Abstract
In resting-state functional MRI (R-fMRI) studies, the global signal (operationally defined as the global average of R-fMRI time courses) is often considered a nuisance effect and commonly removed in preprocessing. This global signal regression method can introduce artifacts, such as false anticorrelated resting-state networks in functional connectivity analyses. Therefore, the efficacy of this technique as a correction tool remains questionable. In this paper, we establish that the accuracy of the estimated global signal is determined by the level of global noise (i.e. non-neural noise that has a global effect on the R-fMRI signal). When the global noise level is low, the global signal resembles the R-fMRI time courses of the largest cluster, but not those of the global noise. Using real data, we demonstrate that the global signal is strongly correlated with the default mode network components, and has biological significance. These results call into question whether or not global signal regression should be applied. We introduce a method to quantify global noise levels. We show that a criteria for global signal regression can be found based on the method. By employing the criteria, one can determine whether to include or exclude the global signal regression in minimizing errors in functional connectivity measures.
Keywords: global noise, global signal, global signal regression, resting-state fMRI
Introduction
Recently, resting-state functional MRI (R-fMRI) has emerged as a powerful tool in investigating brain functional network architecture (1,2) and brain functional network changes in neurological and psychiatric disorders (3–9). R-fMRI data analysis involves the assessment of temporal coherences in the data set. It is commonly found that the data sets are contaminated by various fluctuations not related to neural activity: subject head motion, physiological artifacts (caused by respiration, cardiac pulsation), as well as hardware instabilities and magnetic field drifting. Signal fluctuations from the nonneural processes may introduce false positive coherences and cause an overestimation of functional connectivity strengths. Therefore, sophisticated preprocessing techniques are commonly employed in order to remove such confounding effects prior to the connectivity assessment.
Global signal regression (10), as a preprocessing technique, is intended to eliminate undesired global noise, i.e. non-neural noise that has a global effect on the RfMRI signal. It was hypothesized that R-fMRI signals may cancel each other out in the global average, while the global noise may be additive and dominate the global-averaged signal. Practically, the global signal is obtained by averaging all brain voxel time courses. The technique has been employed in a number of early R-fMRI studies (11–18). However, it was found that global signal regression forces a bell-shaped distribution of functional connectivity measures centered on zero. Therefore, this technique allows for the introduction of spurious negative measures (19,20) and the underestimation of true positive measures.
Techniques to monitor the sources that may induce non-neural noise in the R-fMRI signal have been introduced (21–23). When no measurement was made of these sources, they can be estimated by data-driven methods (24). It has been shown that the low-frequency respiratory volume and cardiac rate regressors had significant shared variance with the global signal (23). Therefore, these non-neural noise correction techniques may reduce the global noise level and diminish the motivation for adopting global signal regression in data preprocessing. However, it is not clear how much global noise level can be reduced, or if it is truly necessary to use global signal regression in further reducing the global noise following these non-neural noise corrections. If the global noise level is high, the global signal may resemble global noise. Therefore, the global signal regression may be beneficial. If the global noise level is low, the global signal may contain a higher portion of signal (most probably from the dominant network component), rather than the global noise. Taking into account that the default mode network has shown high glucose metabolism during rest (25,26), it can be hypothesized that the global signal may be most strongly correlated with the default mode network signal. For this reason, removing the global signal may generate more errors in the functional connectivity measurement, as opposed to reducing them.
The present study aimed to determine the major contributors of the global signal. We focused on whether or not the global signal was significantly correlated with the default mode network signal. We further tested the hypothesis that with the decreased global noise levels, errors in the measured voxelwise cross-correlations may increase, if the global signal regression were performed. On the other hand, errors may decrease, if the global signal regression were not performed. We then introduced a method to quantify global noise levels. We show that a criteria for global signal regression can be found, based on the method. By employing this criteria, one can include or exclude global signal regression to minimize errors in functional connectivity measures.
Materials and Methods
In determining whether or not to apply global signal regression in R-fMRI studies, we conducted experiments, using simulated data and human R-fMRI data.
Simulation
Ten predefined clusters were assigned sinusoidal time courses (180 time points, TR=2000 ms) with distinct frequencies ranging from 0.01 to 0.04 Hz with 0.0037 Hz intervals. The first cluster of 150 voxels was assigned a frequency of 0.01 Hz (with 4% random variations), and a zero-degree initial phase. The second cluster of 50 voxels also was assigned a frequency of 0.01 Hz (with 4% random variations), however it had a 180-degree initial phase (with 4% random variations), so the two clusters were anticorrelated. The rest of the eight clusters, each containing 25 voxels, were assigned unique frequencies from 0.0137 to 0.04 Hz, with 0.0037 Hz intervals (with 4% random variations) and a zero-degree initial phase. Therefore, they were uncorrelated to all other clusters. There are a total of 400 voxel time courses.
The global noise was assigned a sinusoidal time course (180 time points, TR=2000 ms) with a frequency of 0.06 Hz and a zero-degree initial phase. The cross-correlation among the signals of the 400 voxel time courses and the global noise was calculated. The global noise was added to the signal of the 400 voxels with a signal-to global-noise ratio (SGNR, defined as the standard deviation of the signal time course/standard deviation of the global noise time course) of 1, 2 and 10 to generate the mixed time courses. The cross-correlation among the mixed time courses, the global noise and the global average of the mixed time courses (global signal) were calculated for each case.
