in small budded cells that were either untreated or exposed to nocodazole (Figure 2C). Control cells progressed through anaphase as expected (Figure 2C). In all nocodazole treated cells (n=16), the initial nuclear deformation occurred as a flattened extension in the nucleolar region, which then elongated, becoming a loop or hook-shaped extension (Figure 2C, Supplemental Figure S2B, and Supplemental movies S1 and S2). These data show that the nuclear extension forms at NE that is associated with the nucleolus.
The presence of a nuclear extension suggests that during a mitotic delay, the NE expands in the absence of a spindle and despite a block to chromosome segregation. To compare the cell cycle timing of this expansion relative to that of normal NE elongation, we used bud size as a proxy for cell cycle progression, and measured bud length at the first sign of nuclear elongation in control cells (n=26), or nuclear/nucleolar deformation in nocodazole-treated cells (n=16). The relative timing of nuclear elongation in control cells, and the formation of a nuclear extension in nocodazole treated cells, was nearly identical, occurring when the bud lengths were 3.04 ± 0.31 μm and 2.98 ± 0.51 μm, respectively. This suggests that NE expansion occurs in response to a cell cycle cue that is independent of spindle elongation or chromosome segregation.
Phospholipids continue to accumulate during mitotic arrest, leading to the formation of nuclear extensions
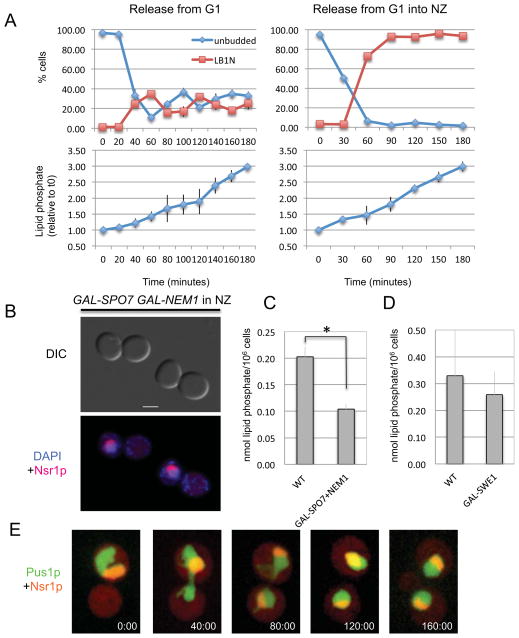
The appearance of a nuclear extension in the presence of DNA or spindle damage suggested that checkpoint activation blocks chromosome segregation but not membrane synthesis. To examine this, cells were synchronized in G1 and released into regular growth media (cycling cells) or the same media containing nocodazole (leading to a mitotic arrest before the first cell division). Samples were taken at various time points and analyzed for phospholipid accumulation. In cycling cells, the total amount of phospholipids rose steadily, reaching a 3-fold increase 180 minutes after release from G1 (Figure 3A, left panels). In cells released into nocodazole, the rate of phospholipid accumulation mirrored that of cycling cells throughout the entire time course, despite the block to cell cycle progression (Figure 3A, right panels). Thus, activation of the spindle checkpoint prevents chromosome segregation but does not affect phospholipid accumulation.
Figure 3. The mitotic nuclear extension depends on continued phospholipid synthesis.
