Abstract
The circadian expression of clock and clock-controlled cognition-related genes in the hippocampus would be essential to achieve an optimal daily cognitive performance. There is some evidence that retinoid nuclear receptors (RARs and RXRs) can regulate circadian gene expression in different tissues. In this study, Holtzman male rats from control and vitamin A-deficient groups were sacrificed throughout a 24-h period and hippocampus samples were isolated every 4 or 5 h. RARα and RXRβ expression level was quantified and daily expression patterns of clock BMAL1, PER1, RORα and REVERB genes, RORα and REVERB proteins, as well as temporal expression of cognition-related RC3 and BDNF genes were determined in the hippocampus of the two groups of rats. Our results show significant daily variations of BMAL1, PER1, RORα and REVERB genes, RORα and REVERB proteins and, consequently, daily oscillating expression of RC3 and BDNF genes in the rat hippocampus. Vitamin A deficiency reduced RXRβ mRNA level as well as the amplitude of PER1, REVERB gene and REVERB protein rhythms, and phase-shifted the daily peaks of BMAL1 and RORα mRNA, RORα protein and RC3 and BDNF mRNA levels. Thus, nutritional factors, such as vitamin A and its derivatives the retinoids, might modulate daily patterns of BDNF and RC3 expression in the hippocampus and they could be essential to maintain an optimal daily performance at molecular level in this learning-and-memory-related brain area.
Keywords: circadian rhythm, BDNF, neurogranin, retinoic acid, brain
INTRODUCTION
A vitamin A derivative, the retinoic acid (RA) has been detected at relatively high levels in the central nervous system. The hippocampus/cortex area has been reported to contain the highest proportion of RA in the brain (Werner and Deluca, 2002) and immunoreactivity for the binding proteins CRBP-I and CRABP-I has been demonstrated in the dendritic layers of the hippocampal formation and dentate gyrus (Zetterström et al., 1999). It is well known that many of the effects of vitamin A are mediated via retinoic acid isomers which bind to specific nuclear receptors (RARs and RXRs) and thus modulate the expression of target genes. RARα, RXRα, -β and –γ nuclear receptors have also been detected in the hippocampus (Zetterstrom et al. 1994 and 1999, Fonzo et al., 2009). Additionally, very recently, Takasaki and collaborators (2011) demonstrated that vitamin A prevents amyloid-β oligomerization and thus it might be a key molecule for prevention of cognitive impairment in aging and Alzheimer Disease.
Long-lasting, activity-dependent changes in synaptic efficacy are described as the cellular mechanisms underlying memory and learning (Pasinelli et al., 1995; Schjetnan and Escobar, 2010). Interestingly, RA signaling has been implicated in changes in both long-term potentiation (LTP) and long-term depression (LTD) of synaptic function in the hippocampus (Lane and Bailey, 2005). It has been also shown that knockout mice for RARβ and RARβ-RARγ display an alteration of LTP, as well as substantial performance deficits in a hippocampal-dependent spatial learning task (Chiang et al., 1998; Mey and McCaffery, 2004; Mao et al., 2006).
Learning and memory processes underlie animals’ predictive and behavioral adaptation to temporal fluctuations in the environment, such as light/dark period, food and water availability, social cues or risk of predation (Chaudhury et al., 2005; Golombek and Rosenstein, 2010). It is well known that the circadian clock synchronizes the phase of the activity/rest cycle, but also it has been shown to control complex physiology including thermoregulation, olfactory sensitivity, hormone secretion, glucose metabolism and even memory processes, among others, in most regions of the brain as well as in the periphery (Eckel-Mahan KL and Storm DR, 2009; Golombek and Rosenstein, 2010). In fact, it has been demonstrated that memory performances for associative learning oscillate in a circadian fashion across time, with high memory retention at multiples of 24 hours post-learning (Holloway and Wansley, 1973). More recently, time-of-day effects on learning and memory have been also observed in human, non-human primates and rats (Winocur and Hasher, 1999, 2002 and 2004; Valentinuzzi et al., 2008). The suprachiasmatic nucleus (SCN) is the anatomical centre that generates circadian rhythms and provides the necessary synchronization of other central nervous system and peripheral tissues (Hastings et al, 2008). A long time ago, Stephan and Kovacevic (1978) observed that SCN lesions impair hippocampus-dependent LTM in rodents, probably implicating the underlying role of the biological clock. However, more recently, Cain & Ralph (2009) showed that circadian modulation of conditioned place avoidance does not require the SCN in hamsters, which would indicate that memory for time of day may involve a circadian oscillator that is distinct from the SCN. Thus, the underlying mechanisms of time-of-day effects on learning and memory, including the possible involvement of the circadian system, have not been carefully investigated.
It has been demonstrated that most tissues show circadian oscillations and have their own cellular clock machinery in mammals (Balsalobre et al., 2000; Yamazaki et al., 2000; Hastings and Maywood, 2000). The heterodimeric basic helix-loop-helix-Per Arnt Sim (bHLH-PAS) transcription factor: BMAL1:CLOCK (from Brain and Muscle ARNT Like protein 1: Circadian Locomoter Output Cycles Kaput protein) drives the expression of clock (PER1, 2 and 3 and CRY1 and 2) and clock-controlled genes by its binding to E-box enhancers on target promoters. REVERB and ROR transcription factors, members of the retinoic acid-related orphan receptor (ROR) family, complete the molecular clock machinery. They bind to REVERB/ROR elements (RORE) on the promoter of target genes, with REVERBα or REVERBβ acting as transcriptional repressors and RORα or RORγ as transcriptional activators (Emery and Reppert, 2004). It has been shown that ROR acts coordinately with REVERB, and their competing activities on the same promoter element, drive the rhythm in BMAL1 transcription (Sato et al., 2004).
There are some evidences that retinoid nuclear receptors (RARs and RXRs) can regulate circadian gene expression, either by interacting with CLOCK and interfering with BMAL:CLOCK transcriptional activity, as seen in the vasculature (McNamara et al., 2001), either regulating at transcriptional level the expression of REVERBα and BMAL1, other key components in the clock machinery. On the other hand, it has been reported that the all-trans RA concentration vary systematically throughout the day in the serum of healthy people, rising to a maximum before lunch (Soderlund et al., 2002). Some other evidence points out that RA regulates the expression of two neuron-specific protein kinase C (PKC) substrates involved in synaptic plasticity: neuromodulin or GAP43 and neurogranin or RC3 (Husson et al., 2004; Iniguez et al., 1994).
Other molecular marker of neural plasticity associated to learning and memory is the brain-derived neurotrophic factor (BDNF). Daily rhythms of BDNF and its receptor TrkB have been demonstrated in different areas of the brain, hippocampus among them (Schaaf et al., 2000; Dolci et al., 2003). Day-night variations of BDNF expression suggest that this gene might be a target of the cellular clock.
Vitamin A deficiency (VAD), the most common micronutrient deficiency worldwide, is currently a risk for over 100 million school-aged children in over 75 countries. Moreover, approximately 127 million preschool-aged children and 7 million pregnant women are vitamin A deficient (Unicef, WHO, 1995, 2009; West KP, 2003). Given that memory and learning are of central importance to the efficient acquisition of cognitive, educational and social skills throughout childhood, it is perhaps not surprising that retinoid signaling has been shown to have an effect on these processes and that a role for vitamin A has been established in phenomena related to higher cognitive function (Misner et al., 2001; Cocco et al., 2002, Etchamendy et al., 2003, Husson et al., 2004). However, at this moment, at least in our knowledge, no report on the effects of the nutritional vitamin A deficiency on daily expression patterns of key cognition-related factors has been published.
Taking into account above background information, we wonder whether feeding a vitamin A-free diet could modify rhythmic endogenous clock activity and, consequently, alter temporal patterns of BDNF and RC3 expression in the hippocampus.
MATERIALS AND METHODS
Animal Model and Diet
Male Holtzman rats were bred in our animal facilities (National University of San Luis, Argentina), and maintained in a 21–23 °C controlled environment with a 12h-light:12h-dark (L:D) cycle. They were weaned at 21 days of age and immediately assigned randomly to either the experimental diet, devoided of vitamin A [vitamin A-deficient (DE) group] or the same diet with 4000 IU of vitamin A (8 mg retinol as retinyl palmitate) per Kg of diet [control (CO) group]. Feeding the animals with a vitamin A-free diet during three months, guarantees subclinical plasma retinol concentration and depleted retinol stores in liver (Anzulovich et al., 2000; Oliveros et al, 2000; Aguilar et al. 2009; Vega et al. 2009). Rats were given free access to food and water throughout the entire experimental period. All experiments were conducted in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals (NIH Publications No. 80-23) and the National University of San Luis Committee’s Guidelines for the Care and Use of Experimental Animals. Diets were prepared according to the AIN-93 for laboratory rodents (Reeves et al, 1993). Both, vitamin A-deficient and control diets had the following composition (g/kg): 397.5 cornstarch, 100 sucrose, 132 dextrinized cornstarch, 200 lactalbumin, 70 soybean oil, 50 cellulose fiber, 35 AIN-93 mineral mix, 10 AIN-93 vitamin mix (devoided of vitamin A for the vitamin A-deficient diet), 3 L-cystine, 2.5 choline bitartrate, and 0,014 tert-butylhydroquinone. After the treatment period, four rats from each group were sacrificed at different time points throughout a 24-h period. Those time points are referred to as zeigeber times (ZT) with ZT=0 when animal room light is on. Hippocampi were removed on an ice-chilled plate, weighed and immediately placed in liquid nitrogen. All the experiments were repeated at least twice.