To determine how the SGNR levels affect the errors of the measured voxelwise Pearson product-moment correlation coefficient (r), we define the r error without using the global regression as
| [1] |
where n is the total number of voxels, rij is the r value between the sinusoidal signals of voxel i and j. r′ij is the measured r value between the mixed time courses (with the added global noise) of voxel i and j without using the global signal regression. We also define the r error using global regression as
| [2] |
where r″ij is the measured r value between the time courses calculated after the global signal regression procedure.
The r errors were measured with discrete SGNR values ranging from 1 to 100. The simulation was repeated 20 times. The average r error, which was established using the global signal regression and not using global signal regression were plotted against the SGNR.
To characterize how different levels of SGNR produce different numbers of voxels that negatively correlate with the global signal, we define a global negative index as
| [3] |
The global negative index was measured with discrete SGNR values ranging from 1 to 100. The simulation was repeated 20 times and the global negative indices were plotted against the SGNR.
Human data
Subjects
Twenty cognitively normal (CN) subjects were recruited through the Memory Disorders Clinic at the Medical College of Wisconsin (MCW). The study was conducted with MCW Institutional Review Board approval and in compliance with HIPAA regulations. Written informed consent was obtained from each participant or the caregiver of the participant. The detailed inclusion and exclusion criteria for the CN subjects have been described previously (8,27). The detailed demographic information of these subjects is listed in Table 1.
Table 1.
Demographics and clinical characteristics of CN subjects.
| Characteristic | CN(n = 20) |
|
|---|---|---|
| M | SD | |
| Gender (female/male) | 9/11 | |
| Age, years | 75.0 | 6.2 |
| Education, years | 14.8 | 2.6 |
| MMSE | 29.5 | 0.8 |
| RAVLT delayed recall | 11.2 | 3.4 |
| GDS | 2.7 | 2.8 |
Note: CN, cognitively normal; M, mean; SD, Standard Deviation; MMSE, Mini-Mental State Examination; RAVLT, Rey Auditory Verbal Learning Test; GDS, Geriatric Depression Scale; Unless otherwise indicated, data are presented as mean ± SD.
Image acquisition
Imaging was performed using a whole-body 3T Signa GE scanner with a standard transmit-receive head coil. During the resting-state acquisitions, no specific cognitive tasks were performed, and the study participants were instructed to close their eyes and relax inside the scanner. Sagittal resting-state functional MRI (fMRI) datasets of the whole brain were obtained in six minutes with a single-shot gradient echo-planar imaging (EPI) pulse sequence. The fMRI imaging parameters were: TE of 25 ms, TR of 2 s, flip angle of 90°; 36 slices were obtained without gap; slice thickness was 4 mm with a matrix size of 64×64 and field of view of 24×24 cm. High-resolution SPGR 3D axial images were acquired for anatomical reference. The parameters were: TE/TR/TI of 4/10/450 ms, flip angle of 12°, number of slices of 144, slice thickness of 1 mm, matrix size of 256×192. A pulse oxymeter and a respiratory belt were employed to measure physiological noise sources to minimize the potential artifacts in the low-frequency spectrum (22,28).
Data preprocessing
A series of preprocessing steps common to most fMRI analyses was conducted, using the Analysis of Functional NeuroImages (AFNI) software (http://afni.nimh.nih.gov/afni/), SPM8 (Wellcome Trust, London) and Matlab (Mathworks, Natick, MA). The preprocessing involves allowing for T1-equilibration effects (removal of the first five volumes of fMRI data); removal of cardiac and respiratory artifacts (3dretroicor); slice-acquisition-dependent time shift correction (3dTshift); motion correction (3dvolreg); detrending (3dDetrend); despiking (3dDespike); segmentation (SPM8) of white matter and CSF components from each subject’s SPGR image; creating the white matter and CSF mask (3dcalc and 3dfractionize); obtaining the averaged white matter and CSF signal (3dROIstats); obtaining the multiple time-shifted respiration volume per unit time (RVT) regressors (22); removing the respiratory-variation-related fluctuations (using the RVT regressors), white matter, CSF signal and motion effect (3dDeconvolve); and low-frequency band-pass filtering (3dFourier). In addition, a whole-brain mask was created for each subject (3dAutomask) and the global signal was extracted by averaging all preprocessed time series inside the whole-brain mask (3dROIstats).
Global signal connectivity pattern
It is conceivable that the global signal may have the strongest correlation with the signals of the dominant resting-state network. Taking into account that the default mode network showed high glucose metabolism during rest (25,26), we hypothesized that the global signal may be most strongly correlated with the signal of the default mode network. To test this, a method that is similar to the seed-based analysis method (8) was employed. In this case, the seed region is the entire brain, and the average of the seed-time courses is the global signal. The r values between the global signal and each brain voxel's time series were calculated. The Fisher transformation (m=0.5ln[(1+r)/(1−r)]), which yielded variants of approximately normal distribution (29), was applied to the individual r maps to generate the m maps. The m maps were then transformed into Talairach space (adwarp), resampled to 2-mm isotropic voxels and smoothed with a Gaussian kernel (6-mm full width at half maximum) using AFNI (3dmerge). The results are the normalized m maps, which represent the connectivity distribution of the global signal. One sample t-test was performed on the normalized m maps of the 20 CN subjects, to identify regions that significantly correlated to the global signal.