(A) Wild type cells (KW913) were arrested in G1 with alpha factor mating pheromone and released from the arrest into regular growth media (left panels) or into media containing nocodazole (right panels). Samples for cell cycle distribution (upper panels) and phospholipid analysis (lower panels) were taken at the indicated time points. For cell cycle distribution, shown are unbudded cells (blue curves) or large budded cells with a single nucleus (LB1N, red curves). For phospholipid analysis, shown are the amounts of lipid phosphate relative to time 0, immediately before the release from the G1 arrest. (B) Examples of cells over-expressing SPO7 and NEM1 and treated with nocodazole. DNA (DAPI) is in blue and Nsr1p-CR is shown in red. (C) Analysis of lipid phosphate in the same cells shown in panel B. Cells were arrested in G1, induced for galactose-induced expression and released from the arrest into media containing galactose and nocodazole. Samples were taken at time 0 and 3 hours, and the graph shows the difference in the amount of lipid phosphate between these two time points per 106 cells. Error bars indicate SEM. (D) Analysis of lipid phosphate for control and GAL-SWE1 cells, as described in panel C. The difference in lipid phosphate between the two strains is not statistically significant. The presence of nocodazole did not affect phospholipid accumulation (data not shown). (E) An example of a wild type cell (KW926), as it is released from a nocodazole induced mitotic arrest. Green: Pus1p-GFP, red: Nsr1p-CR. Images were taken at the indicated time points (minutes).
If phospholipid synthesis contributes to the formation of the nuclear extension in mitotically arrested cells, then inhibiting phospholipid synthesis should decrease the number of mitotic cells with a nuclear extension. Over-expression of the Pah1p-activating proteins Spo7p and Nem1p blocks nuclear elongation [6] and reduced phospholipid accumulation (Figure 3C). We therefore examined whether over-expression of Spo7p and Nem1p affected the formation of nuclear extensions in nocodazole-arrested cells. Control cells and cells over-expressing SPO7 and NEM1 from a galactose inducible promoter were grown in galactose for 1 hour and then treated with nocodazole. Cells in both cultures arrested efficiently in mitosis (Table 1). While control cells exhibited the typical increase in nuclear extensions, cells over-expressing SPO7 and NEM1 had mostly round nuclei (Table 1 and Figure 3B). Consistent with a role for phospholipid synthesis in NE expansion during a mitotic delay, cells over-expressing OPI1, a transcription inhibitor of phospholipid synthesis genes, also had a reduced number of cells with nuclear extensions, albeit to a lesser extent than the reduction seen in cells over-expressing SPO7 and NEM1 (Table 1). Thus, Spo7p and Nem1p may affect nuclear morphology via pathways that are different from those regulated by Opi1p [10]. This possibility is also consistent with appearance of nuclear extensions in spo7Δ and nem1Δ mutants, but not in opi1Δ mutants [10]. Interestingly, cells arrested in G2 by SWE1 over-expression did not exhibit nuclear extensions, and yet the accumulation of phospholipids in these cells was similar to that of mitotically arrested cells (Figure 3D). Thus, phospholipid synthesis is necessary, but not sufficient, for nuclear extension formation. The absence of nuclear extensions in cells over-expressing SWE1 suggests that mitotic Cdk activity may also be required.
In contrast to mutants that exhibit constitutive elevated phospholipid synthesis, such as spo7Δ or opi1Δ mutants, in which the ER exhibits dramatic structural changes [12, 27], mitotic delay was not accompanied by gross changes to ER morphology ([28] and our data not shown). The reason for this difference is not known; as cells delay in mitosis they continue to grow in size, and perhaps continued phospholipid synthesis contributes to the formation of new ER membrane that is not altered in shape. Alternatively, cells may have a mechanism that controls membrane allocation to the nucleus during mitosis, such that there is a preferential increase in nuclear surface area relative to the ER. Finally, it is possible that the structure of the ER is altered during a mitotic delay, but these changes are too subtle to detect by light microscopy.
Nuclear extensions are resolved after release from mitotic arrest
During normal growth conditions, cells may delay temporarily in mitosis due to an inhibitory signal generated by an intracellular defect, and then resume cell cycle progression once the inhibitory signal is extinguished. To examine what happens to the nuclear extension once cells are allowed to re-enter the cell cycle, cells were released from a nocodazole-induced mitotic arrest and followed by microscopy. Of the 47 cells that were followed, 91.5 % proceeded through mitosis (i.e. underwent chromosome segregation and initiated a new cell cycle as determined by bud formation), and in the majority of cases (75.7%) one or both nuclei regained a round shape following nuclear division (for example, see Figure 3C), before the next cell division. Thus, the nuclear extension is resolved in the subsequent cell cycle.
