Abstract
Women with high-grade cervical intraepithelial neoplasia (HGCIN) frequently present with multiple cervical lesions, and multiple concomitant Human papillomavirus (HPV) genotype infections. To elucidate HPV genotype attribution in different regions on the cervix, we performed molecular mapping of cervical disease in women with HGCIN. Thirteen subjects referred to colposcopy for abnormal cervical cancer screening results were included. A cervical smear and biopsies from four different areas on the cervix were collected. HPV genotyping using Linear Array (for cytology) or SPF-10-LiPA25 (for histology) were performed in 13 smears, 52 whole sections from biopsies, and 138 tissue regions isolated with laser capture microdissection (LCM). Twelve subjects had a diagnosis of CIN3 and one subject had a diagnosis of CIN2 based on the worst histology found in four biopsies. Eight of the 13 smears (62%) showed multiple genotype infections. Four of 13 women (31%) had multiple HPV infections in their biopsies. After performing LCM-PCR, only one woman (8%) had two different carcinogenic HPV types in morphologically distinct, but colliding HGCIN lesions. HPV16 was identified as the causal type in all women with HPV16 in cytology. A large proportion of other HPV types found in cervical smears were not detected at the tissue level. Using tissue-based genotyping and LCM-PCR analysis, we were able to attribute an individual HPV type to each area of CIN lesions. We demonstrate that HPV16 is even more etiologically dominant than previously thought, based on various genotype attribution models.
Keywords: CIN, HPV, colposcopy, LCM, genotyping
Introduction
Molecular and epidemiological studies have established the association between persistent infections with HPV and the development of cervical precancer and cancer(1;2). A group of approximately a dozen carcinogenic HPV types comprises the main etiologic factor for the development of neoplasms of the lower genital tract and it is estimated that 70-80% of all women are infected with cervical HPV at least once during their lifetime(3). Approximately 10% of women fail to clear HPV infections, resulting in long term persistent infections, which increase the risk of developing precancerous lesions that have a 30–50% risk of invasion over the remainder of a woman's life(4;5).
Multiple infections with carcinogenic HPV types are common, especially among younger women(6). It is generally assumed that HPV genotypes act independently and that each lesion is caused by a single genotype infection(7). In 15-50% of women with prevalent CIN of all grades, infection with more than one HPV genotype has been found in cervical smears (6;8;9), complicating the attribution of carcinogenic types to disease stages. Precise estimates of type attribution are important to understand the heterogeneous carcinogenicity of different HPV types. Furthermore, knowledge about type attribution to precancer is important in order to decide which HPV genotypes should be included in HPV detection assays and in preventive vaccines. HPV genotyping from cervical smears gives the composite view of all infections present on the cervical surface, including transforming infections associated with HGCIN, and transient infections in low grade lesions. In addition, vaginal infections completely unrelated to the cervix can be picked-up with the cervical smear. Thus, attribution of HPV genotypes to individual cervical lesions can only be determined by targeted tissue-based genotyping.
Laser Capture Microdissection (LCM) combined with sensitive PCR is considered the gold standard to study HPV genotype attribution in cervical lesions, but has only been used to a very limited degree to investigate cervical precancer(10;11). Here, we performed meticulous HPV genotyping of cervical lesions at three levels of resolution: on the entire cervix by cytology (covering several centimeters), on individual biopsies (covering several millimeters), and on sub-regions processed with LCM (covering several 100 micrometers). We used this unique approach to resolve HPV genotype attribution and to analyze the clonal relationship of multiple HGCIN lesions on the cervix.
Materials & Methods
Study population and clinical investigations
We conducted our analysis in the NIH-OUHSC Biopsy Study, a population-based study of women referred to colposcopy for abnormal cervical cancer screening results at the University of Oklahoma Health Sciences Center (OUHSC). Design and methodology have been described in depth elsewhere(6;12;13). In the current phase of the study, an extended colposcopic biopsy protocol with up to four biopsies was used. We successively selected women enrolled in the NIH-OUHSC Biopsy Study who had 4 biopsies taken, who had at least one biopsy with a result of CIN2 or greater, and formalin-fixed, paraffin-embedded tissue available for all four biopsies. Subjects were not selected by HPV genotypes present in cytology.
Prior to colposcopic biopsy procedures, cervical cytology was collected using a broom device and transferred to PreservCyt™ solution (Hologic, Boxborough, MA, USA). The cytology specimen was used for Thinprep® liquid based cytology and for HPV DNA analysis. Up to four biopsies were taken from distinct acetowhite lesions or large heterogeneous lesions extending over two quadrants. If fewer than four directed biopsies were taken, a biopsy from a quadrant without any visible CIN (random biopsy) was added. Digital images were taken during the examination from representative cervical views. Dedicated annotation software, based on the Boundary Marking Tool (BMT), a web-based application, allowed us to mark boundaries and biopsy sites on digital images. Annotations of the overall impression, as well as boundaries and biopsy sites related to the graphical data, were stored in a database. As per standard practice, all histologically confirmed CIN3 and most CIN2 were treated by loop electrosurgical excision procedure (LEEP) of the transformation zone.