Criteria to perform global signal regression
The relationship between the SGNR, r error and global negative index may be affected by the cluster size, negative connectivity strength between clusters, etc., in the simulation using synthetic data. To validate this relationship, artificial global noise (GNt was randomly selected from the preprocessed time series of a different subject to provide global noise in the same frequency range while satisfying randomness) was added to each subject's preprocessed time series Si,t
| [5] |
Where Si,t denotes the preprocessed time series of the i-th voxel at time t, GNi,t denotes the artificial global noise added to the i-th voxel. GNi,t was normalized; therefore, its standard deviation is equal to the standard deviation of Si,t:
| [6] |
Global signal (global average of the mixed time courses ) was then regressed out from , the result is .
The r values were calculated using and and compared with the r value calculated from Si,t. The r errors were calculated using Eq. 1 and 2. As a result, for each case, each voxel had n-1 r errors (n is the total number of voxels). These n-1 r errors were averaged for each voxel and saved as a distribution map in original space. The distribution map was then transformed into Talairach space (adwarp) to display the averaged group pattern.
The whole-brain averaged r errors determined by using the global signal regression (calculated using and Si,t) was plotted against the SGNR. The r error, which was determined by not using the global signal regression (with added global noise, calculated using and Si,t ) , was also plotted against SGNR. The global negative index was calculated using the mixed time courses and were plotted against the SGNR.
Results
Characteristic relationship between the simulated signal, global noise and global signal
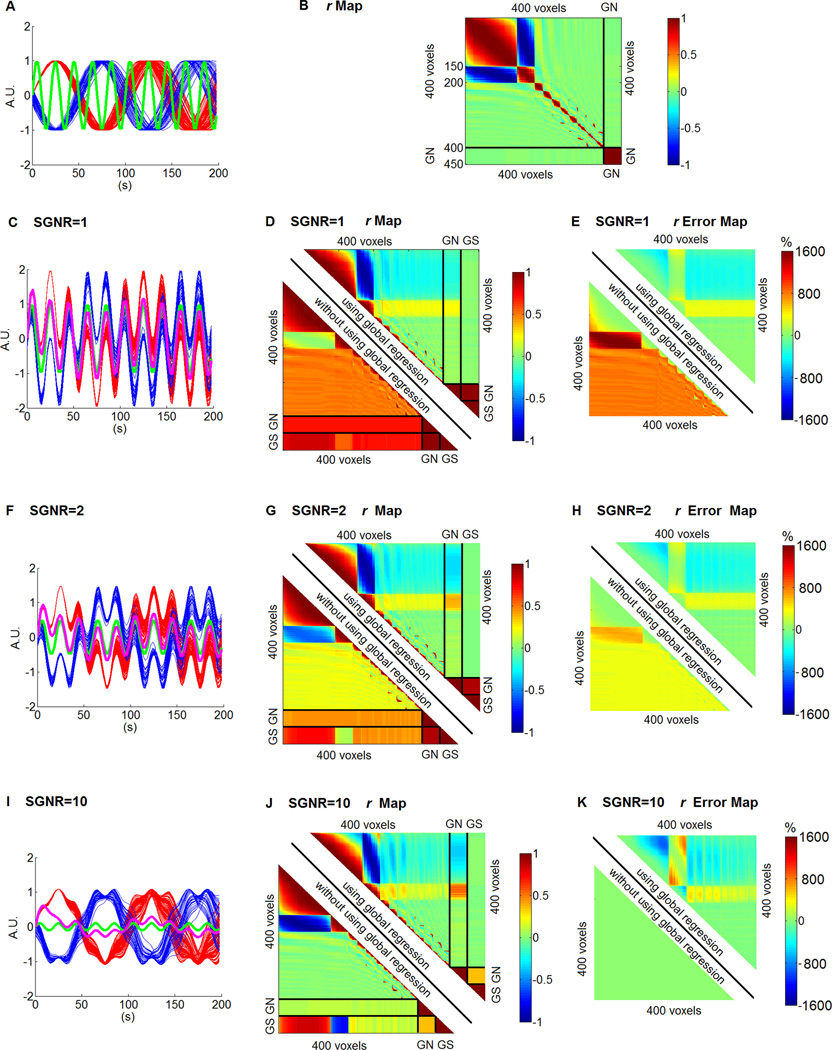
Figure 1A shows the signals in the first cluster (150 voxels) in red and the signals in the second cluster (50 voxels) in blue. The global noise is shown in green. Figure 1B shows the r value among the 400 signals and the global noise (replicated 50 times for better illustration). The signals in the first two clusters are negatively correlated. The global noise is not correlated with the signals. It was added to the signal with the SGNR of 1, 2 and 10 to generate the mixed time courses, as shown in Figures 1C, 1F and 1I. The mixed time courses in the first cluster are shown in red and the mixed time courses in the second cluster are shown in blue. The global noise is shown in green and the global signal is shown in purple. Figures 1D, 1G and 1J show the r value among the 400 mixed time courses, the global noise (GN) and the global signal (GS) using (top right half) and without using global regression (bottom left half), with the SGNR of 1, 2 and 10, respectively. Figures 1E, 1H and 1K show the r error among the 400 voxels calculated using (top right half) and without using global regression (bottom left half), with an SGNR of 1, 2 and 10, respectively.
Figure 1.