mRNA isolation and RT-PCR
Total RNA was extracted from three pools of two hippocampi each. Hippocampus samples were isolated every 4 or 5 h starting at ZT2, from control and vitamin A-deficient groups. All RNA isolations were performed using the Trizol reagent (Invitrogen Co) as directed by the manufacturers. Gel electrophoresis and ethidium bromide staining confirmed the integrity of the samples. Quantification of RNA was based on spectrophotometric analysis at 260 nm. 3 μg of total RNA were reverse-transcribed with 200 units of MMLV Reverse Transcriptase (Promega Inc.) using random hexamers in a 25 μl reaction mixture and following manufacturer’s instructions. Transcript levels of BMAL1, PER1, RORα, REVERB, BDNF and RC3 were determined by RT-PCR and normalized to β-ACTIN as endogenous control. Fragments coding for those genes were amplified by PCR in 50 μl of reaction solution containing 0.2 mM dNTPs, 1.5 mM MgCl2, 1.25 U of Taq polymerase, 50 pmol of each rat specific oligonucleotide primer and RT-generated cDNA (1/5 of RT reaction). The sequences of the specific primers are shown on Table 1. For RC3 cDNA amplification, samples were heated in a thermalcycler (My Cycler, BioRad) at 94°C during 4 min followed by 28 cycles of: (1) denaturation, 94 °C for 45 seg; (2) annealing, 59°C during 45 seg; (3) extension, 72°C for 45 seg; After 28 reaction cycles, the extension reaction was continued for another 10 min. In the case of BDNF, samples were heated under the following conditions: 94°C during 2 min, followed by 35 cycles of: (1) denaturation, 94°C for 1 min; (2) annealing, 59°C during for 1 min; (3) extension, 72°C for 1 min. After 35 reaction cycles, the extension reaction was continued for another 5 minutes. For clock BMAL1, PER1, RORα and REVERB genes the thermalcycling conditions were similar but following 40 cycles of denaturation-annealing-extension. PCR products were then electrophoresed on 2,5% (w/v) agarose gel with 0.01% (w/v) ethidium bromide. The amplified fragments were visualized under ultraviolet (UV) transillumination and photographed using a Cannon PowerShot A75 3.2MP digital camera. The mean of gray value for each band was measured using the NIH ImageJ software (Image Processing and Analysis in Java from http://rsb.info.nih.gov/ij/) and the relative abundance of each band was normalized according to the housekeeping β-ACTIN gene, calculated as the ratio of the mean of gray value of each product to that of β-ACTIN.
Table 1.
Primer pairs used for RT-PCR (clock and Rc3 and Bdnf genes) and Real Time PCR (retinoid receptors genes).
| Gene name | GenBank Accession No | Forward primer 5′-3′ | Reverse primer 5′-3′ | Fragment size |
|---|---|---|---|---|
| RARα | NM_031528 | CGCCTGTGAGGGCTGTAAG | ATGCCCACTTCGAAGCATTT | 150 bp |
| RXRβ | NM_206849 | CGAAGCTCAGGCAAGCACTA | TCCTGTACCGCCTCCCTTTT | 200 bp |
| Bmal1 | AB012600 | GGGAAATACGGGTGAAGTCTATGG | ATGCCTGGAAGAGTGGGATGAGTC | 460 bp |
| Per1 | AB092976 | ATTCCGCCTAACCCCATATGT | GTGTGCCGTGTGGTGAAGAT | 150 bp |
| Rorα | XM_001056334 | GAGACAAATCGTCAGGAATCCAT | CCACAGCCAGGCACTTCTG | 180 bp |
| RevErb | AY336126 | GGTGCCTAGAATCCTGATTGTGA | TCCGCTGGAGCCAATGTAG | 200 bp |
| Rc3 | NM_024140 | GCCAGACGACGATATTCTAGACATC | CACACTCTCCACTCTTTATCTTCTTCCT | 122 bp |
| Bdnf | NM_012513 | GGGTCACAGCGGCAGATAAA | CGATTGGGTAGTTCGGCATT | 200 bp |
Real Time PCR
Relative quantification of RARα and RXRβ mRNA levels was performed by Real-Time PCR using the ABI Prism® 7500 thermocycler (Applied Biosystems, USA) as described previously (Fonzo et al., 2009). Briefly, cDNA obtained by RT-PCR was diluted to 20ng/μl with nuclease-free water. The diluted cDNA (5 μl) was amplified in a 25 μl final volume reaction mix containing 1X SYBR Green I fluorescent dye (Applied Biosystems, USA) and 500 nM gene-specific primers (Table 1). Reactions were subjected to one step of 95°C for 5 min followed by 40 cycles of 95°C for 15 sec and 60°C for 1 min. Relative expression of the real-time PCR products was determined by the ΔΔCt method. In this case, β-ACTIN was chosen as housekeeping gene and one of the control samples as calibrator. Each sample was run in triplicate, and the mean Ct was used in the ΔΔCt equation. Data for the normalized transcript levels of RARα and RXRβ are shown as means ± S.E.M.
Immunobloting
Protein extracts were prepared from three pools of two hippocampi each, obtained from each group of rats at the different ZTs during a 24-h period, in buffer C (20 mM HEPES, pH 7.9, 0.42 M NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 0.5 mM dithiothreitol, 0.5 mM phenylmethylsulfonyl fluoride, 1 μg/ml leupeptin, 1 μg/ml of pepstatin, 1 mM sodium fluoride, 5 μM sodium orthovanadate, and 25% glycerol). Aliquots containing 40 μg of total protein were subjected to electrophoresis in 4–12% NuPageTM Bis-Tris gels (Invitrogen Life Technologies, Carlsbad, CA), and then transferred to Immobilon-PTM transfer membranes (Millipore, Bedford, MA). Immunoblot analyses were performed as described in the manufacturers’ protocols for the detecting antibodies. Briefly, membranes were blocked in Blotto (5% nonfat dry milk, 10 mM Tris-HCl, pH 8.0, and 150 mM NaCl) followed by 12h overnight incubation at 10°C with either goat anti-RORα, anti-REVERB and anti-ACTIN antibodies (Santa Cruz Biotechnology, Santa Cruz, CA) in Blotto containing 0.05% thimerosal. After incubation with primary antibody, the membranes were washed in TBS (10 mM Tris-HCl, pH 8.0, and 150 mM NaCl) containing 0.05% Tween-20, before incubation with horseradish-peroxide-conjugated donkey anti-goat IgG (Santa Cruz Biotechnology, Santa Cruz, CA) diluted 1:10,000 in Blotto for one and a half hour at room temperature. After washing, antibody/protein complexes on membranes were detected using Vectastain DAB Peroxidase Substrate kit from Vector Laboratories (Burlingame, CA) and following the manufacturers indications. The mean of intensity of each band was measured using the NIH ImageJ software (Image Processing and Analysis in Java from http://rsb.info.nih.gov/ij/). RORα and REVERB protein levels were normalized against ACTIN (endogenous control).
Scanning of clock and clock-controlled genes upstream regions for putative RARE, RXRE, E-box and RORE sites
To identify putative RAR, RXR, E-box and/or ROR DNA consensus regulatory sites, BMAL1 (NCBI Gene ID # 29657), PER1 (NCBI Gene ID # 287422), RORa (NCBI Gene ID # 300807), REVERB (NCBI Gene ID # 252917), BDNF (NCBI Gene ID # 24225) and RC3 (NCBI Gene ID # 64356) gene regulatory regions, up to 1500 bp upstream of the translation start codon, were scanned for significant matches using the MatInspector software from Genomatix (http://www.genomatix.de, Quandt et al., 1995).
Statistical Analysis
Time point data were expressed as means ± standard errors of the mean (SE) and pertinent curves were drawn. Time series were computed by one-way ANOVA followed by Tukey’s post-hoc test for specific comparisons. A P<0.05 was considered to be significant. When mesor, amplitude or phase was required, a fitting technique was applied. Data were fitted by the following function: c + a cos[2π(t – ø)/24], where c is the mesor, a is the amplitude of the cosine wave, t is time in hours, and ø is the phase in hours from ZT 0 in the imposed light cycle. The fitting was performed using Nonlinear Regression from GraphPad Prism 3.0 software (CA, USA). The routine also estimates the standard error of the fit parameters. The standard error arises from scatter in the data and from deviations of the data from cosine form. Note that the frequency was taken as the 1 cycle per 24 h of the light regime.
RESULTS
RARα and RXRβ expression levels in the hippocampus of vitamin A deficient rats
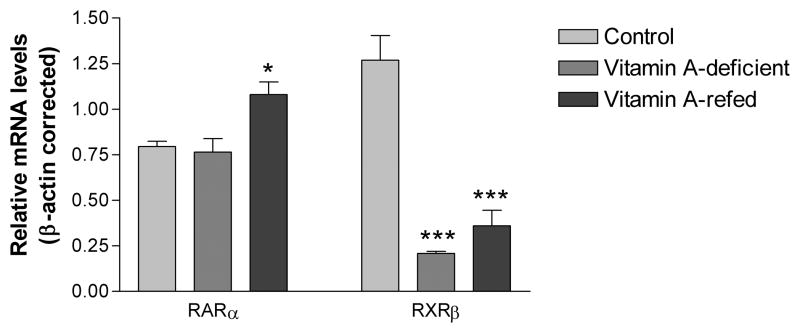
As we previously showed in Fonzo et al. (2009) vitamin A deficiency decreased the RXRβ transcript level (P<0.001), but had no effect on the RARα mRNA expression in the hippocampus (Figure 1).