Histological processing and laser capture microdissection
Four μm thick sections were cut from the biopsies, stained with hematoxylin and eosin (H&E) and evaluated by a pathologist. From every biopsy, additional consecutive tissue sections were used for H&E and p16INK4a immunostaining. P16INK4a immunohistochemistry was used to support diagnosis(14). Review diagnosis on these additional slides was done by a second pathologist and if discordance occurred, a third pathologist reviewed the discordances. Consensus diagnosis was determined by the agreement of two of the three interpretations. Immunostaining with p16INK4a was done using mouse anti-p16INK4a (Immunologic, Duiven, the Netherlands) and the EnVision™ kit (Dako A/S, Glostrup, Denmark) for signal visualization. A third consecutive slide was used for whole tissue HPV analysis.
In cases where multiple HPV infections were detected in whole tissue specimens, LCM-PCR analysis was used to study HPV genotypes on the individual lesional level. Samples of CIN lesions and normal cervix were obtained for analysis on the additional H&E and p16-stained slides. All slides were scanned using digital microscopy (Aperio Technologies Inc, Vista, Ca, USA). We compared the consensus histology result of the slides used for HPV genotyping to the clinical diagnosis; 40 of 52 biopsies (77%) had the same diagnosis in both slides, 8 biopsies were downgraded compared to the original result, while 4 were upgraded. A pathologist annotated areas of CIN grades 1, 2, and 3 and normal tissue on both H&E and p16-stained slides. Several regions spread over the total biopsy epithelium and random regions of epithelium negative for CIN, or debris surrounding the tissue, were selected. The sample size ranged from 14,000 – 193,000 square micrometers. Using the annotated digital image of the slide, the selected regions were extracted with the Zeiss P.A.L.M. microbeam ultraviolet (UV) laser microdissection and catapulting system and transferred to an Adhesive Cap500 opaque tube (Zeiss Microimaging GmbH, Jena, Germany). In addition, LCM-PCR was performed on a negative control (human placenta) before each sample analysis to control and check for potential contamination of the patient samples, which was not found.
Cytology-based genotyping
HPV detection and genotyping of cytology specimens was done using the Linear Array (LA) HPV Genotyping System (Roche Molecular Diagnostics, Branchburg, NJ, USA). The LA assay is a type-specific PGMY09/11 L1 primer-based PCR assay for 37 HPV types (6, 11, 16, 18, 26, 31, 33, 35, 39, 40, 42, 45, 51, 52, 53, 54, 55, 56, 58, 59, 61, 62, 64, 66, 67, 68, 69, 70, 71, 72, 73, 81, 82, 83, 84, 89). LA genotyping was performed according to the manufacturer's instructions with slight modifications, as previously described(6). In brief, the procedure followed recommendations of the manufacturer with the variation that 10 μL of template DNA was amplified and the amplified products were hybridized and detected using an automated Auto-LiPA staining system (Innogenetics N.V., Gent, Belgium) using 2.5 mL of each reagent per strip (as compared to 4.0 mL in manual processing). The LA results were evaluated by unmagnified examination of the strips by two independent observers. An unambiguous, continuous band was judged to indicate that biotinylated amplicons had hybridized to complementary sequences of probes bound to the strips, and was considered a positive result. The evaluators also subjectively graded the intensity of each band, as strong (S), moderate (M), weak (W), very weak (VW) or extremely weak (EW).
Tissue-based genotyping
The tissue was scraped by a sterile cotton swab from the slide after deparaffinization and dissolving the tissue in proteinase-K dilution buffer. Next, the tissue was resuspended in 100 μL proteinase-K solution (1 mg/mL). DNA isolation was performed overnight at 70°C. Proteinase K was inactivated at 95°C for 10 min, and 10 μL of isolated DNA was used directly for amplification by broad-spectrum primers that amplify a 65 bp region of the L1 gene(15-17). The amplification products were detected by the HPV SPF-10 PCR DNA enzyme immunoassay (DEIA) system (Labo Bio-medical Products, Rijswijk, the Netherlands), which detects at least 54 different HPV genotypes. DEIA-positive SPF-10 amplimers were used to identify the HPV genotype by reverse hybridization on a line probe assay (SPF-10 HPV LiPA version 1, Labo Bio-medical Products), which detects 25 HPV genotypes (6, 11, 16, 18, 31, 33, 34, 35, 39, 40, 42, 43, 44, 45, 51, 52, 53, 56, 58, 59, 66, 68, 70, and 74)(18). The comparison of multiple HPV genotypes in cytology specimens and tissue is limited by the different type coverage of LA and LiPa25. HPV types included in the LA assay, but not in the LiPA25 assay (HPV26, 54, 55, 61, 62, 64, 67, 69, 71, 72, 73, 81, 82, 83, 84 and 89) could not be evaluated on the tissue level in this analysis.