Simulation results. (A) The signals in the first cluster (150 voxels) in red, the signals in the second cluster (50 voxels) in blue. The signals of the rest of the 200 voxels are not shown. The global noise is shown in green. (B) The r value among the 400 signals and the global noise (GN) (replicated 50 times for better illustration). (C, F and I) The mixed time courses in the first cluster in red, the mixed time courses in the second cluster in blue, the global noise is shown in green and the global signal is shown in purple with SGNR of 1, 2 and 10 respectively. (D, G and J) The r value among the global noise (GN), global signal (GS), and the 400 voxels' time courses using (top right half) and without using global regression (bottom left half), with SGNR of 1, 2 and 10 respectively. (E, H and K) The r error among the 400 voxels calculated using (top right half) and without using global regression (bottom left half) with SGNR of 1, 2 and 10 respectively.
Several important characteristics were observed in the simulation results. First, it was found that the global signal has the strongest correlation with the signals in the largest cluster. With the increase of the SGNR, the correlation between the global signal and signals in the largest cluster remains higher, although the correlation between the global signal and the signals of the other clusters becomes less. This suggests that the dominant intrinsic resting-state network may be the major contributor of the global signal.
In addition, comparing the r values between global noise and the global signal (represented by the small square near the bottom right corner in Figures 1D, 1F and 1H), it is found that the r values between global noise and the global signal decrease with the SGNR increase. In other words, the discrepancy between global noise and global signal becomes larger with the SGNR increase. It is also found that the number of voxels that are negatively connected with the global signal is increasing with the SGNR increase.
Also, in comparing Figures 1E, 1H and 1K, it is found that the r errors are generally lower using global regression (top right) than without using global regression (bottom left) at lower SGNR (1E and 1H); and the r errors are generally higher using global regression (top right) than without using global regression (bottom left) at higher SGNR (1K, note the r errors are near 0% without using global regression [bottom left]).
Global signal connectivity pattern and r error distribution pattern
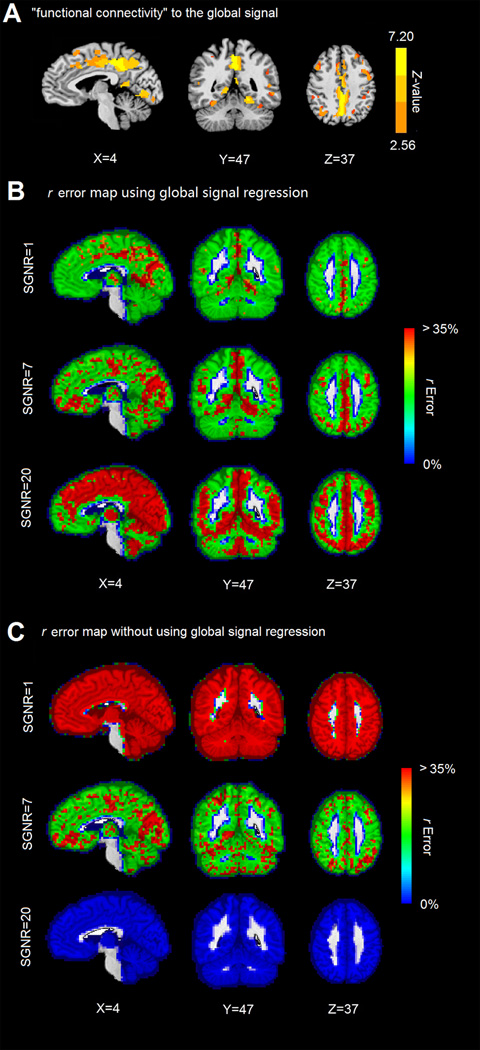
It was observed in the simulation that the global signal has the strongest correlation with the signals in the largest cluster. Taking into account that the default mode network showed high glucose metabolism during rest (25,26), we hypothesize that the global signal may be most strongly correlated with the signal of the default mode network when the SGNR level is high. Indeed, the global signal was found to be strongly correlated with the default mode network components PCC and precuneus, as shown in Figure 2A. The distribution of the correlation is localized (rather than diffused and covering the entire brain). The results suggest that the major contributors of the global signal are the dominant intrinsic networks rather than globally distributed non-neural noise.
Figure 2.
Global signal connectivity pattern and r error map. (A) Regions that strongly correlated global signal. (B) r error map using global signal regression with SGNR of 1, 7 and 20 respectively. (C) r error map without using global signal regression with SGNR of 1, 7 and 20 respectively.
The averaged r error pattern using global signal regression and without using global signal regression are shown in Figures 2B and 2C. The result confirmed the findings obtained from the synthetic data. The r errors are generally lower using global regression than without using global regression at lower SGNR (SGNR=1); the r errors are generally higher using global regression than without using global regression at higher SGNR (SNR=20, note the r errors are near 0% without using global regression).