Figure 1.
Transcript levels of RARα and RXRβ in the hippocampus of control and vitamin A-deficient rats. Basal mRNA levels were determined by Real-Time PCR and normalized to β-actin. Each bar represents the mean ± SE of 4 samples in triplicates with ***P<0.001 in comparison to controls.
Putative RARE and/or RXRE sites on clock genes upstream region
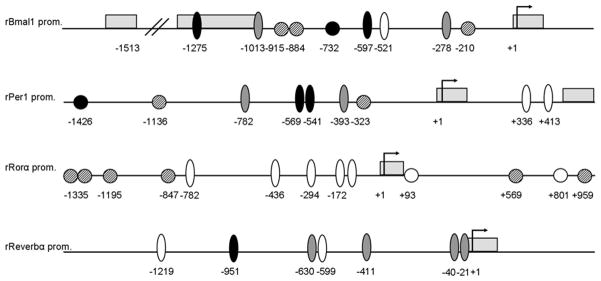
Looking for putative RA responsive sites on clock genes regulatory regions, we scanned 1500 bp upstream of the translation start codon of BMAL1, PER1, RORα and REVERB genes in the MatInspector database from Genomatix. The search revealed one RARE (AGGTCANNNNNAGGTCA) and three RXREs (AGGTCANAGGTCA) on the BMAL1 regulatory region, one RARE and two RXREs on PER1, and four RXREs on RORα 5′ upstream region (Figure 2). Additionally, two RAREs and two RXREs sites were found on the first intron of RORα gene. Interestingly, we also found two RORα responsive elements (RORE, (A/G)GGTCA preceded by 6 pb A/T) on the BMAL1 and PER1 genes and four ROREs on REVERBα promoter (Figure 2).
Figure 2.
Schematic representation of RARE, RXRE, E-box and RORE sites on the 5’ regulatory region of BMAL1, PER1, RORα and REVERB genes. The Gene ID # for the sequences taken from the NCBI Gene database are: rBMAL1 (NCBI Gene ID # 29657), rPER1 (NCBI Gene ID # 287422), rRORa (NCBI Gene ID # 300807) and rREVERB (NCBI Gene ID # 252917). Arrows indicate the first translation codon, gray boxes represent exons, dashed circles are RXRE sites, black circles are RAREs, black ovals represent perfect E-boxes, white ovals Ebox-like elements and grey ovals are RORE sites. Negative (−) numbers indicate regulatory sites positions relative to the start of translation (+1).
Daily rhythms of clock genes expression in the hippocampus of vitamin A-deficient rats
Clock genes BMAL1, PER1, RORα and REVERB, mRNA expression oscillates during a 24h period in the rat hippocampus (P<0.05, P<0.05, P<0.01 and P<0.001, respectively; Figure 3A–E). Knowing the presence of RA responsive sites on BMAL1, PER1 and RORα genes’ regulatory region, we continued to test to which extent vitamin A deficiency could affect the daily expression of key clock factors. Feeding rats with a vitamin A-free diet phase shifted BMAL1 mRNA peak from ZT 15:30±00:10 to ZT 06:20±00:10 (P<0.05; Figure 3A and Table 2) and abolished daily rhythmicity of PER1 transcript levels in comparison to controls (P=0.72; Figure 3B). On the other hand, VAD modified the acrophase of RORα oscillating expression (ZT21:08 ± 01:20 vs ZT07:48 ± 00:04, P<0.01; Figure 3C and Table 2) while it attenuated daily rhythmicity of REVERBα mRNA (P=0.45; Figure 3D).
Figure 3.
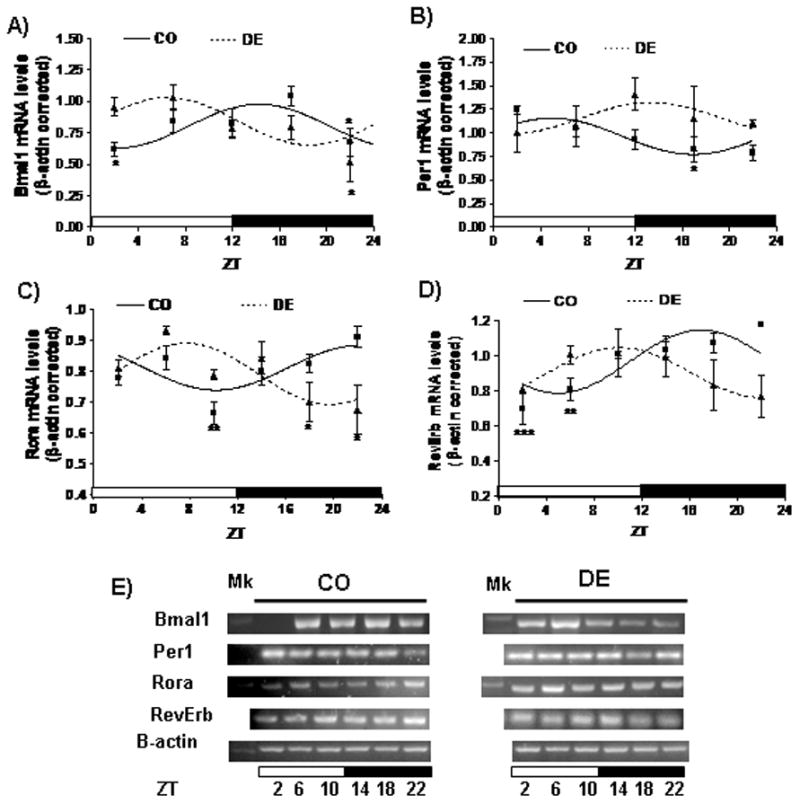
Daily rhythms of clock genes expression in the hippocampus of control and vitamin A-deficient rats. (A–D) Cosine fitting curves for normalized BMAL1 (A), PER1 (B), RORα (C) and REVERB (D) mRNA levels throughout a day. Horizontal bars represent the distribution of light (open) and dark (closed) phases of a 24-h (ZT0-ZT24) photoperiod. Each point on the curves represents the mean ± SE of three pools of two hippocampus samples each at a given ZT (with ZT=0 when light is on). Significant daily variation was evaluated using one-way ANOVA followed by Tukey test with *P<0.05, **P<0.01 and ***P<0.001 when indicated means were compared to the corresponding maximal value in each group. (E) Representative patterns of PCR products at different ZTs throughout a day-night cycle.
Table 2.
Rhythm’s parameters of daily Bmal1, Per1, Rorα and Reverb mRNA levels in the hippocampus of control and vitamin A-deficient rats.
| Gene name | Mesor (mean ± SE) | Amplitude (mean ± SE) | Acrophase (hh:mm; mean ± SE) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| CO | DE | P | CO | DE | P | CO | DE | P | |
| Bmal1 | 0.78 ± 0.0 | 0.83 ± 0.4 | ns | 0.17 ± 0.0 | 0.21 ± 0.0 | ns | 15:30 ± 00:10 | 06:20 ± 00:10 | <0,05 |
| Per1 | 0.90 ± 0.0 | NA | - | 0.14 ± 0.0 | NA | - | 05:70 ± 00:20 | NA | - |
| Rorα | 0.81 ± 0.02 | 0.76 ± 0.01 | ns | 0.08 ± 0.01 | 0.13 ± 0.04 | ns | 21:08 ± 01:20 | 07:48 ± 00:04 | <0,01 |
| Reverb | 0.97 ± 0.04 | NA | - | 0.18 ± 0.01 | NA | - | 16:56 ± 00:05 | - | |
Note: NA means “not apply” since daily mRNA levels became arrhythmic.
Daily rhythms of clock proteins expression in the hippocampus of vitamin A-deficient rats
Previously, we observed BMAL1 and PER1 proteins oscillate throughout a day in the rat hippocampus. Amplitude of BMAL1 and PER1 rhythms was reduced in the hippocampus of vitamin A-deficient animals in comparison to controls (Fonzo et al, 2009). Additionally, here, we analyzed the daily patterns of other molecular clock components, RORα and REVERB protein levels, in the hippocampus of control and vitamin A-deficient rats. We observed RORα and REV-ERB levels vary significantly throughout a day (P<0.01 and P<0.05, respectively) in the rat hippocampus, with maximal protein levels occurring at ZT 14:21±00:06 and ZT 03:08±02:12, respectively (Figure 4A–C, Table 3). Three months of vitamin A deficiency, exerted differential effects on clock proteins rhythms. In the case of these BMAL1 expression regulators, VAD phase shifted RORα protein rhythmicity (acrophase: ZT 14:21±00:06 vs ZT 18:48±00:08, P<0.01, Figure 4A and Table 3) and reduced REVERB oscillation’s mesor and amplitude (P<0.01 and P<0.02, respectively; Figure 4B and Table 3).
Figure 4.