Each run contained negative and internal and external positive controls to monitor for efficiency of DNA isolation, PCR amplification, hybridization, and genotyping procedures. Contamination or failure of analyses was not encountered. DNA isolation and PCR for HPV DNA from LCM samples was performed as described for whole tissue biopsy specimens, with an exception of the proteinase-K incubation temperature, which was 56°C.
Results
HPV genotype distribution in cytology samples and biopsy histology results
A cytology result of high-grade squamous intraepithelial lesion (HSIL) or worse was reported in 8 women, atypical squamous cells - cannot exclude HSIL (ASC-H) in 3 women, and low-grade squamous intraepithelial lesion (LSIL) in 2 women. Five of 13 cases (38%) were considered as a single HPV genotype infection and 8 cases (62%) were considered as infected with multiple (between 2 and 6) HPV genotypes. LA signal intensity for each detected HPV genotype showed at least one strong signal for a genotype in each woman (Table 1). Among the 52 individual biopsy results (13 women each with four biopsies), 10 were diagnosed as normal, 8 with CIN1, 8 with CIN2, and 26 with CIN3. The number of biopsies with a CIN3 result obtained from an individual woman ranged from one to four.
Table 1.
Cytology, histology and LCM-PCR results
| Cytology | Histology | LCM | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Case | Diagnosis | HPV (LA signal intensity)† | Clinical diagnosis | Consensus tissue | p16 staining* | HPV | Total no. LCM regions | No. LCM regions:negative | Single HPV types by LCM (# areas) | Multiple HPV types by LCM (# areas) |
| Single HPV infections in cytology | ||||||||||
| 1 | ASC-H | 16(s) | CIN 1 | Neg | 0 | Neg | n.a. | n.a. | n.a. | n.a. |
| CIN 2 | CIN 2 | 4 | 16 | n.a. | n.a. | n.a. | n.a. | |||
| CIN 2 | CIN 2 | 4 | 16 | n.a. | n.a. | n.a. | n.a. | |||
| CIN 3 | CIN 3 | 4 | 16 | 1 | 0 | 16 (1) | 0 | |||
| 2 | HSIL | 35(s) | CIN 3 | CIN 3 | 4 | 35 | n.a. | n.a. | n.a. | n.a. |
| CIN 3 | CIN 3 | 4 | 35 | n.a. | n.a. | n.a. | n.a. | |||
| CIN 3 | CIN 3 | 4 | 35 | n.a. | n.a. | n.a. | n.a. | |||
| CIN 3 | CIN 3 | 4 | 35 | 1 | 0 | 35 (1) | 0 | |||
| 3 | HSIL | 16(s) | Neg | CIN 2 | 3 | 16 | n.a. | n.a. | n.a. | n.a. |
| CIN 3 | CIN 3 | 4 | 16 | n.a. | n.a. | n.a. | n.a. | |||
| CIN 3 | CIN 3 | 4 | 16 | n.a. | n.a. | n.a. | n.a. | |||
| CIN 3 | CIN 3 | 4 | 16 | 1 | 0 | 16 (1) | 0 | |||
| 4 | Inv. ca.** | 18(s) | CIN 3 | CIN 3 | 4 | 18 | 1 | 0 | 18 (1) | 0 |
| CIN 3 | CIN 3 | 4 | 18 | n.a. | n.a. | n.a. | n.a. | |||
| CIN 3 | CIN 2 | 3 | 18 | n.a. | n.a. | n.a. | n.a. | |||
| CIN 3 | CIN 2 | 4 | 18 | n.a. | n.a. | n.a. | n.a. | |||
| 5 | HSIL | 16(s) | CIN 3 | CIN 3 | 4 | 16 | 1 | 0 | 16 (1) | 0 |
| CIN 3 | CIN 3 | 4 | 16 | n.a. | n.a. | n.a. | n.a. | |||
| CIN 3 | Neg | 1 | 16 | n.a. | n.a. | n.a. | n.a. | |||
| Neg | Neg | 1 | Neg | n.a. | n.a. | n.a. | n.a. | |||
| Multiple HPV infections in cytology | ||||||||||
| 6 | ASC-H | 16(s), 52(ew), 55(vw) | Neg | Neg | 0 | Neg | 2 | 2 | n.a.*** | n.a. |
| CIN 3 | CIN 3 | 4 | 16 | 8 | 0 | 16 (8) | 0 | |||
| CIN 3 | CIN 3 | 4 | 16 | 9 | 1 | 16 (8) | 0 | |||
| CIN 1 | Neg | 0 | 16 | 8 | 8 | n.a. | n.a. | |||
| 7 | HSIL | 42(m), 53(s), 55(s), 66(ew), 67(s), 73(s) | Neg | Neg | 0 | 51 | 4 | 4 | n.a. | n.a. |
| Neg | Neg | 0 | 73 | 2 | 2 | n.a. | n.a. | |||
| CIN 2 | CIN 2 | 1 | 73,53 | 6 | 0 | 73 (6) | 0 | |||
| CIN 1 | CIN 1 | 3 | 73 | 6 | 0 | 73 (6) | 0 | |||