Criteria to perform global signal regression
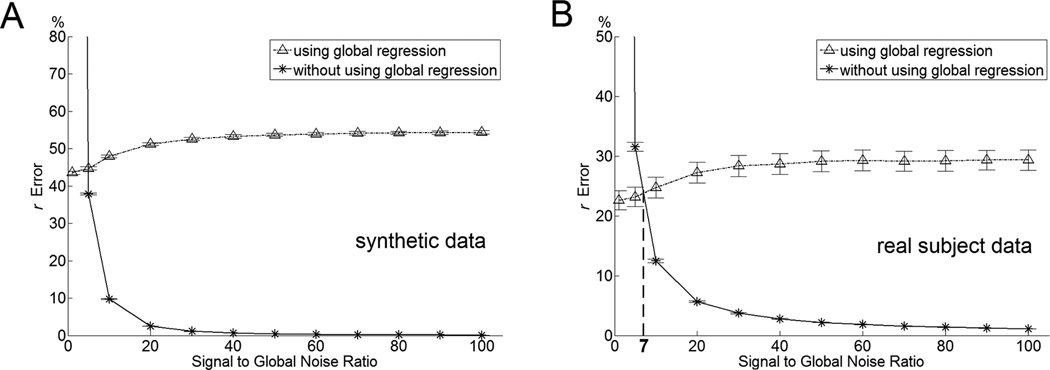
It was found in the simulation that the increase of the SGNR global signal regression may induce more errors, and not performing the global signal regression may generate less errors. Figure 3 shows voxelwise r errors with (dot-dashed line) and without (solid line) global signal regression. In using global signal regression, the r errors monotonically increase with the increase of SGNR. Without using the global signal regression, the r errors monotonically decrease with the increase of SGNR.
Figure 3.
Relationship between SGNR and mean r error simulated using synthetic data (A) and real subject data (B). In Figure 3B, the dot-dashed line and solid line cross each other at SGNR value of 7.03. Below this SGNR, performing global signal regression induces less errors as opposed to not performing global signal regression. And above this SGNR, performing global signal regression induces more errors.
An important fact evidenced by Figure 3 is that the dot-dashed line and solid line cross each other at a certain SGNR value (i.e., 7.03 in Figure 3B). Below this SGNR, performing the global signal regression induces less error as opposed to not performing it. Above this SGNR, performing global signal regression induces more errors. This result suggests that the SGNR value can be employed as a criteria in deciding whether or not to perform the global signal regression. We define the crossover SGNR value as the criterial SGNR.
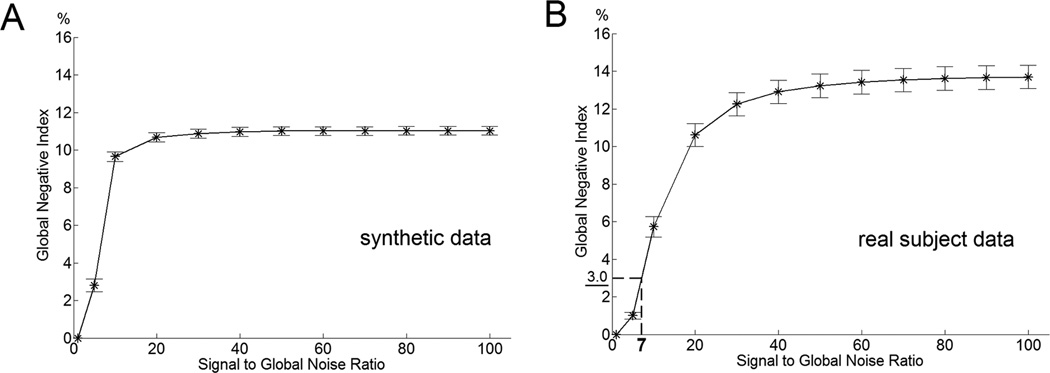
It was found that the number of voxels that negatively correlated with the global signal may be used to estimate the SGNR in real subject data sets. Figure 4 shows that the global negative index monotonically increases with the increase of SGNR in simulations using synthetic and real subject data.
Figure 4.
Relationship between SGNR and the mean global negative index simulated using synthetic data (A) and real subject data (B). The global negative index measurement of the criteria in doing and not doing global signal regression is 3.0 at the SGNR level of 7.03 determined in Figure 3B.
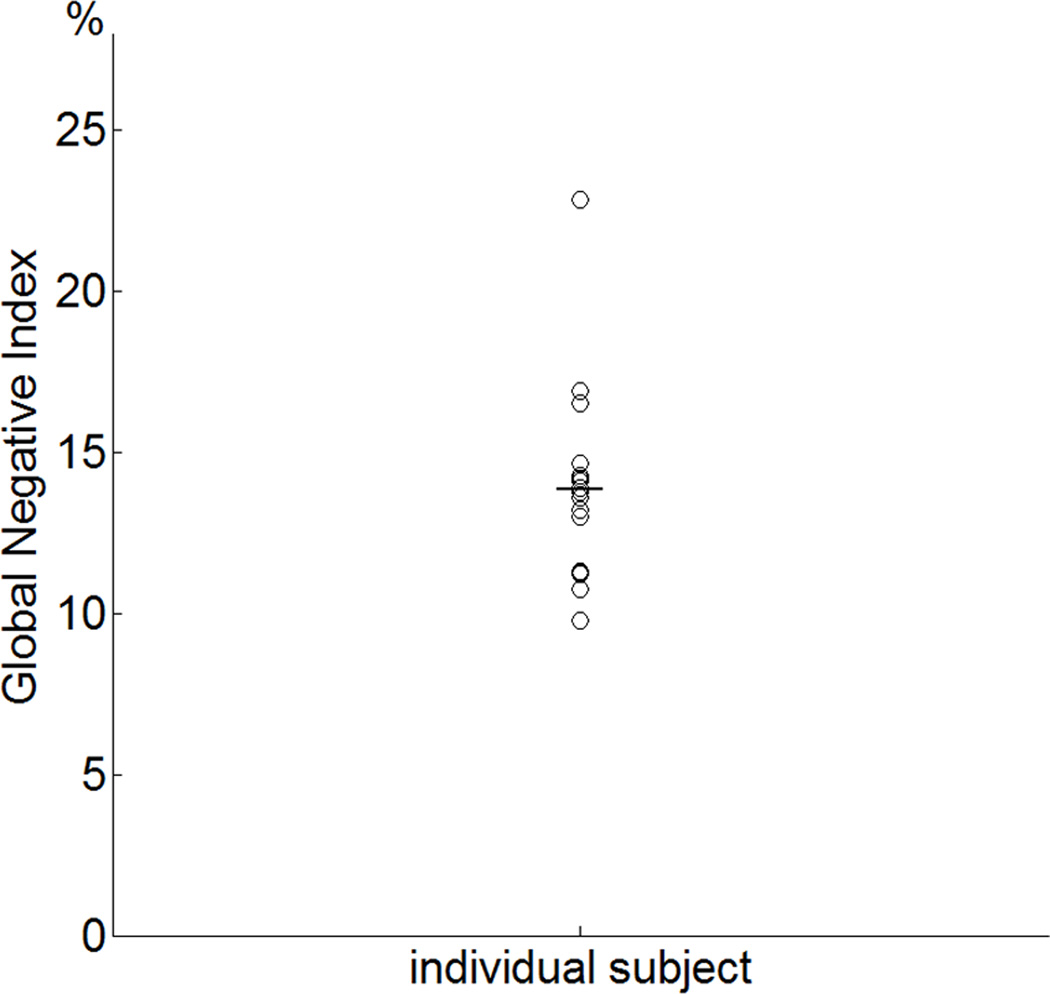
The criteria SGNR used in Figure 3B was found to be 7.03. In Figure 4B, with preprocessed data sets, it corresponds to the criterial global negative index of 3.0. Figure 5 shows the global negative indices of all 20 subjects with mean and standard deviation of 13.8±2.8. The global negative indices of all 20 subjects are all above the criterial value. The results suggest that not performing the global signal regression will yield less of a voxelwise r error than performing the global signal regression for every subject.
Figure 5.
The global negative indices of the 20 subjects. Their GNI is well above the criterial value of 3.0.
Discussion
In the present study, we demonstrated that the accuracy of the global signal regression is determined by the level of global noise by using simulation. When the global noise level is high, the global signal resembles global noise. When the global noise level is low, the global signal resembles the R-fMRI time courses of the largest cluster, but not global noise. We then showed, using real data, that the global signal is strongly correlated with the default mode network components PCC and precuneus. To answer the question as to whether or not the global signal should be considered a nuisance effect to be removed, we introduced a global negative index to quantify global noise levels. We demonstrated the monotonic relationship between the signal-to-global noise ratio and the global negative index. Finally, we discovered that there is a criterial global negative index associated with a criterial SGNR. Below this criterial global negative index, performing global signal regression induced less errors, therefore, it is highly suggested that the global regression be performed. Above this criterial global negative index, performing global signal regression induced more errors. Therefore, we suggest that the global signal regression not be performed. One can decide whether or not to apply this technique for each individual data set by comparing the global negative index of the data set to the criterial global negative index.