Closed mitosis imposes restrictions on nuclear membrane expansion during chromosome segregation. In Schizosaccharomyces japonicus, this problem is solved by mitotic NE rupture, which is independent of microtubule-induced forces but allows anaphase spindle elongation to occur [29, 30]. Budding yeast circumvent the need for NE rupture by adding membrane to the NE during mitosis. However, this solution is challenged during mitotic delay. Our data show that a mitotic delay/arrest leads to the formation of a nuclear extension that is dependent on phospholipid synthesis. This suggests that the increase in NE surface area during mitosis is not caused by spindle elongation, but rather by a cell cycle regulated process that promotes phospholipid synthesis and/or allocates membrane to the NE. The fact that G2 arrested cells accumulate phospholipids but do not exhibit a nuclear extension suggests that this regulatory process is dependent, at least in part, on mitotic Cdk activity. It is currently unclear whether new membrane is added evenly throughout the nuclear envelope, or whether membrane addition occurs at a specific region, such as the nucleolus. Also, the properties of the NE that allow membrane accumulation adjacent to the nucleolus, but not elsewhere in the NE, remain to be discovered. Finally, why during a mitotic delay is NE expansion confined to the nucleolar region? Accumulating data suggest that there is higher-level nuclear organization, including distinct chromosome positioning and specific intra-nuclear domains [31]. Increase the surface area of the entire NE during a mitotic delay could increase the volume in which the chromatin resides and lead to disruption of the intra-nuclear spatial organization. By using the NE associated with the nucleolus as a “membrane sink,” cells may be able to preserve chromosomal organization during mitotic delay.
Experimental Procedures
Can be found in the Supplemental Material
Supplementary Material
Highlights.
In budding yeast, a mitotic delay induces the formation of a nuclear extension.
The nuclear extension forms adjacent to the nucleolus, which fills the extension.
Phospholipid synthesis continues in mitotically arrested cells.
Phospholipid synthesis is necessary, but not sufficient, for extension formation.
Acknowledgments
We thank Kevin O'Connell, Dimitris Liakopoulos, Danny Lew, Kerry Bloom, Chris May and Mohammad Rahman for advice and comments on the manuscript. We also thank Danny Lew, Steve Bell, Oscar Aparicio and Symeon Siniossoglou for strains and plasmids. Y.L., J.L.Y.K. and B.J.A were funded by a grant from the Canadian Institutes of Health Research (CIHR) MOP-97939 (GMX-211012). K.L.W., S.S., M.T.W., S.L., A.D.W., B.D., W.A.P. and O.C.F. were funded by intramural NIH grants from NIDDK to W.A.P and O.C.F.