| 8 | LSIL | 16(s), 53(s), 68(s) | CIN 3 | CIN 2 | 4 | 16,68 | 10 | 0 | 16 (10) | 0 |
| CIN 3 | CIN 2 | 4 | 16,53,68 | 8 | 1 | 16 (7) | 0 | |||
| CIN 2 | CIN 2 | 4 | 16,68 | 10 | 0 | 16 (10) | 0 | |||
| Neg | Neg | 1 | 16,53,68 | 4 | 4 | n.a. | n.a. | |||
| 9 | HSIL | 16(s), 31(s), 42(s) | CIN 3 | CIN 3 | 4 | 16,31 | 12 | 0 | 16 (4), 31 (7) | 16+31 (1) |
| CIN 2 | CIN 2 | 4 | 31 | n.a. | n.a. | n.a. | n.a. | |||
| CIN 3 | CIN 3 | 4 | 16 | n.a. | n.a. | n.a. | n.a. | |||
| CIN 3 | CIN 3 | 4 | 16,31 | 8 | 0 | 16 (8) | 0 | |||
| 10 | HSIL | 16(s), 40(s), 59(s) | CIN 1 | CIN 1 | 2 | 16,40 | 16 | 1 | 16 (7), 40 (1) | 16+40 (7) |
| CIN 1 | CIN 1 | 2 | 16 | n.a. | n.a. | n.a. | n.a. | |||
| CIN 3 | CIN 2 | 3 | 16 | n.a. | n.a. | n.a. | n.a. | |||
| Neg | Neg | 2 | 16 | n.a. | n.a. | n.a. | n.a. | |||
| 11 | LSIL | 16(m), 51(s) | CIN 3 | CIN 2 | 4 | 16,51 | 10 | 1 | 16 (9) | 0 |
| Neg | CIN 1 | 2 | 51 | 1 | 0 | 51 (1) | 0 | |||
| CIN 1 | CIN 1 | 3 | 51 | 1 | 0 | 51 (1) | 0 | |||
| Neg | Neg | 1 | 51 | 3 | 1 | 51 (1) | 16+51 (1) | |||
| 12 | ASC-H | 16(s), 84(vw)# | CIN 3 | CIN 3 | 4 | 16 | 1 | 0 | 16 (1) | 0 |
| CIN 1 | CIN 1 | 2 | 16 | n.a. | n.a. | n.a. | n.a. | |||
| CIN 2 | CIN 2 | 3 | 16 | n.a. | n.a. | n.a. | n.a. | |||
| CIN 2 | CIN 2 | 3 | 16 | n.a. | n.a. | n.a. | n.a. | |||
| 13 | HSIL | 16(s), 61(m)# | CIN 1 | Neg | 0 | 16 | 3 | 1 | 16 (2) | 0 |
| CIN 2 | CIN 3 | 4 | 16 | 1 | 0 | 16 (1) | 0 | |||
| CIN 3 | CIN 3 | 4 | 16 | n.a. | n.a. | n.a. | n.a. | |||
| Neg | Neg | 2 | 16 | n.a. | n.a. | n.a. | n.a. | |||
The figures in parenthesis indicate the number of LCM areas in which single or multiple HPV type(s) were detected in each case
LA signal intensity: (s)= strong; (m)= medium; (w)= weak; (vw)=very weak; (ew)= extremely weak
0= negative, 1= patchy staining, 2= diffuse staining first 1/3 basal, 3= diffuse staining first 2/3 basal 4= diffuse staining whole epithelium
Inv. ca.= Invasive carcinoma
n.a.= Not analyzed
HPV type not detected by both HPV DNA assays (LA and LiPA25)
HPV genotype attribution in whole biopsies
We performed tissue-based genotyping using DNA extracted from a whole tissue section of each individual biopsy. Among the 5 women with single HPV infections in cytology, the same genotype found in cytology was detected in all 17 biopsies showing a cervical lesion (CIN2 or CIN3). Two of three biopsy specimens with normal epithelium in these women did not harbor any HPV DNA, while one was positive for HPV16. Of the 32 biopsies obtained from the 8 women with multiple infections in cytology, 22 (69%) were found to have a single HPV infection in the biopsy, 9 (28%) were positive for two or three HPV genotypes and one biopsy was negative for HPV (3%). For each biopsy except one, the HPV genotypes identified in the tissues were a subset of those found in the corresponding cytology specimen. HPV51 was found as a single type in one biopsy without features of CIN, but this type was not detected in the corresponding cytology specimen. In women with multiple HPV genotype infections, 10 of 24 total HPV genotypes (42%) detected in cytology samples were not detected in the corresponding biopsies. These included mainly infections with non-carcinogenic types, such as HPV42, 55, 61, 67, and 84. Of note, in two women with two infections on the cytology level, one of the two types was not covered by LiPA25 (HPV84 in subject #12 and HPV61 in subject #13) and therefore could not be assessed on the tissue level (Table 1).