The findings of the monotonic relationship between SGNR and GNI is one of the most important components of the study. However, their relationship was obtained empirically, and it is important to understand the mechanisms. Theoretically, when SGNR is +∞, the global signal resembles the R-fMRI time courses of the largest cluster. Therefore, the GNI will approximate the percentage of the number of voxels that are negatively correlated with the largest cluster. When the SGNR is 0, the global signal will resemble the global noise, which will be positively connected to every voxel. Therefore, the GNI will be 0. When the SGNR is between 0 and +∞, the global signal will resemble a mixture of R-fMRI time courses of the largest cluster and global noise. Therefore, the GNI will depend on the ratio of the two components in this mixture, which is determined by the SGNR. As a result, the GNI may be used to estimate SGNR and gauge the r error.
The complete knowledge of global non-neural noise sources remains unclear. However, it has been demonstrated that the low-frequency respiratory volume, cardiac rate, white matter ROI signal and CSF ROI signal regressors had significant shared variances with the global signal (23). Therefore, these non-neural noise correction techniques that were employed may generate the high SGNR observed here. It was observed that the SGNR varies from subject to subject. As a result, it is recommended that one determine the global negative index on a case-by-case basis, rather than simply excluding the global regression in data preprocessing. It also is possible to monitor the SGNR in a realtime manner and repeat scans when the abnormally low SGNR occurs.
This study has focused on the correlation-based analyses, one of the most popular methods employed in investigating resting-state functional interactions. Independent component analysis (ICA) is another technique that is frequently used to study functional interactions (30–32). Often, the global signal is removed in preprocessing before the ICA is employed. This may produce undesirable errors, as discussed in this study. To solve the problem, the effect of global signal regression in the ICA can be quantified using similar approaches outlined in the present study.
R-fMRI is an expanding field of research that has great potential in investigating neurological and psychiatric disorders. However, an essential consensus has not been reached in regard to using the global signal regression in preprocessing. While some studies emphasize the pitfalls of the correction technique (19,20), others (22,33) have shown the benefit of global regression. Because different imaging and preprocessing techniques have been used, direct comparison to the studies will be difficult. Nonetheless, the current study is a timely report demonstrating that the effect of the global signal regression is determined by the SGNR of the data. The SGNR can be estimated from the data based on the global negative index. By comparing the global negative index of the data set and the criterial global negative index, one can determine whether to include or exclude the global signal regression in minimizing errors in functional connectivity measures.