The authors declare that they have no financial interest that might affect the results or the interpretation of this manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kutay U, Hetzer MW. Reorganization of the nuclear envelope during open mitosis. Curr Opin Cell Biol. 2008;20:669–677. doi: 10.1016/j.ceb.2008.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hartwell LH. Saccharomyces cerevisiae cell cycle. Bacteriol Rev. 1974;38:164–198. doi: 10.1128/br.38.2.164-198.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Webster M, Witkin KL, Cohen-Fix O. Sizing up the nucleus: nuclear shape, size and nuclear-envelope assembly. J Cell Sci. 2009;122:1477–1486. doi: 10.1242/jcs.037333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Capell BC, Collins FS. Human laminopathies: nuclei gone genetically awry. Nat Rev Genet. 2006;7:940–952. doi: 10.1038/nrg1906. [DOI] [PubMed] [Google Scholar]
- 5.Zink D, Fischer AH, Nickerson JA. Nuclear structure in cancer cells. Nat Rev Cancer. 2004;4:677–687. doi: 10.1038/nrc1430. [DOI] [PubMed] [Google Scholar]
- 6.Santos-Rosa H, Leung J, Grimsey N, Peak-Chew S, Siniossoglou S. The yeast lipin Smp2 couples phospholipid biosynthesis to nuclear membrane growth. EMBO J. 2005;24:1931–1941. doi: 10.1038/sj.emboj.7600672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Han GS, Wu WI, Carman GM. The Saccharomyces cerevisiae Lipin homolog is a Mg2+-dependent phosphatidate phosphatase enzyme. J Biol Chem. 2006;281:9210–9218. doi: 10.1074/jbc.M600425200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Han GS, Siniossoglou S, Carman GM. The cellular functions of the yeast lipin homolog PAH1p are dependent on its phosphatidate phosphatase activity. J Biol Chem. 2007;282:37026–37035. doi: 10.1074/jbc.M705777200. [DOI] [PubMed] [Google Scholar]
- 9.Choi HS, Su WM, Han GS, Plote D, Xu Z, Carman GM. Pho85p-Pho80p Phosphorylation of Yeast Pah1p Phosphatidate Phosphatase Regulates Its Activity, Location, Abundance, and Function in Lipid Metabolism. J Biol Chem. 2012 doi: 10.1074/jbc.M112.346023. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.O'Hara L, Han GS, Peak-Chew S, Grimsey N, Carman GM, Siniossoglou S. Control of phospholipid synthesis by phosphorylation of the yeast lipin Pah1p/Smp2p Mg2+-dependent phosphatidate phosphatase. J Biol Chem. 2006;281:34537–34548. doi: 10.1074/jbc.M606654200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Siniossoglou S, Santos-Rosa H, Rappsilber J, Mann M, Hurt E. A novel complex of membrane proteins required for formation of a spherical nucleus. EMBO J. 1998;17:6449–6464. doi: 10.1093/emboj/17.22.6449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Campbell JL, Lorenz A, Witkin KL, Hays T, Loidl J, Cohen-Fix O. Yeast nuclear envelope subdomains with distinct abilities to resist membrane expansion. Mol Biol Cell. 2006;17:1768–1778. doi: 10.1091/mbc.E05-09-0839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hellmuth K, Lau DM, Bischoff FR, Kunzler M, Hurt E, Simos G. Yeast Los1p has properties of an exportin-like nucleocytoplasmic transport factor for tRNA. Mol Cell Biol. 1998;18:6374–6386. doi: 10.1128/mcb.18.11.6374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thrower DA, Stemple J, Yeh E, Bloom K. Nuclear oscillations and nuclear filament formation accompany single-strand annealing repair of a dicentric chromosome in Saccharomyces cerevisiae. J Cell Sci. 2003;116:561–569. doi: 10.1242/jcs.00251. [DOI] [PubMed] [Google Scholar]
- 15.Castagnetti S, Oliferenko S, Nurse P. Fission yeast cells undergo nuclear division in the absence of spindle microtubules. PLoS Biol. 2010;8:e1000512. doi: 10.1371/journal.pbio.1000512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clarke DJ, Bachant J. Kinetochore structure and spindle assembly checkpoint signaling in the budding yeast, Saccharomyces cerevisiae. Front Biosci. 2008;13:6787–6819. doi: 10.2741/3189. [DOI] [PubMed] [Google Scholar]