HPV genotype attribution in microdissected lesions
We performed LCM on all 9 biopsy samples that showed multiple HPV genotypes on the whole section analysis (including 2 CIN3, 5 CIN2, one CIN1, and one normal biopsy), on a random selection of 17 biopsies that revealed only one HPV genotype, and on one HPV-negative biopsy.
Of the 9 biopsies that showed multiple type infections on analysis of the whole tissue section, 6 were found to be positive for only one HPV genotype in LCM. Two biopsies had multiple types in LCM regions (one CIN3 and one CIN1 biopsy), and one had no HPV detected by LCM analyses (normal tissue biopsy). In the 40 biopsies with a single HPV genotype infection in the whole tissue specimen, 52 LCM regions were analysed. Of these regions, 17 were HPV-negative, 34 showed a single HPV infection and in one LCM region, a double HPV infection (HPV16 and 51) was observed. This region consisted of fragmented debris that was not associated with the normal epithelium obtained in the biopsy.
Overall, from all 138 selected LCM regions, only 9 LCM regions showed multiple genotype infections in the same LCM region (Table 1). Only one woman (Table 1; case 9) had two different carcinogenic HPV types (HPV16 and 31) that were attributed to HGCIN in one biopsy.
In all cases with a multiple HPV infection in cytology an HPV type with a strong signal intensity on the reverse line blot strip was retrieved on lesion level by LCM, except for case 11 where both HPV16 (medium intensity) and HPV51 (strong intensity) were retrieved (Table 1).
Table 2 gives an overview of all histological diagnoses of selected LCM regions and respective HPV genotype distributions. All CIN1+ regions were positive for at least one HPV genotype. HPV analysis showed negativity in 25 regions (18%) that all were histologically normal epithelium. Of all regions with multiple genotypes detected, one was negative for CIN, 6 consisted of CIN1 regions, one consisted of CIN2 and one of cellular debris.
Table 2.
Diagnosis and HPV positivity on LCM level
| Pathological diagnosis LCM level | LCM regions | ||||
|---|---|---|---|---|---|
| Total no. | Negative | Positive | HPV single type | HPV multiple types | |
| Negative | 34* | 25 | 9* | 7* | 2* |
| CIN 1 | 20 | 0 | 20 | 14 | 6 |
| CIN 2 | 54 | 0 | 54 | 53 | 1 |
| CIN 3 | 30 | 0 | 30 | 30 | 0 |
| Total | 138 | 25 | 113 | 104 | 9 |
2 areas of debris selected in biopsy negative for CIN
We compared LCM-genotyping between H&E and p16-stained slides in 57 randomly selected regions from CIN1, 2 or 3 and normal tissue. An almost perfect agreement was found between HPV positivity in H&E- and p16-stained slides (kappa 0.89) (Table 3). All regions positive for HPV were positive for the same genotype in H&E- and p16-stained slides.
Table 3.
Concordance HPV positivity on HE & p16 slides; number LCM regions
| p16 | ||||
|---|---|---|---|---|
| HPV type concordance | HPV negative | Total | ||
| HE | HPV type concordance | 45 | 1 | 46 |
| HPV negative | 1 | 10 | 11 | |
| Total | 46 | 11 | 57 | |
Topography of cervical lesions with multiple HPV genotypes
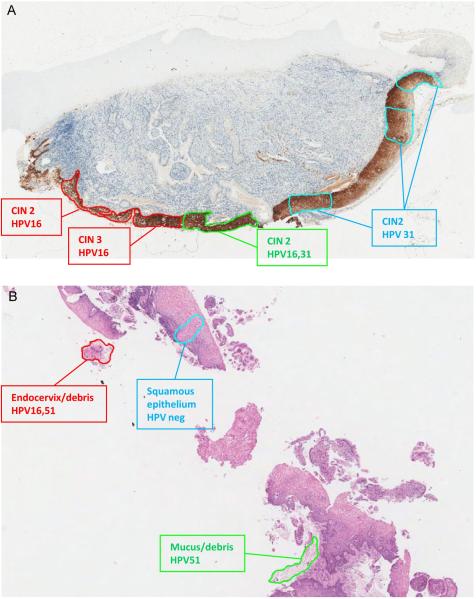
Two cases with multiple HPV genotype infections on individual LCM regions are shown in Figure 1. Figure 1A (case 9) shows a cervical biopsy of CIN3 associated with 2 HPV genotypes. The H&E slide showed an extended continuous, but morphologically heterogeneous lesion, a pattern confirmed by p16 staining. LCM-PCR revealed that the two HPV types detected in the whole section (HPV16, 31) were present as two separate but colliding high-grade CIN lesions, each associated with one HPV type. The HPV16-associated lesion extended towards the endocervix, while the HPV31 related lesion extended towards the ectocervix. In case 11, one LCM region showed infection with HPV16 and 51 in a biopsy that was negative for CIN. The analyzed region consisted of debris with (endo)cervical cells detached from the cervical epithelium. An LCM region from the adjacent normal squamous epithelium was negative for HPV (Figure 1B).