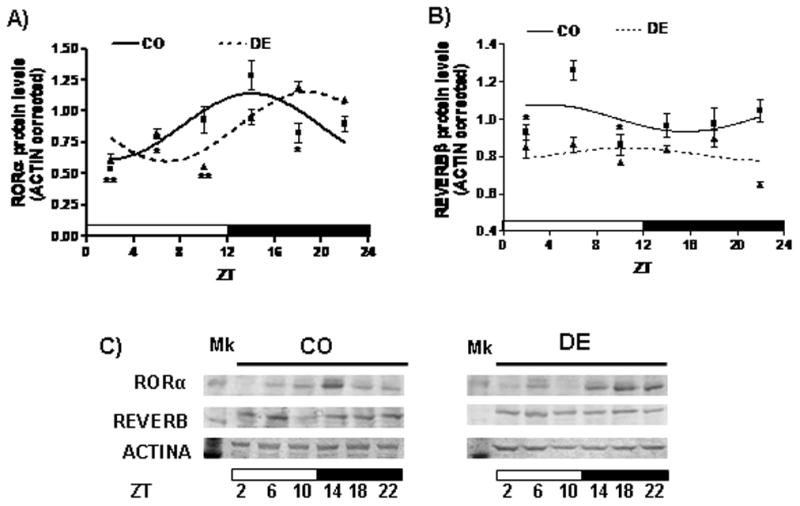
Daily rhythms of RORα and REVERB protein levels in the rat hippocampus. (A–B) Cosine fitting curves for normalized RORα (A) and REVERB (B) protein levels throughout a day, obtained from the densitometric quantitation of the Immunoblots. Horizontal bars represent the distribution of light (open) and dark (closed) phases of a 24-h (ZT0-ZT24) photoperiod. Each value on the curves represents the mean ± SE of three pools of two hippocampus samples each at a given ZT (with ZT=0 when light is on). Significant daily variation was evaluated using one-way ANOVA followed by Tukey test with *P<0.05 and **P<0.01 when compared indicated means with the corresponding maximal value in each group. (C) Representative Immunoblot analysis of protein extracted from CO and DE rat hippocampi isolated at ZT2, ZT6, ZT10, ZT14, ZT18, and ZT22.
Table 3.
Rhythm’s parameters of daily RORa and REVERB proteins levels in the hippocampus of control and vitamin A-deficient rats.
| Protein name | Mesor (mean ± SE) | Amplitude (mean ± SE) | Acrophase (hh:mm; mean ± SE) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| CO | DE | P | CO | DE | P | CO | DE | P | |
| RORα | 0.88 ± 0.06 | 0.87 ± 0.05 | ns | 0.26 ± 0.05 | 0.27 ± 0.00 | ns | 14:21 ± 00:06 | 18:48 ± 00:08 | <0.01 |
| REVERB | 1.00 ± 0.03 | 0.81 ± 0.02 | <0.01 | 0.14 ± 0.02 | 0.06 ± 0.00 | <0.02 | 03:08 ± 02:12 | 12:27 ± 01:24 | ns |
Note: ns means “not significant”.
Putative E-box and RARE sites on BDNF and RC3 genes upstream region
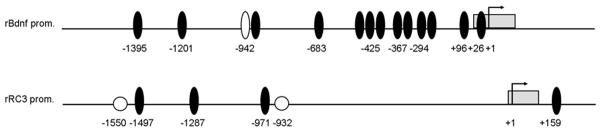
Once we had knowledge that clock proteins rhythms were altered in the hippocampus of vitamin A-deficient rats, we wondered whether key molecular factors involved in memory and learning could be under the endogenous clock control and have clock responsive, E-box, and/or RA responsive, RARE, sites on their gene promoters. Scanning of 1500 bp upstream of the translation start codon of BDNF and RC3 genes in the Genomatix database, revealed thirteen perfect, CACGTG, E-box and one, CANNTG, Ebox-like sites in the BDNF regulatory region while three perfect E-box elements and two RAREs (AGGTCANNNNNAGGTCA) were found on the RC3 gene upstream region (Figure 5).
Figure 5.
Schematic representation of E-box and RARE sites on the 5′ regulatory region of the BDNF and RC3 genes. Genes ID # are: 24225 and 64356 for the rBDNF and rRC3 sequences taken from the NCBI database, respectively. Arrow indicates the first translation codon, gray box represents first exon, black ovals are perfect E-boxes, the white oval is an E-box-like and white circles represent RARE elements. Negative (−) numbers indicate clock-and retinoic acid-responsive sites positions relative to the start of translation (+1).
Daily BDNF and RC3 expression in the rat hippocampus
We found RC3 mRNA expression displays a 24-h rhythm in the rat hippocampus (P<0.05, Figure 6A and C). Daily RC3 mRNA levels peak at ZT 23:25±00:03 in the control rats (Figure 6A and Table 4). As it has already been reported (Schaaf et al., 2000), BDNF expression also display a robust daily rhythmicity in the hippocampus of our control rats (P<0.01) peaking at ZT 00:21± 00:30 (Figure 6B and Table 4). VAD phase shifted RC3 oscillating expression (acrophase: ZT 23:25±00:03 vs ZT 03:56±01:13, P<0,001) without affecting the amplitude or mesor parameters (Figure 6A and Table 4). Similarly, BDNF maximal expression was delayed to ZT 08:24±00:50 (P<0.002, Figure 6B and Table 4) in the vitamin A-deficient animals.
Figure 6.
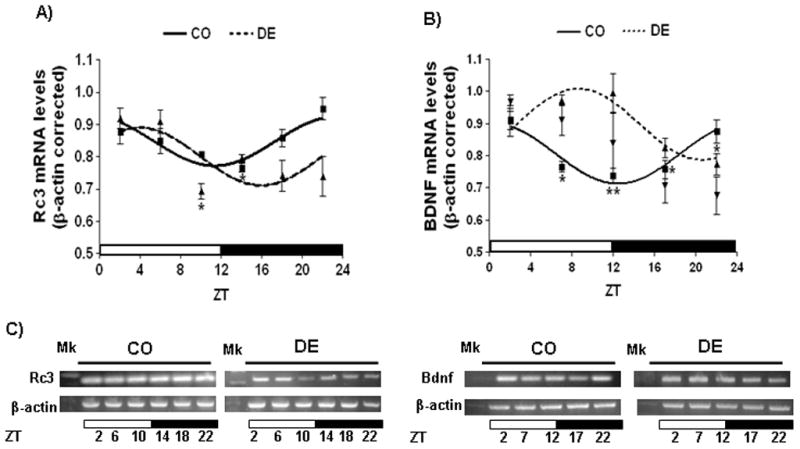
Daily RC3 and BDNF expression in the hippocampus of control and vitamin A-deficient rats. (A–B) Cosine fitting curves for normalized RC3 (A) and BDNF (B) mRNA levels throughout a day. Horizontal bars represent the distribution of light (open) and dark (closed) phases of a 24-h (ZT0-ZT24) photoperiod. Each point on the curves represents the mean ± SE of three pools of two hippocampus samples each at a given ZT (with ZT=0 when light is on). Significant daily variation was evaluated using one-way ANOVA followed by Tukey test with *P<0.05 and **P<0.01 when indicated means were compared to the corresponding maximal value in each group. (C) Representative patterns of PCR products at different ZTs throughout a day-night cycle.
Table 4.
Rhythm’s parameters of daily Bdnf and Rc3 mRNA levels in the hippocampus of control and vitamin A-deficient rats.
| Gene name | Mesor (mean ± SE) | Amplitude (mean ± SE) | Acrophase (hh:mm; mean ± SE) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| CO | DE | P | CO | DE | P | CO | DE | P | |
| Rc3 | 0,80 ± 0,02 | 0,80 ± 0,00 | ns | 0.07 ± 0,02 | 0.10 ± 0.01 | ns | 23:25 ± 00:03 | 03:56 ± 01:13 | <0.001 |
| Bdnf | 0.81 ± 0.01 | 0.90 ± 0,01 | <0.05 | 0.10 ± 0.02 | 0.12 ± 0.02 | ns | 00:21 ± 00:30 | 08:24 ± 00:50 | <0.002 |
Note: ns means “not significant”.
DISCUSSION
Differences between day and night in the expression of gene transcripts have been observed in the hippocampus, cerebral cortex and cerebellum (Cirelli et al. 2004). In the cerebral cortex, about 10% (1,564) of 15,459 transcribed sequences, are differentially expressed between day and night (Katoh-Semba et al., 2008). A disturbance of the light-induced phase shift is known to induce neuronal degeneration in the rat brain, and also in the human brain, as it is evident by the atrophy of the temporal cortex resulting from chronic jet lag (Cho, 2001). This indicates the importance of normal oscillation of some transcript levels to maintain normal physiological activity.
Micronutrients, such as vitamin A, might be essential to maintain the circadian expression of clock and clock-controlled genes in peripheral oscillators such as the hippocampus. Here we observed, for the first time at our knowledge, that a nutritional vitamin deficiency modifies daily rhythmicity of endogenous molecular clock and cognition-related clock-controlled factors in the brain. Previously, we observed VAD dampened rats’ daily locomotor activity rhythm and modified daily oscillation of BMAL1 and PER1 protein levels in the rat hippocampus and liver (Fonzo et al., 2009; Ponce et al., 2011) which indicate a putative role for vitamin A in the regulation of the endogenous clock activity. Here, we additionally study the temporal expression of BMAL1 regulators, RORα and REVERBα, at the mRNA and protein levels, as well as oscillating BMAL1 and PER1 transcripts levels and daily BDNF and RC3 expression in the hippocampus.