- 17.Wang H, Liu D, Wang Y, Qin J, Elledge SJ. Pds1 phosphorylation in response to DNA damage is essential for its DNA damage checkpoint function. Genes Dev. 2001;15:1361–1372. doi: 10.1101/gad.893201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sanchez Y, Bachant J, Wang H, Hu F, Liu D, Tetzlaff M, Elledge SJ. Control of the DNA damage checkpoint by chk1 and rad53 protein kinases through distinct mechanisms. Science. 1999;286:1166–1171. doi: 10.1126/science.286.5442.1166. [DOI] [PubMed] [Google Scholar]
- 19.Cohen-Fix O, Koshland D. The anaphase inhibitor of Saccharomyces cerevisiae Pds1p is a target of the DNA damage checkpoint pathway. Proc Natl Acad Sci U S A. 1997;94:14361–14366. doi: 10.1073/pnas.94.26.14361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Irniger S, Piatti S, Michaelis C, Nasmyth K. Genes involved in sister chromatid separation are needed for B-type cyclin proteolysis in budding yeast. Cell. 1995;81:269–278. doi: 10.1016/0092-8674(95)90337-2. [DOI] [PubMed] [Google Scholar]
- 21.Icho T, Wickner RB. Metal-binding, nucleic acid-binding finger sequences in the CDC16 gene of Saccharomyces cerevisiae. Nucleic Acids Res. 1987;15:8439–8450. doi: 10.1093/nar/15.20.8439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cohen-Fix O, Peters JM, Kirschner MW, Koshland D. Anaphase initiation in Saccharomyces cerevisiae is controlled by the APC-dependent degradation of the anaphase inhibitor Pds1p. Genes Dev. 1996;10:3081–3093. doi: 10.1101/gad.10.24.3081. [DOI] [PubMed] [Google Scholar]
- 23.Stone EM, Heun P, Laroche T, Pillus L, Gasser SM. MAP kinase signaling induces nuclear reorganization in budding yeast. Curr Biol. 2000;10:373–382. doi: 10.1016/s0960-9822(00)00413-9. [DOI] [PubMed] [Google Scholar]
- 24.Wood JS, Hartwell LH. A dependent pathway of gene functions leading to chromosome segregation in Saccharomyces cerevisiae. J Cell Biol. 1982;94:718–726. doi: 10.1083/jcb.94.3.718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hartwell LH, Culotti J, Pringle JR, Reid BJ. Genetic control of the cell division cycle in yeast. Science. 1974;183:46–51. doi: 10.1126/science.183.4120.46. [DOI] [PubMed] [Google Scholar]
- 26.Booher RN, Deshaies RJ, Kirschner MW. Properties of Saccharomyces cerevisiae wee1 and its differential regulation of p34CDC28 in response to G1 and G2 cyclins. EMBO J. 1993;12:3417–3426. doi: 10.1002/j.1460-2075.1993.tb06016.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schuck S, Prinz WA, Thorn KS, Voss C, Walter P. Membrane expansion alleviates endoplasmic reticulum stress independently of the unfolded protein response. The Journal of cell biology. 2009;187:525–536. doi: 10.1083/jcb.200907074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Prinz WA, Grzyb L, Veenhuis M, Kahana JA, Silver PA, Rapoport TA. Mutants affecting the structure of the cortical endoplasmic reticulum in Saccharomyces cerevisiae. J Cell Biol. 2000;150:461–474. doi: 10.1083/jcb.150.3.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yam C, He Y, Zhang D, Chiam KH, Oliferenko S. Divergent strategies for controlling the nuclear membrane satisfy geometric constraints during nuclear division. Curr Biol. 2011;21:1314–1319. doi: 10.1016/j.cub.2011.06.052. [DOI] [PubMed] [Google Scholar]
- 30.Aoki K, Hayashi H, Furuya K, Sato M, Takagi T, Osumi M, Kimura A, Niki H. Breakage of the nuclear envelope by an extending mitotic nucleus occurs during anaphase in Schizosaccharomyces japonicus. Genes Cells. 2011;16:911–926. doi: 10.1111/j.1365-2443.2011.01540.x. [DOI] [PubMed] [Google Scholar]
- 31.Van de Vosse DW, Wan Y, Wozniak RW, Aitchison JD. Role of the nuclear envelope in genome organization and gene expression. Wiley Interdiscip Rev Syst Biol Med. 2011;3:147–166. doi: 10.1002/wsbm.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.