Figure 1.
HPV genotype map in 2 cases. A: Case 9 P16 stained slide. Apparently single high- grade lesion associated with HPV16 and 31. B: Case 11 H&E stained slide. HPV16 and 51 in debris with (endo)cervical cells, but not in normal squamous epithelium.
Attribution of HPV16 to high grade CIN
For all cases positive for HPV16 in cytology (n=10; 3 in single and 7 in multiple genotype infections), HPV16 was retrieved in whole tissue analysis of the biopsy and from LCM regions representative for the highest grade histology result. Only two HPV16-positive cases had additional carcinogenic types in the whole section genotyping analysis. In one case positive for HPV16 and 51 in cytology, both types were retrieved at the lesional level; HPV16 was found in the CIN2 lesion while HPV51 was found in a separate CIN1 lesion. In another case, HPV16 and HPV31 were found in two distinct CIN3 lesions on the same tissue section.
Discussion
Cervical studies using HPV, cytology, and histology can yield the incorrect impression of homogeneity. A woman is said to have cervical infection with certain HPV types, and to have a cytological impression or histological diagnosis of a certain grade. In fact, as we show in this analysis, the topography of cervical HPV infection and resultant neoplasia is much more complex.
Characterization of HPV genotype attribution in individual cervical intraepithelial lesions is crucial to understanding the heterogeneous biology of individual HPV genotypes. HPV type attribution also has important translational relevance, e.g. to decide which types should be included in HPV detection assays and to estimate the efficacy of preventive HPV vaccines. Multiple genotype HPV infections occur in 15-40% of women with CIN of all grades and even in a small percentage of invasive cervical cancers(8;19). Type attribution is currently mainly based on assumptions; actual measures of HPV genotype attribution can only be obtained by tissue genotyping. Recent data have suggested that each morphologically independent area of CIN is caused by one HPV genotype(7). However, these analyses have so far been restricted to individual biopsies from the cervical surface.
In this analysis, the heterogeneous presentation of HGCIN was studied by histology evaluation and genotyping of 13 cervical smears, 52 whole tissue sections, and 138 LCM regions. We were able to attribute individual ‘causal’ HPV genotypes to all lesions. Among five women with single-genotype infections in the cytology specimen, the same genotype was found in the whole section and in the individual corresponding LCM regions, confirming that the genotypes found in cytology caused the CIN3 or CIN2 lesions. We demonstrate that LCM-PCR is a technique that can be used to investigate HPV genotype patterns in relation to histological findings in routine H&E- and p16-stained cervical biopsies. Using LCM, the transforming HPV infection in HGCIN lesions can be distinguished from transient HPV infections. We found HPV genotypes in all regions with CIN1 or greater, while the majority of normal tissue areas were negative for HPV, suggesting that even transient infections may frequently be associated with CIN1 morphology.
So far, our understanding of the biological relationship of multiple HPV genotypes and multiple lesions present on the cervix has been limited. In this study, only one case with multiple HGCIN biopsies had two carcinogenic types that were found in two independent HGCIN lesions. All others had the same carcinogenic type even when multiple carcinogenic types were found in cytology. Our data support that multiple high grade lesions on the cervix are often caused by a single carcinogenic genotype while other carcinogenic HPV types detected in cervical smears of the same patients are related to independent transient infections. Our findings support the “one virus-one lesion” hypothesis and speak against biological interactions of multiple infections on the lesion level(7).
Whole tissue genotyping of colposcopically-guided biopsies reduces the proportion of multiple HPV genotype infections substantially compared to cytology specimens and LCM-PCR assigns an individual HPV type to each morphologically distinct area of CIN. In all cases of HGCIN, LCM-PCR identified one of the genotypes detected in cytology, supporting that LCM-PCR is both sensitive and specific in identifying the causal HPV genotype for HGCIN. Due to the small areas targeted, LCM-PCR is not effective in retrieving all HPV genotypes present on the complete surface of the cervix, which would require hundreds of LCM-PCRs. The results from the different HPV detection methods used to test cytology and histology specimens in this study were highly concordant for the types present in the lesions, supporting previous data from van Doorn et al.(16), demonstrating that the genotyping results obtained by LA (PGMY primer set) and SPF-10 LiPA25 are highly comparable. In detail, HPV types 55, 61, 67 and 84 were detected by the LA in some cases with multiple HPV infection in cytology. These genotypes are not present in the LiPA25 assay and therefore could not be detected in biopsy and LCM-PCR samples. This could have caused an underestimation of HPV infection in these specimens, although all of these LA detected HPV types are low-risk types.