As expected, and similar to Feillet et al. (2008) and Wang et al. (2009), we observed BMAL1 and PER1 transcript levels vary throughout a 24h period, peaking at the beginning of the night and at the first half of the day, respectively, in the hippocampus (Figure 3). Even though there are many reports that associate vitamin deficiencies with altered daily expression patterns, just a very few report the effect of nutritional deficiencies on circadian clock gene expression; only two of them determine the effect of vitamin A deficiency on the oscillating BMAL1 and PER1 or 2 expression in the liver (Shirai et al., 2006; Ponce et al., 2011). It is known that the central clock in the SCN controls the daily activity-rest cycles via a direct route (Schibler and Sassone-Corsi 2002). It is well established that the hippocampus is critically involved in memory formation and that synaptic connections within the hippocampus are known to undergo activity-dependent changes in synaptic strength, referred to as long-term potentiation (LTP, Chaudhury et al., 2005). Noteworthy, we previously observed VAD dampened rats’ daily locomotor activity without affecting the phase of the activity/rest rhythm (Ponce et al., 2011). VAD also reduced the amplitude of BMAL1 and PER1 protein rhythms in the rat hippocampus (Fonzo et al., 2009). Interestingly, we found one RARE and three RXREs on the BMAL1 regulatory region and one RARE and two RXRE sites on the PER1 promoter. This would probably indicate these clock genes expression might be susceptible to the VAD. In agreement with recently results observed in liver (Ponce et al., 2011) in the present work, VAD modified BMAL1 and PER1 expression at transcriptional level in the hippocampus. The nutritional deficiency phase shifted circadian BMAL1 mRNA rhythm and dampened daily PER1 mRNA and protein levels (Figure 3A and B, Table 2). Several studies suggest an association between cellular molecular clock disruptions and impaired cognitive functions (Garcia et al., 2000; Abarca et al., 2002; Hampp et al., 2008; Jilg et al., 2009). Changes in BMAL1 and PER1 mRNA rhythms could probably be explained by changes in the daily expression patterns of transcriptional regulators RORα and/or REVERB.
ROR and REVERB expression has been documented in the rat hippocampus and related to synapses formation and developmental plasticity (Peña-de-Ortiz and Jamieson 1997; Paganoni et al., 2010). Interestingly, and for the first time at our knowledge, we show RORα and REVERB proteins and transcripts oscillate in a daily pattern in the rat hippocampus (Figures 3C and D and 4A and B). It has been demonstrated these two factors regulate BMAL1 rhythmic expression and at the same time RORα and REVERB genes are target of the BMAL1:CLOCK transcription factor in the mammalian circadian clock (Preitner et al., 2002; Emery and Reppert, 2004; Gillaumond et al., 2005). In this study, peaks of REVERB and RORα mRNA levels occur at the middle and at the end of the night, respectively, following BMAL1 expression acrophase (Figures 3A, C and D). As seen by Preitner et al. (2002), our MatInspector analysis revealed the presence of ROR/REVERB responsive sites on the BMAL1 gene regulatory region. As expected, we observed BMAL1 mRNA peak (ZT 15:30 ± 00:10) follows maximal levels of activating RORα protein (ZT 14:21 ± 00:06) while REVERB acrophase (ZT 03:08 ± 02:12) concurs with the nadir in the oscillating BMAL1 expression. Others also reported the presence of RORE sites in other clock core factors such as CRY and the CLOCK gene homologous, NPAS2, and demonstrated RORα and REV-ERBα coordinately regulate not only BMAL1 expression but also other molecular clock’s components (Etchegaray et al., 2003; Ueda et al., 2005; Crumbley et al., 2010). In fact, we also found RORE sites in the PER1 and REVERB genes promoter (Figure 2). The last is also an example of a very well coordinated circadian regulation observed in this study. Thus, we observed REVERB mRNA rhythm acrophase (ZT 16:56 ± 00:05) follows RORα protein peak (ZT14:21 ± 00:06) and precedes its own protein maximal expression (ZT 03:08 ± 02:12). Consistently, REVERB transcription is repressed at the beginning of the day by its own protein daily peak. All above observations would suggest a putative reciprocal control between core clock factors and the retinoic acid-related orphan receptors (RORs) family in the hippocampus.
As in Fonzo et al. (2009) here, we observed VAD significantly reduced the levels of RXRβ in the rat hippocampus. This is consistent with ours and others previous work (Etchamendy et al., 2003; Mao et al., 2006; Aguilar et al. 2009; Vega et al. 2009; Ponce et al., 2011) who demonstrated nutritional VAD reduced RARβ, RXRα and/or RXRβ expression in different tissues of rat and mouse. It has been shown that RORa-mediated transcription is activated synergistically by RAR in Pukinje cerebellar cells (Matsui, 1997), evidencing a functional interaction between RORα and RAR transcription factors. RARs operate as heterodimers with RXRs (Soprano et al., 2004). In these studies, vitamin A deficiency had no effect on the RARα mRNA expression but significantly reduced the RXRβ transcript level (Figure 1). Taking into account the presence of RXR responsive sites on clock BMAL1, PER1 and RORα genes upstream region (Figure 2), the lower availability of RXRβ might affect some necessary transcriptional interactions to maintain rhythmic expression patterns of clock and consequently, clock-controlled genes; for example, affecting either RXR heterodimerization, either RORα mediated transcriptional activation of clock (BMAL1) or clock-controlled (RC3 and/or BDNF) cycling genes in the hippocampus of vitamin A-deficient animals.
Since BMAL1 is a RORα target gene (Akashi and Takumi, 2005), changes in the RORα rhythm acrophase would be responsible for the phase shifting in the BMAL1 expression pattern observed in the vitamin A-deficient rats. On the other hand, attenuation of BMAL1 protein rhythmicity observed by Fonzo et al. (2009) would explain dampened RevErb mRNA and protein expression observed in this study in the VAD (Figures 3D and 4B).
At this point, we observed VAD desynchronizes the molecular clock machinery in the rat hippocampus. It has been reported the circadian system can influence the daily performance of cognitive functions (Valentinuzzi et al., 2004 and 2008; Chaudhury and Colwell, 2002; Eckel-Mahan et al., 2008) and a variety of studies suggest that learning and memory processes are sensitive to disruptions in sleep and circadian rhythms (Peigneux et al., 2004; Ellenbogen et al., 2006; Ruby et al., 2008; Jilg et al., 2009). For example, mutations in at least some of the genes responsible for the generation of circadian oscillations can alter learning in mice (Garcia et al., 2000; Abarca et al., 2002; Hampp et al., 2008; Jilg et al., 2009).
All above observations led us to question whether the clock alterations observed in the VAD could affect the temporal organization of hippocampal cognitive functions at molecular level. Thus, looking for some putative clock-controlled genes involved in cognition processes, we continue to study the effect of VAD on the day/night expression of BDNF and RC3, two key molecular factors involved in synaptic plasticity underlying memory and learning processes. First, we recognize the presence of clock-, retinoid- and/or ROR/REVERB-responsive sites (Ebox, RXREs and ROREs, respectively) in their regulatory regions (Figure 5). Second, we analyzed their daily mRNA expression in the hippocampus of control and vitamin A-deficient groups.
Diurnal rhythms on BDNF mRNA and protein levels have been reported in the cerebellum, hippocampus and cerebral cortex (Berchtold et al., 1999; Pollock et al., 2001; Katoh-Semba et al., 2008). Consistently, herein we found not only BDNF but also RC3 gene display rhythmic expression patterns in the rat hippocampus (Figure 6A and B). As expected, they are in phase with circadian BMAL1 and PER1 protein levels reported in Fonzo et al. (2009) with a maximal RC3 mRNA expression occurring at the end of the night and preceding, as in the known cellular events occurs for postsynaptic activation, BDNF mRNA peak at ZT 00:21 ± 00:30. Given that it takes more than 6 (six) hours for the protein level to increase following mRNA peak (Katoh-Semba et al., 2008) we expect hippocampal BDNF-related synaptic plasticity to be maximal during the day (rest period for rats) participating in memory-related processes during sleep. These observations lead us to propose the existence of a very well temporally orchestrated RC3 and BDNF cycle underlying temporal patterns of synaptic plasticity in the hippocampus.