Acknowledgments
The authors thank Carrie M. O’Connor, M.A., for editorial assistance, Judi Zaferos-Pylant, B.S.M., and Yu Liu, M.S., for MRI technical support. This work was supported by National Institutes of Health grants: R01 AG20279, NIHNCRR CTSA program grant 1UL1RR031973.
Footnotes
Disclosure Statement: No authors of this paper have reported any possible conflict of interests.
References
- 1.Biswal BB, Mennes M, Zuo XN, Gohel S, Kelly C, Smith SM, Beckmann CF, Adelstein JS, Buckner RL, Colcombe S, Dogonowski AM, Ernst M, Fair D, Hampson M, Hoptman MJ, Hyde JS, Kiviniemi VJ, Kotter R, Li SJ, Lin CP, Lowe MJ, Mackay C, Madden DJ, Madsen KH, Margulies DS, Mayberg HS, McMahon K, Monk CS, Mostofsky SH, Nagel BJ, Pekar JJ, Peltier SJ, Petersen SE, Riedl V, Rombouts SA, Rypma B, Schlaggar BL, Schmidt S, Seidler RD, Siegle GJ, Sorg C, Teng GJ, Veijola J, Villringer A, Walter M, Wang L, Weng XC, Whitfield-Gabrieli S, Williamson P, Windischberger C, Zang YF, Zhang HY, Castellanos FX, Milham MP. Toward discovery science of human brain function. Proc Natl Acad Sci U S A. 2010;107(10):4734–4739. doi: 10.1073/pnas.0911855107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Buckner RL, Vincent JL. Unrest at rest: default activity and spontaneous network correlations. Neuroimage. 2007;37(4):1091–1096. doi: 10.1016/j.neuroimage.2007.01.010. discussion 1097–1099. [DOI] [PubMed] [Google Scholar]
- 3.Fox MD, Raichle ME. Spontaneous fluctuations in brain activity observed with functional magnetic resonance imaging. Nat Rev Neurosci. 2007;8(9):700–711. doi: 10.1038/nrn2201. [DOI] [PubMed] [Google Scholar]
- 4.Chen G, Ward BD, Xie C, Li W, Wu Z, Jones JL, Franczak M, Antuono P, Li SJ. Classification of Alzheimer disease, mild cognitive impairment, and normal cognitive status with large-scale network analysis based on resting-state functional MR imaging. Radiology. 2011;259(1):213–221. doi: 10.1148/radiol.10100734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fox MD, Greicius M. Clinical applications of resting state functional connectivity. Front Syst Neurosci. 2010;4:19. doi: 10.3389/fnsys.2010.00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li SJ, Li Z, Wu G, Zhang MJ, Franczak M, Antuono PG. Alzheimer Disease: evaluation of a functional MR imaging index as a marker. Radiology. 2002;225(1):253–259. doi: 10.1148/radiol.2251011301. [DOI] [PubMed] [Google Scholar]
- 7.Rosen BR, Napadow V. Quantitative markers for neuropsychiatric disease: give it a rest. Radiology. 2011;259(1):17–19. doi: 10.1148/radiol.10102253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xie C, Goveas J, Wu Z, Li W, Chen G, Franczak M, Antuono PG, Jones JL, Zhang Z, Li SJ. Neural basis of the association between depressive symptoms and memory deficits in nondemented subjects: resting-state fMRI study. Hum Brain Mapp. 2011 doi: 10.1002/hbm.21291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xie C, Li SJ, Shao Y, Fu L, Goveas J, Ye E, Li W, Cohen AD, Chen G, Zhang Z, Yang Z. Identification of hyperactive intrinsic amygdala network connectivity associated with impulsivity in abstinent heroin addicts. Behav Brain Res. 2011;216(2):639–646. doi: 10.1016/j.bbr.2010.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Macey PM, Macey KE, Kumar R, Harper RM. A method for removal of global effects from fMRI time series. Neuroimage. 2004;22(1):360–366. doi: 10.1016/j.neuroimage.2003.12.042. [DOI] [PubMed] [Google Scholar]
- 11.Kelly AM, Uddin LQ, Biswal BB, Castellanos FX, Milham MP. Competition between functional brain networks mediates behavioral variability. Neuroimage. 2008;39(1):527–537. doi: 10.1016/j.neuroimage.2007.08.008. [DOI] [PubMed] [Google Scholar]
- 12.Fair DA, Cohen AL, Dosenbach NU, Church JA, Miezin FM, Barch DM, Raichle ME, Petersen SE, Schlaggar BL. The maturing architecture of the brain's default network. Proc Natl Acad Sci U S A. 2008;105(10):4028–4032. doi: 10.1073/pnas.0800376105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fox MD, Snyder AZ, Vincent JL, Corbetta M, Van Essen DC, Raichle ME. The human brain is intrinsically organized into dynamic, anticorrelated functional networks. Proc Natl Acad Sci U S A. 2005;102(27):9673–9678. doi: 10.1073/pnas.0504136102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fransson P. Spontaneous low-frequency BOLD signal fluctuations: an fMRI investigation of the resting-state default mode of brain function hypothesis. Hum Brain Mapp. 2005;26(1):15–29. doi: 10.1002/hbm.20113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fransson P. How default is the default mode of brain function? Further evidence from intrinsic BOLD signal fluctuations. Neuropsychologia. 2006;44(14):2836–2845. doi: 10.1016/j.neuropsychologia.2006.06.017. [DOI] [PubMed] [Google Scholar]