Our findings demonstrate that genotyping data from cervical cytology specimens may lead to attribution of less-carcinogenic types to disease; these types are mostly present in concomitant, transient infections. In addition, Castle et al.(20) showed that non-carcinogenic HPV types of the α3/α15 phylogenetic species have a tropism for vaginal epithelium but are routinely detected in cervical smears, potentially by contamination of the cervical smear with vaginal cells. They concluded that many HPV genotypes detected by standard cytology might not be involved in the development of CIN on the cervical surface.
Our data demonstrate that HPV16 was the most predominant causal genotype, because in all cases positive for HPV16 (n=10; 77%) by cytology, this genotype was retrieved in whole tissue analysis of the biopsy and confirmed as the causal type for each CIN lesion in LCM regions of the same lesion. The unique role of HPV16 as a carcinogenic agent has been widely recognized. The attribution of HPV16 to cervical cancers, where multiple type infections are far less common than in CIN, is over 50% with a big distance to other carcinogenic types(21). Our data support the notion that HPV16 is more etiologically dominant in HGCIN than what can be appreciated when analyzing genotype attribution in cervical smears with frequent multiple infections.
As we continue our molecular studies in the NIH-OUHSC Biopsy Study, we will further enhance the understanding of the molecular basis of HPV natural history. A wider application to other techniques, such as identifying host and viral markers, could provide new insights into the complex biology of concurrent HPV infections in the cervix. This may allow us to gain a better understanding of the issue of biological heterogeneity of CIN lesions and thus improve our diagnostic tools for patients with CIN.
Novelty and impact statement.
Multiple high-grade lesions on the cervix are often caused by a single carcinogenic HPV genotype while other types detected in cervical smears are related to independent transient infections. HPV16 is more etiologically dominant in HGCIN than what can be interpreted from genotype attribution in cervical smears.
Acknowledgments
Funding source: This research was funded by the Intramural Research Program of the National Cancer Institute.
Abbreviations
- HGCIN
High-grade cervical intraepithelial neoplasia
- HPV
Human papillomavirus
- LCM
Laser capture microdissection
- OUHSC
Oklahoma University Health Sciences Center
- BMT
Boundary Marking Tool
- LEEP
Loop electrosurgical excision procedure
- H&E
Hematoxylin and eosin
- UV
Ultraviolet
- LA
Linear Array
- DEIA
DNA enzyme immuno assay
- SPF
Short PCR fragment
- HSIL
High-grade squamous intraepithelial lesion
- LSIL
Low-grade squamous intraepithelial lesion
- ASC-H
Atypical squamous cells - cannot exclude HSIL
Reference List
- 1.Galani E, Christodoulou C. Human papilloma viruses and cancer in the post-vaccine era. Clin Microbiol Infect. 2009;15:977–81. doi: 10.1111/j.1469-0691.2009.03032.x. [DOI] [PubMed] [Google Scholar]
- 2.Bosch FX, Lorincz A, Munoz N, Meijer CJ, Shah KV. The causal relation between human papillomavirus and cervical cancer. J Clin Pathol. 2002;55:244–65. doi: 10.1136/jcp.55.4.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baseman JG, Koutsky LA. The epidemiology of human papillomavirus infections. J Clin Virol. 2005;32(Suppl 1):S16–S24. doi: 10.1016/j.jcv.2004.12.008. [DOI] [PubMed] [Google Scholar]
- 4.Bodily J, Laimins LA. Persistence of human papillomavirus infection: keys to malignant progression. Trends Microbiol. 2011;19:33–9. doi: 10.1016/j.tim.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McCredie MR, Sharples KJ, Paul C, Baranyai J, Medley G, Jones RW, Skegg DC. Natural history of cervical neoplasia and risk of invasive cancer in women with cervical intraepithelial neoplasia 3: a retrospective cohort study. Lancet Oncol. 2008;9:425–34. doi: 10.1016/S1470-2045(08)70103-7. [DOI] [PubMed] [Google Scholar]
- 6.Wentzensen N, Schiffman M, Dunn ST, Zuna RE, Walker J, Allen RA, Zhang R, Sherman ME, Wacholder S, Jeronimo J, Gold MA, Wang SS. Grading the severity of cervical neoplasia based on combined histopathology, cytopathology, and HPV genotype distribution among 1,700 women referred to colposcopy in Oklahoma. Int J Cancer. 2009;124:964–9. doi: 10.1002/ijc.23969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Quint W, Jenkins D, Molijn A, Struijk L, Sandt van de M, Doorbar J, Mols J, Hoof van C, Hardt K, Struyf F, Colau B. One virus one lesion - Individual components of CIN lesions contain a specific HPV type. J Pathology. 2011 doi: 10.1002/path.3970. DOI:10.1002/path.3970. [DOI] [PubMed] [Google Scholar]