Even though it is difficult to link cellular/molecular research to behavioral endpoints, we would like to discuss the possible functional significance of VAD effects on daily rhythms of clock and clock-controlled synaptic-plasticity-related genes, in the hippocampus. Among other functions, the hippocampus has been clearly linked to memory formation, especially declarative memories (i.e., the encoding, storage, and recall of memories for specific events) (Chaudhury et al., 2005). Particularly, hippocampal BDNF is involved in formation, fixation and recall of episode memory, as revealed by decreased levels in Alzheimer’s cases with memory dysfunction (Phillips et al. 1991). Thus, it is conceivable that daily rhythms of BDNF and RC3 underlie daily rhythms in hippocampal-dependent learned behaviors. More broadly, there is evidence that the time of day is an aspect of the physical environment that can be learned much in the same way that an organism can learn spatial parameters (Chaudhury et al., 2005). A number of studies have documented the ability of organisms to learn to go to specific locations at specific times of the day (Daan, 2000). These learned associations of time and place may not involve the SCN-based circadian system (as seen by Cain and Ralph, 2009) but would be expected to be dependent on an intact clock in the hippocampus. The results presented here show that the expression of memory-and-learning-related genes is in phase with daily patterns of key molecular clock factors, however, such temporal organization would be susceptible to the nutritional vitamin A deficiency. Even though, it has been reported vitamin A, as well as other vitamin deficiencies, reduce BDNF and RC3 expression in the hippocampus (Husson et al., 2004, Sable et al., 2011), no study describes the consequences of nutritional deficiencies on temporal profiles of synaptic-plasticity-related genes. Here, we observed BDNF and RC3 oscillating expression was phase shifted and, in the case of BDNF, also attenuated, in the hippocampus of vitamin A-deficient rats. Probably, these changes are consequence of alterations seen in the daily patterns of key molecular clock factors. Thus, our results suggest nutritional VAD might affect the daily cognitive performance by disrupting daily rhythms of molecular clock and, consequently, temporal profiles of clock-controlled cognition-related factors in the hippocampus. However, we can not omit to discuss other putative effects of VAD. For instance, it is known VAD affects the retina (night blindness), the conjunctiva (conjunctival xerosis, with or without Bitot spots), and the cornea (corneal xerosis) (Diniz Ada and Santos, 2000; McBain et al., 2007). The diurnal changes in environmental illumination are conveyed from the retina to the brain to entrain circadian rhythms throughout the body. In sighted people, light triggers several changes in retinal neurochemistry that are transmitted via the retinohypothalamic tract to the SCN (Golombek and Rosenstein, 2010). In particular, retinal ganglion cells containing a vitamin Abased chromophore, melanopsin, play a role in photic synchronization and it has been observed that targeted destruction of photoreceptive ganglion cells attenuated circadian responses to light (Goz et al., 2008). Contrary to expectations, Thompson et al. (2001) demonstrated that light signaling to the SCN is preserved in vitamin A-deficient mice, as determined by PER gene photoinduction in SCN cells. However, as discussed above and similar to the observed by Goz and collaborators (2008), we have shown that feeding animals with a vitamin A-free diet dampened their daily motor activity without affecting its day-night pattern, which is naturally synchronized to the nocturnal phase in most rodents (Ponce et al., 2011). Probably, integrity of light signaling pathway is not completely affected by three months of feeding a vitamin A-free diet or, other possibility is that, in the presence of less efficient retinal photoreceptors, other non-photic stimuli are entraining peripheral clocks, such as the hippocampus, in the vitamin A-deficient rats. The last possibility would explain that rhythms of BMAL1, RORα, BDNF and RC3, although phase shifted, remain in the hippocampus of vitamin A-deficient animals. On the other hand, changes in the pineal and eye indoleamine metabolism have been observed in vitamin A-deficient rats and birds (Herbert and Reiter, 1985; Fu et al., 2001). Since melatonin is also a circadian signal which synchronizes several physiological functions and, as a number of animal studies demonstrated, effects of melatonin have been seen in the hippocampus, particularly suggesting involvement in synaptic plasticity (Pandi-Perumal et al., 2008), the effects of VAD reported here could be also a consequence of alterations in melatonin rhythm. Thus, VAD could affect the circadian timing system at different levels such as retina, SCN, pineal and/or directly at the peripheral clocks, such as the hippocampus. The last would be supported by the significant reduction in the retinoic acid receptor, RXRβ, mRNA levels and changes in the daily expression of RXR and RAR target genes, such as BMAL1, PER1, RORα and RC3, observed in the hippocampus of vitamin A deficient-animals.
Above observations raise the possibility that nutritional factors and food processing products, such as vitamin A and its derivatives the retinoids, might modulate daily patterns of BDNF and RC3 expression in the hippocampus and thus, vitamin A could be essential to maintain an optimal daily performance at molecular level in this learning-and-memory-related brain area.
Learning how vitamin A deficiency affects the circadian expression of genes involved synaptic plasticity may have an impact on the neuro and chronobiology fields, emphasizing the importance of nutritional factors such as dietary micronutrients in the regulation of circadian parameters in brain. While numerous epidemiological and experimental studies have provided strong evidence that in both humans and animals, a lack or excess of vitamin A is harmful to the embryo, the present studies will strengthen the hypothesis that a deregulation of retinoid-mediated molecular events may be a potential factor for circadian cognitive deterioration indicating that sufficient levels of vitamin A are also fundamental for the maintenance of mature brain function and a daily cognitive performance. VAD is a serious concern and has a clinical and socio-economical significance worldwide. We would expect that the emerging data from this study will also highlight retinoid signaling pathways as potential novel therapeutic targets for cognitive deficits and/or clock-related disorders.
Acknowledgments
This work was supported by NIH Res. Grant # R01-TW006974 funded by the Fogarty International Center, National Institutes of Health (USA). We thank Dr. Ana Rastrilla and LABIR (UNSL) for providing us with Holtzman rats. We acknowledge Mr Mario Quiroga for his assistance in making diets and taking care of animals. The dextrinized cornstarch was generously provided by Productos de Maíz SRL (Bragado, BA, Argentina).
Abbreviations
- 9cisRA
9-cis-retinoic acid
- ANOVA
analysis of variance
- ATRA
all-trans-retinoic acid
- BDNF
brain-derived neurothrophic factor
- RAR
retinoic acid receptor
- RARE
retinoic acid responsive element
- RC3
neurogranin
- RORE
retinoid-related orphan receptor responsive element
- RXR
retinoid X receptor
- RXRE
retinoid X responsive element
- SCN
suprachiasmatic nucleus
- ZT
zeitgeber time
References
- Abarca C, Albrecht U, Spanagel R. Cocaine sensitization and reward are under the influence of circadian genes and rhythm. Proc Natl Acad Sci U S A. 2002;99:9026–9030. doi: 10.1073/pnas.142039099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilar RP, Genta S, Oliveros L, Anzulovich A, Gimenez MS, Sanchez S. Vitamin A deficiency injures liver parenchyma and alters the expression of hepatic extracellular matrix. J Appl Toxicol. 2009;29:214–222. doi: 10.1002/jat.1399. [DOI] [PubMed] [Google Scholar]
- Anzulovich AC, Oliveros LB, Muñoz EM, Martinez LD, Gimenez MS. Nutritional vitamin A deficiency alters antioxidant defenses and modifies the liver histoarchitecture in rat. J Tr Elem Exp Med. 2000;13:343–357. [Google Scholar]
- Akashi M, Takumi T. The orphan nuclear receptor RORalpha regulates circadian transcription of the mammalian core-clock Bmal1. Nat Struct Mol Biol. 2005;12:441–448. doi: 10.1038/nsmb925. [DOI] [PubMed] [Google Scholar]
- Balsalobre A, Brown SA, Marcacci L, Tronche F, Kellendonk C, Reichardt HM, Schutz G, Schibler U. Resetting of circadian time in peripheral tissues by glucocorticoid signaling. Science. 2000;289:2344–2347. doi: 10.1126/science.289.5488.2344. [DOI] [PubMed] [Google Scholar]
- Berchtold NC, Oliff HS, Isackson P, Cotman CW. Hippocampal BDNF mRNA shows a diurnal regulation, primarily in the exon III transcript. Brain Res Mol Brain Res. 1999;71:11–22. doi: 10.1016/s0169-328x(99)00137-0. [DOI] [PubMed] [Google Scholar]
- Cain SW, Ralph MR. Circadian modulation of conditioned place avoidance in hamsters does not require the suprachiasmatic nucleus. Neurobiol Learn Mem. 2009;91:81–84. doi: 10.1016/j.nlm.2008.10.005. [DOI] [PubMed] [Google Scholar]