- 16.Tian L, Jiang T, Liu Y, Yu C, Wang K, Zhou Y, Song M, Li K. The relationship within and between the extrinsic and intrinsic systems indicated by resting state correlational patterns of sensory cortices. Neuroimage. 2007;36(3):684–690. doi: 10.1016/j.neuroimage.2007.03.044. [DOI] [PubMed] [Google Scholar]
- 17.Uddin LQ, Kelly AM, Biswal BB, Xavier Castellanos F, Milham MP. Functional connectivity of default mode network components: correlation, anticorrelation, and causality. Hum Brain Mapp. 2009;30(2):625–637. doi: 10.1002/hbm.20531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang K, Liang M, Wang L, Tian L, Zhang X, Li K, Jiang T. Altered functional connectivity in early Alzheimer's disease: a resting-state fMRI study. Hum Brain Mapp. 2007;28(10):967–978. doi: 10.1002/hbm.20324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murphy K, Birn RM, Handwerker DA, Jones TB, Bandettini PA. The impact of global signal regression on resting state correlations: are anti-correlated networks introduced? Neuroimage. 2009;44(3):893–905. doi: 10.1016/j.neuroimage.2008.09.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weissenbacher A, Kasess C, Gerstl F, Lanzenberger R, Moser E, Windischberger C. Correlations and anticorrelations in resting-state functional connectivity MRI: a quantitative comparison of preprocessing strategies. Neuroimage. 2009;47(4):1408–1416. doi: 10.1016/j.neuroimage.2009.05.005. [DOI] [PubMed] [Google Scholar]
- 21.Anderson JS, Druzgal TJ, Lopez-Larson M, Jeong EK, Desai K, Yurgelun-Todd D. Network anticorrelations, global regression, and phase-shifted soft tissue correction. Hum Brain Mapp. 2011;32(6):919–934. doi: 10.1002/hbm.21079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Birn RM, Diamond JB, Smith MA, Bandettini PA. Separating respiratory-variation-related fluctuations from neuronal-activity-related fluctuations in fMRI. Neuroimage. 2006;31(4):1536–1548. doi: 10.1016/j.neuroimage.2006.02.048. [DOI] [PubMed] [Google Scholar]
- 23.Chang C, Glover GH. Effects of model-based physiological noise correction on default mode net work anti-correlations and correlations. Neuroimage. 2009;47(4):1448–1459. doi: 10.1016/j.neuroimage.2009.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beall EB, Lowe MJ. Isolating physiologic noise sources with independently determined spatial measures. Neuroimage. 2007;37(4):1286–1300. doi: 10.1016/j.neuroimage.2007.07.004. [DOI] [PubMed] [Google Scholar]
- 25.Buckner RL, Andrews-Hanna JR, Schacter DL. The brain's default network: anatomy, function, and relevance to disease. Ann N Y Acad Sci. 2008;1124:1–38. doi: 10.1196/annals.1440.011. [DOI] [PubMed] [Google Scholar]
- 26.Raichle ME, MacLeod AM, Snyder AZ, Powers WJ, Gusnard DA, Shulman GL. A default mode of brain function. Proc Natl Acad Sci U S A. 2001;98(2):676–682. doi: 10.1073/pnas.98.2.676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goveas J, Xie C, Wu Z, Douglas Ward B, Li W, Franczak MB, Jones JL, Antuono PG, Yang Z, Li SJ. Neural correlates of the interactive relationship between memory deficits and depressive symptoms in nondemented elderly: resting fMRI study. Behav Brain Res. 2011;219(2):205–212. doi: 10.1016/j.bbr.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Glover GH, Li TQ, Ress D. Image-based method for retrospective correction of physiological motion effects in fMRI: RETROICOR. Magn Reson Med. 2000;44(1):162–167. doi: 10.1002/1522-2594(200007)44:1<162::aid-mrm23>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 29.Zar JH. Biostatistical analysis. Englewood Cliffs, N.J: Prentice-Hall; 1996. x, 662 p.p. [Google Scholar]
- 30.Calhoun VD, Adali T, Stevens MC, Kiehl KA, Pekar JJ. Semi-blind ICA of fMRI: A method for utilizing hypothesis-derived time courses in a spatial ICA analysis. Neuroimage. 2005;25(2):527–538. doi: 10.1016/j.neuroimage.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 31.Greicius MD, Srivastava G, Reiss AL, Menon V. Default-mode network activity distinguishes Alzheimer's disease from healthy aging: evidence from functional MRI. Proc Natl Acad Sci U S A. 2004;101(13):4637–4642. doi: 10.1073/pnas.0308627101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McKeown MJ, Makeig S, Brown GG, Jung TP, Kindermann SS, Bell AJ, Sejnowski TJ. Analysis of fMRI data by blind separation into independent spatial components. Hum Brain Mapp. 1998;6(3):160–188. doi: 10.1002/(SICI)1097-0193(1998)6:3<160::AID-HBM5>3.0.CO;2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fox MD, Zhang D, Snyder AZ, Raichle ME. The global signal and observed anticorrelated resting state brain networks. J Neurophysiol. 2009;101(6):3270–3283. doi: 10.1152/jn.90777.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]