- 8.Cuschieri KS, Cubie HA, Whitley MW, Seagar AL, Arends MJ, Moore C, Gilkisson G, McGoogan E. Multiple high risk HPV infections are common in cervical neoplasia and young women in a cervical screening population. J Clin Pathol. 2004;57:68–72. doi: 10.1136/jcp.57.1.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sherman ME, Wang SS, Tarone R, Rich L, Schiffman M. Histopathologic extent of cervical intraepithelial neoplasia 3 lesions in the atypical squamous cells of undetermined significance low-grade squamous intraepithelial lesion triage study: implications for subject safety and lead-time bias. Cancer Epidemiol Biomarkers Prev. 2003;12:372–9. [PubMed] [Google Scholar]
- 10.Kalantari M, Garcia-Carranca A, Morales-Vazquez CD, Zuna R, Montiel DP, Calleja-Macias IE, Johansson B, Andersson S, Bernard HU. Laser capture microdissection of cervical human papillomavirus infections: copy number of the virus in cancerous and normal tissue and heterogeneous DNA methylation. Virology. 2009;390:261–7. doi: 10.1016/j.virol.2009.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chew K, Rooney PH, Cruickshank ME, Murray GI. Laser capture microdissection and PCR for analysis of human papilloma virus infection. Methods Mol Biol. 2005;293:295–300. doi: 10.1385/1-59259-853-6:295. [DOI] [PubMed] [Google Scholar]
- 12.Wang SS, Zuna RE, Wentzensen N, Dunn ST, Sherman ME, Gold MA, Schiffman M, Wacholder S, Allen RA, Block I, Downing K, Jeronimo J, et al. Human papillomavirus cofactors by disease progression and human papillomavirus types in the study to understand cervical cancer early endpoints and determinants. Cancer Epidemiol Biomarkers Prev. 2009;18:113–20. doi: 10.1158/1055-9965.EPI-08-0591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wentzensen N, Schiffman M, Dunn T, Zuna RE, Gold MA, Allen RA, Zhang R, Sherman ME, Wacholder S, Walker J, Wang SS. Multiple human papillomavirus genotype infections in cervical cancer progression in the study to understand cervical cancer early endpoints and determinants. Int J Cancer. 2009;125:2151–8. doi: 10.1002/ijc.24528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Klaes R, Benner A, Friedrich T, Ridder R, Herrington S, Jenkins D, Kurman RJ, Schmidt D, Stoler M, von Knebel DM. p16INK4a immunohistochemistry improves interobserver agreement in the diagnosis of cervical intraepithelial neoplasia. Am J Surg Pathol. 2002;26:1389–99. doi: 10.1097/00000478-200211000-00001. [DOI] [PubMed] [Google Scholar]
- 15.Kleter B, van Doorn LJ, ter SJ, Schrauwen L, van KK, Burger M, ter HB, Quint W. Novel short-fragment PCR assay for highly sensitive broad-spectrum detection of anogenital human papillomaviruses. Am J Pathol. 1998;153:1731–9. doi: 10.1016/S0002-9440(10)65688-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van Doorn LJ, Quint W, Kleter B, Molijn A, Colau B, Martin MT, Kravang I, Torrez-Martinez N, Peyton CL, Wheeler CM. Genotyping of human papillomavirus in liquid cytology cervical specimens by the PGMY line blot assay and the SPF(10) line probe assay. J Clin Microbiol. 2002;40:979–83. doi: 10.1128/JCM.40.3.979-983.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Quint WG, Scholte G, van Doorn LJ, Kleter B, Smits PH, Lindeman J. Comparative analysis of human papillomavirus infections in cervical scrapes and biopsy specimens by general SPF(10) PCR and HPV genotyping. J Pathol. 2001;194:51–8. doi: 10.1002/path.855. [DOI] [PubMed] [Google Scholar]
- 18.Kleter B, van Doorn LJ, Schrauwen L, Molijn A, Sastrowijoto S, ter SJ, Lindeman J, ter HB, Burger M, Quint W. Development and clinical evaluation of a highly sensitive PCR-reverse hybridization line probe assay for detection and identification of anogenital human papillomavirus. J Clin Microbiol. 1999;37:2508–17. doi: 10.1128/jcm.37.8.2508-2517.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clifford GM, Smith JS, Plummer M, Munoz N, Franceschi S. Human papillomavirus types in invasive cervical cancer worldwide: a meta-analysis. Br J Cancer. 2003;88:63–73. doi: 10.1038/sj.bjc.6600688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Castle PE, Rodriguez AC, Porras C, Herrero R, Schiffman M, Gonzalez P, Hildesheim A, Burk RD. A comparison of cervical and vaginal human papillomavirus. Sex Transm Dis. 2007;34:849–55. doi: 10.1097/OLQ.0b013e318064c8c5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li N, Franceschi S, Howell-Jones R, Snijders PJ, Clifford GM. Human papillomavirus type distribution in 30,848 invasive cervical cancers worldwide: Variation by geographical region, histological type and year of publication. Int J Cancer. 2011;128:927–35. doi: 10.1002/ijc.25396. [DOI] [PubMed] [Google Scholar]