- Chaudhury D, Colwell CS. Circadian modulation of learning and memory in fear-conditioned mice. Behav Brain Res. 2002;133:95–108. doi: 10.1016/s0166-4328(01)00471-5. [DOI] [PubMed] [Google Scholar]
- Chaudhury D, Wang LM, Colwell CS. Circadian Regulation of Hippocampal Long-Term Potentiation. J Biol Rhythm. 2005;20:225–236. doi: 10.1177/0748730405276352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang MY, Misner D, Kempermann G, Schikorski T, Giguere V, Sucov HM, Gage FH, Stevens CF, Evans RM. An Essential Role for Retinoid Receptors RARb and RXRg In Long-Term Potentiation and Depression. Neuron. 1998;21:1353–1361. doi: 10.1016/s0896-6273(00)80654-6. [DOI] [PubMed] [Google Scholar]
- Cho K. Chronic ‘jet lag’ produces temporal lobe atrophy and spatial cognitive deficits. Nat Neurosci. 2001;4:567–568. doi: 10.1038/88384. [DOI] [PubMed] [Google Scholar]
- Cirelli C, Gutierrez CM, Tononi G. Extensive and divergent effects of sleep and wakefulness on brain gene expression. Neuron. 2004;41:35–43. doi: 10.1016/s0896-6273(03)00814-6. [DOI] [PubMed] [Google Scholar]
- Cocco S, Diaz G, Stancampiano R, Diana A, Carta M, Curreli R, Sarais L, Fadda F. Vitamin A deficiency produces spatial learning and memory impairment in rats. Neuroscience. 2002;115:475–482. doi: 10.1016/s0306-4522(02)00423-2. [DOI] [PubMed] [Google Scholar]
- Crumbley C, Wang Y, Kojetin DJ, Burris TP. Characterization of the core mammalian clock component, NPAS2, as a REV-ERBalpha/RORalpha target gene. J Biol Chem. 2010;285:35386–35392. doi: 10.1074/jbc.M110.129288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daan S. Learning and Circadian Behavior. J Biol Rhythms. 2000;4:296–299. doi: 10.1177/074873000129001396. [DOI] [PubMed] [Google Scholar]
- Diniz Ada S, Santos LM. Vitamin A deficiency and xerophtalmia. J Pediatr (Rio J) 2000;76:S311–322. doi: 10.2223/jped.169. [DOI] [PubMed] [Google Scholar]
- Dolci C, Montaruli A, Roveda E, Barajon I, Vizzotto L, Grassi Zucconi G, Carandente F. Circadian variations in expression of the trkB receptor in adult rat hippocampus. Brain Res. 2003;994:67–72. doi: 10.1016/j.brainres.2003.09.018. [DOI] [PubMed] [Google Scholar]
- Eckel-Mahan KL, Phan T, Han S, Wang H, Chan GC, Scheiner ZS, Storm DR. Circadian oscillation of hippocampal MAPK activity and cAmp: implications for memory persistence. Nat Neurosci. 2008;11:1074–1082. doi: 10.1038/nn.2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckel-Mahan KL, Storm DR. Circadian rhythms and memory: not so simple as cogs and gears. EMBO reports. 2009;10:584–591. doi: 10.1038/embor.2009.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellenbogen JM, Payne JD, Stickgold R. The role of sleep in declarative memory consolidation: passive, permissive, active or none? Curr Opin Neurobiol. 2006;16:716–722. doi: 10.1016/j.conb.2006.10.006. [DOI] [PubMed] [Google Scholar]
- Emery P, Reppert SM. A rhythmic Ror. Neuron. 2004;43:443–446. doi: 10.1016/j.neuron.2004.08.009. [DOI] [PubMed] [Google Scholar]
- Etchamendy N, Enderlin V, Marighetto A, Pallet V, Higueret P, Jaffard R. Vitamin A deficiency and relational memory deficit in adult mice: relationships with changes in brain retinoid signaling. Behav Brain Res. 2003;145:37–49. doi: 10.1016/s0166-4328(03)00099-8. [DOI] [PubMed] [Google Scholar]
- Etchamendy N, Enderlin V, Marighetto A, Vouinba RM, Pallet V, Jaffard R, Higueret P. Alleviation of a selective age-related relational memory deficit in mice by pharmacologically induced normalization of brain retinoid signaling. J Neurosc. 2001;21:6423–6429. doi: 10.1523/JNEUROSCI.21-16-06423.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etchegaray JP, Lee C, Wade PA, Reppert SM. Rhythmic histone acetylation underlies transcription in the mammalian circadian clock. Nature. 2003;421:177–182. doi: 10.1038/nature01314. [DOI] [PubMed] [Google Scholar]
- Feillet CA, Mendoza J, Albrecht U, Pevet P, Challet E. Forebrain oscillators ticking with different clock hands. Mol Cell Neurosci. 2008;37:209–221. doi: 10.1016/j.mcn.2007.09.010. [DOI] [PubMed] [Google Scholar]
- Fonzo LS, Golini RS, Delgado SM, Ponce IT, Bonomi MR, Rezza IG, Gimenez MS, Anzulovich AC. Temporal patterns of lipoperoxidation and antioxidant enzymes are modified in the hippocampus of vitamin A-deficient rats. Hippocampus. 2009;19:869–880. doi: 10.1002/hipo.20571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Z, Kato H, Kotera N, Noguchi T, Sugahara K, Kubo T. Regulation of hydroxyindole-O-methyltransferase gene expression in Japanese quail (Coturnix coturnix japonica) Biosci Biotechnol Biochem. 2001;65:2504–2511. doi: 10.1271/bbb.65.2504. [DOI] [PubMed] [Google Scholar]
- Garcia JA, Zhang D, Estill SJ, Michnoff C, Rutter J, Reick M, Scott K, Diaz-Arrastia R, McKnight SL. Impaired cued and contextual memory in NPAS2-deficient mice. Science. 2000;288:2226–2230. doi: 10.1126/science.288.5474.2226. [DOI] [PubMed] [Google Scholar]
- Golombek DA, Rosenstein RE. Physiology of Circadian Entrainment. Physiol Rev. 2010;90:1063–1102. doi: 10.1152/physrev.00009.2009. [DOI] [PubMed] [Google Scholar]
- Göz D, Studholme K, Lappi DA, Rollag MD, Provencio I, Morin LP. Targeted destruction of photosensitive retinal ganglion cells with a saporin conjugate alters the effects of light on mouse circadian rhythms. PLoS One. 2008;3:e3153. doi: 10.1371/journal.pone.0003153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillaumond F, Dardente H, Giguère V, Cermakian N. Differential control of Bmal1 circadian transcription by REV-ERB and ROR nuclear receptors. J Biol Rhythms. 2005;20:391–403. doi: 10.1177/0748730405277232. [DOI] [PubMed] [Google Scholar]
- Hastings M, Maywood ES. Circadian clocks in the mammalian. Brain Bioessays. 2000;22:23–31. doi: 10.1002/(SICI)1521-1878(200001)22:1<23::AID-BIES6>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- Hastings MH, Maywood ES, Reddy AB. Two decades of circadian time. J Neuroendocrinol. 2008;20:812–819. doi: 10.1111/j.1365-2826.2008.01715.x. [DOI] [PubMed] [Google Scholar]
- Hampp G, Albrecht U. The circadian clock and mood-related behavior. Commun Integr Biol. 2008;1:1–3. doi: 10.4161/cib.1.1.6286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbert DC, Reiter RJ. Changes in pineal indoleamine metabolism in vitamin A deficient rats. Life Sci. 1985;37:2515–2522. doi: 10.1016/0024-3205(85)90609-5. [DOI] [PubMed] [Google Scholar]
- Holloway FA, Wansley RA. Behav Biol. 1973;9:1–14. doi: 10.1016/s0091-6773(73)80164-6. [DOI] [PubMed] [Google Scholar]
- Husson M, Enderlin V, Alfos S, Boucheron C, Pallet V, Higueret P. Expression of neurogranin and neuromodulin is affected in the striatum of vitamin A-deprived rats. Brain Res Mol Brain Res. 2004;123:7–17. doi: 10.1016/j.molbrainres.2003.12.012. [DOI] [PubMed] [Google Scholar]
- Iñiguez MA, Morte B, Rodriguez-Peña A, Muñoz A, Gerendasy D, Sutcliffe JG, Bernal J. Characterization of the promoter region and flanking sequences of the neuron-specific gene RC3 (neurogranin) Brain Res Mol Brain Res. 1994;27:205–214. doi: 10.1016/0169-328x(94)90002-7. [DOI] [PubMed] [Google Scholar]
- Jilg A, Lesny S, Peruzki N, Schwegler H, Selbach O, Dehghani F, Stehle JH. Temporal dynamics of mouse hippocampal clock gene expression support memory processing. Hippocampus. 2010;20(3):377–388. doi: 10.1002/hipo.20637. [DOI] [PubMed] [Google Scholar]
- Katoh-Semba R, Tsuzuki M, Miyazaki N, Matsuda M, Nakagawa C, Ichisaka S, Sudo K, Kitajima S, Hamatake M, Hata Y, Nagata K. A phase advance of the light-dark cycle stimulates production of BDNF, but not of other neurotrophins, in the adult rat cerebral cortex: association with the activation of CREB. J Neurochem. 2008;106:2131–2142. doi: 10.1111/j.1471-4159.2008.05565.x. [DOI] [PubMed] [Google Scholar]
- Lane MA, Bailey SJ. Role of retinoid signalling in the adult brain. Prog Neurobiol. 2005;75:275–293. doi: 10.1016/j.pneurobio.2005.03.002. [DOI] [PubMed] [Google Scholar]
- Mao CT, Li TY, Qu P, Zhao Y, Wang R, Liu YX. Effects of early intervention on learning and memory in young rats of marginal vitamin A deficiency and it’s mechanism. Zhonghua Er Ke Za Zhi. 2006;44:15–20. [PubMed] [Google Scholar]
- Matsui T. Transcriptional regulation of a Purkinje cell-specific gene through a functional interaction between RORa and RAR. Genes Cells. 1997;2:263–272. doi: 10.1111/j.1365-2443.1997.119gc0317.x. [DOI] [PubMed] [Google Scholar]
- McBain VA, Egan CA, Pieris SJ, Supramaniam G, Webster AR, Bird AC, Holder GE. Functional observations in vitamin A deficiency: diagnosis and time course of recovery. Eye (Lond) 2007;21:367–376. doi: 10.1038/sj.eye.6702212. [DOI] [PubMed] [Google Scholar]
- McNamara P, Seo SP, Rudic RD, Sehgal A, Chakravarti D, FitzGerald GA. Regulation of CLOCK and MOP4 by nuclear hormone receptors in the vasculature: a humoral mechanism to reset a peripheral clock. Cell. 2001;105:877–889. doi: 10.1016/s0092-8674(01)00401-9. [DOI] [PubMed] [Google Scholar]
- Mey J, McCaffery P. Retinoic acid signaling in the nervous system of adult vertebrates. Neuroscientist. 2004;10:409–421. doi: 10.1177/1073858404263520. [DOI] [PubMed] [Google Scholar]
- Misner DL, Jacobs S, Shimizu Y, de Urquiza AM, Solomin L, Perlmann T, De Luca LM, Stevens CF, Evans RM. Vitamin A deprivation results in reversible loss of hippocampal long-term synaptic plasticity. Proc Natl Acad Sci U S A. 2001;98:11714–11719. doi: 10.1073/pnas.191369798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveros L, Vega V, Anzulovich A, Ramirez D, Gimenez MS. Vitamin A deficiency modifies antioxidant defenses and essential element contents in rat heart. Nutr Res. 2000;20:1139–1150. [Google Scholar]
- Paganoni S, Bernstein J, Ferreira A. Ror1-Ror2 complexes modulate synapse formation in hippocampal neurons. Neuroscience. 2010;165:1261–1274. doi: 10.1016/j.neuroscience.2009.11.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandi-Perumal SR, Trakht I, Srinivasan V, Spence DW, Maestroni GJ, Zisapel N, Cardinali DP. Physiological effects of melatonin: role of melatonin receptors and signal transduction pathways. Prog Neurobiol. 2008;85:335–353. doi: 10.1016/j.pneurobio.2008.04.001. [DOI] [PubMed] [Google Scholar]
- Pasinelli P, Ramakers GM, Urban IJ, Hens JJ, Oestreicher AB, de Graan PN, Gispen WH. Long-term potentiation and synaptic protein phosphorylation. Behav Brain Res. 1995;66:53–59. doi: 10.1016/0166-4328(94)00124-x. [DOI] [PubMed] [Google Scholar]
- Peigneux P, Laureys S, Fuchs S, Collette F, Perrin F, Reggers J, Phillips C, Degueldre C, Del Fiore G, Aerts J, Luxen A, Maquet P. Are spatial memories strengthened in the human hippocampus during slow wave sleep? Neuron. 2004;44:535–545. doi: 10.1016/j.neuron.2004.10.007. [DOI] [PubMed] [Google Scholar]
- Peña-de-Ortiz S, Jamieson GA., Jr Molecular cloning and brain localization of HZF-2 alpha, a new member of the Rev-erb subfamily of orphan nuclear receptors. J Neurobiol. 1997;32:341–358. doi: 10.1002/(sici)1097-4695(199703)32:3<341::aid-neu7>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- Phillips HS, Hains JM, Armanini M, Laramee GR, Johnson SA, Winslow JW. BDNF mRNA is decreased in the hippocampus of individuals with Alzheimer’s disease. Neuron. 1991;7:695–702. doi: 10.1016/0896-6273(91)90273-3. [DOI] [PubMed] [Google Scholar]
- Pollock GS, Vernon E, Forbes ME, Yan Q, Ma YT, Hsieh T, Robichon R, Frost DO, Johnson JE. Effects of early visual experience and diurnal rhythms on BDNF mRNA and protein levels in the visual system, hippocampus, and cerebellum. J Neurosci. 2001;21:3923–3931. doi: 10.1523/JNEUROSCI.21-11-03923.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponce IT, Rezza IG, Delgado SM, Navigatore LS, Bonomi MR, Golini RL, Gimenez MS, Anzulovich AC. Daily oscillation of glutathione redox cycle is dampened in the nutritional vitamin A deficiency. Accepted in Biol Rhythm Res. 2011 doi: 10.1080/09291016.2011.593847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preitner N, Damiola F, Lopez-Molina L, Zakany J, Duboule D, Albrecht U, Schibler U. The orphan nuclear receptor REV-ERBalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell. 2002;110:251–260. doi: 10.1016/s0092-8674(02)00825-5. [DOI] [PubMed] [Google Scholar]
- Quandt K, Frech KK, Karas H, Wingender E, Werner T. MatInd and MatInspector: new fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res. 1995;23:4878–4884. doi: 10.1093/nar/23.23.4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves PG, Nielsen FH, Fahey GC., Jr AIN-93 purified diets for laboratory rodents: final report of the American Institute of Nutrition ad hoc writing committee on the reformulation of the AIN-76A rodent diet. J Nutr. 1993;123:1939–1951. doi: 10.1093/jn/123.11.1939. [DOI] [PubMed] [Google Scholar]
- Ruby NF, Hwang CE, Wessells C, Fernandez F, Zhang P, Sapolsky R, Heller HC. Hippocampal-dependent learning requires a functional circadian system. Proc Natl Acad Sci USA. 2008;105:15593–15598. doi: 10.1073/pnas.0808259105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sable P, Dangat K, Kale A, Joshi S. Altered brain neurotrophins at birth: consequence of imbalance in maternal folic acid and vitamin B(12) metabolism. Neuroscience. 2011 doi: 10.1016/j.neuroscience.2011.05.010. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- Sato TK, Panda S, Miraglia LJ, Reyes TM, Rudic RD, McNamara P, Naik KA, FitzGerald GA, Kay SA, Hogenesch JB. A functional genomics strategy reveals Rora as a component of the mammalian circadian clock. Neuron. 2004;43:527–537. doi: 10.1016/j.neuron.2004.07.018. [DOI] [PubMed] [Google Scholar]
- Schaaf MJ, Duurland R, de Kloet ER, Vreugdenhil E. Circadian variation in BDNF mRNA expression in the rat hippocampus. Brain Res Mol Brain Res. 2000;75:342–354. doi: 10.1016/s0169-328x(99)00314-9. [DOI] [PubMed] [Google Scholar]
- Schjetnan AG, Escobar ML. In vivo BDNF modulation of hippocampal mossy fiber plasticity induced by high frequency stimulation. Hippocampus. 2010 doi: 10.1002/hipo.20866. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- Shirai H, Oishi K, Ishida N. Circadian expression of clock genes is maintained in the liver of Vitamin A-deficient mice. Neurosci Lett. 2006;398:69–72. doi: 10.1016/j.neulet.2005.12.055. [DOI] [PubMed] [Google Scholar]
- Söderlund MB, Sjöberg A, Svärd G, Fex G, Nilsson-Ehle P. Biological variation of retinoids in man. Scand J Clin Lab Invest. 2002;62:511–519. doi: 10.1080/003655102321004521. [DOI] [PubMed] [Google Scholar]
- Stephan FK, Kovacevic NS. Multiple retention deficit in passive avoidance in rats is eliminated by suprachiasmatic lesions. Behav Biol. 1978;22:456–462. doi: 10.1016/s0091-6773(78)92565-8. [DOI] [PubMed] [Google Scholar]
- Takasaki J, Ono K, Yoshiike Y, Hirohata M, Ikeda T, Morinaga A, Takashima A, Yamada M. Vitamin A has Anti-Oligomerization Effects on Amyloid-β In Vitro. J Alzheimers Dis. 2011 doi: 10.3233/JAD-2011-110455. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- Ueda HR, Hayashi S, Chen WB, Sano M, Machida M, Shigeyoshi Y, Iino M, Hashimoto S. System level identification of transcriptional circuits underlying mammalian circadian clocks. Nat Genet. 2005;37:187–192. doi: 10.1038/ng1504. [DOI] [PubMed] [Google Scholar]
- Valentinuzzi VS, Menna-Barreto L, Xavier GF. Effect of circadian phase on performance of rats in the Morris water maze task. J Biol Rhythms. 2004;19:312–324. doi: 10.1177/0748730404265688. [DOI] [PubMed] [Google Scholar]
- Valentinuzzi VS, Neto SP, Carneiro BT, Santana KS, Araújo JF, Ralph MR. Memory for time of training modulates performance on a place conditioning task in marmosets. Neurobiol Learn Mem. 2008;89:604–607. doi: 10.1016/j.nlm.2007.08.002. [DOI] [PubMed] [Google Scholar]
- Vega VA, Anzulovich AC, Varas SM, Bonomi MR, Gimenez MS, Oliveros LB. Effect of nutritional vitamin A deficiency on lipid metabolism in the rat heart: its relation to PPAR gene expression. Nutrition. 2009;25(7–8):828–838. doi: 10.1016/j.nut.2009.01.008. [DOI] [PubMed] [Google Scholar]
- Wang LM, Dragich JM, Kudo T, Odom IH, Welsh DK, O’Dell TJ, Colwell CS. Expression of the circadian clock gene Period2 in the hippocampus: possible implications for synaptic plasticity and learned behaviour. ASN Neuro1. 2009;(3):e00012. doi: 10.1042/AN20090020. pii. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner EA, Deluca HF. Retinoic acid is detected at relatively high levels in the CNS of adult rats. Am J Physiol Endocrinol Metab. 2002;282:E672–678. doi: 10.1152/ajpendo.00280.2001. [DOI] [PubMed] [Google Scholar]
- WHO. Document WHO/NUT/95.1 available from the Nutritional Program. Geneva, Switzerland: 1995. WHO global prevalence of Vitamin A deficiency. MDIS working paper #2. [Google Scholar]
- WHO. WHO Global Database on Vitamin A Deficiency. Geneva: World Health Organization; 2009. Global prevalence of vitamin A deficiency in populations at risk 1995–2005. [Google Scholar]
- Winocur G, Hasher L. Aging and time-of-day effects on cognition in rats. Behav Neurosci. 1999;113:991–997. doi: 10.1037//0735-7044.113.5.991. [DOI] [PubMed] [Google Scholar]
- Winocur G, Hasher L. Circadian rhythms and memory in aged humans and animals. In: Squire LR, Schacter DL, editors. Neuropsychology of memory. 3. New York: Guilford Press; 2002. pp. 273–285. [Google Scholar]
- Winocur G, Hasher L. Age and time-of-day effects on learning and memory in a non-matching-to-sample test. Neurobiol Aging. 2004;25:1107–1115. doi: 10.1016/j.neurobiolaging.2003.10.005. [DOI] [PubMed] [Google Scholar]
- Yamazaki S, Numano R, Abe M, Hida A, Takahashi R, Ueda M, Block GD, Sakaki Y, Menaker M, Tei H. Resetting central and peripheral circadian oscillators in transgenic rats. Science. 2000;288:682–685. doi: 10.1126/science.288.5466.682. [DOI] [PubMed] [Google Scholar]
- Zetterström RH, Simon A, Giacobini MM, Eriksson U, Olson L. Localization of cellular retinoid-binding proteins suggests specific roles for retinoids in the adult central nervous system. Neuroscience. 1994;62:899–918. doi: 10.1016/0306-4522(94)90482-0. [DOI] [PubMed] [Google Scholar]
- Zetterström RH, Lindqvist E, Mata de Urquiza A, Tomac A, Eriksson U, Perlmann T, Olson L. Role of retinoids in the CNS: differential expression of retinoid binding proteins and receptors and evidence for presence of retinoic acid. Eur J Neurosci. 1999;11:407–416. doi: 10.1046/j.1460-9568.1999.00444.x. [DOI] [PubMed] [Google Scholar]