Abstract
In vivo imaging of transplanted hematopoietic stem and progenitor cells (HSPCs) was developed to investigate the relationship between HSPCs and components of their microenvironment in the bone marrow. In particular, it allows a direct observation of the behavior of hematopoietic cells during the first few days after transplantation, when the critical events in homing and early engraftment are occurring. By directly imaging these events in living animals, this method permits a detailed assessment of functions previously evaluated by crude assessments of cell counts (homing) or after prolonged periods (engraftment). This protocol offers a new means of investigating the role of cell-intrinsic and cell-extrinsic molecular regulators of hematopoiesis during the early stages of transplantation, and it is the first to allow the study of cell-cell interactions within the bone marrow in three dimensions and in real time. In this paper, we describe how to isolate, label and inject HSPCs, as well as how to perform calvarium intravital microscopy and analyze the resulting images. A typical experiment can be performed and analyzed in ~1 week.
INTRODUCTION
Development of the protocol
Hematopoietic stem cells (HSCs) are responsible for the maintenance of blood and immune cell turnover, both in physiological conditions and in response to injury. They accomplish this because of a tight balance between quiescence, self-renewal and differentiation. The mechanisms regulating HSC fate are a combination of cell-intrinsic and cell-extrinsic molecular signals, most of which are still unknown. In particular, the molecular and cellular components of the HSC microenvironment, or niche, are objects of intense study and the origin of numerous controversies. Knockout and transgenic mouse models with altered osteoblast numbers and function indicate that these bone-making cells are a major component of the HSC niche1–4; however, several other bone marrow stromal cells have been observed in the vicinity of HSCs, and the full nature of the HSC niche remains elusive5–11.
In vivo imaging of mouse calvarium was first performed by von Andrian and co-workers12, using fluorescence microscopy to detect various hematopoietic cell populations rolling within the bone marrow microvasculature. Following the same principles, we used confocal microscopy to observe homing of a leukemia cell line in proximity to calvarium bone marrow vasculature expressing high amounts of stromal cell-derived factor-1 (SDF-1)13. We then combined confocal and two-photon microscopy to simultaneously observe up to five different cellular and extracellular components in the same area (bone collagen was observed through second harmonic generation (SHG)14; osteoblasts through lineage-specific EGFP expression15; hematopoietic stem and progenitor cell (HSPC) populations through ex vivo DiD (1,1′-dioctadecyl-3,3,3′,3′-tetramethylindodicarbocyanine perchlorate) labeling; autofluorescent cells through confocal microscopy; and vasculature through injection of nontargeted quantum dots)16,17. We have been using calvarial sutures as reliable and consistent spatial reference points, allowing us to scan the entire bone marrow area without bias and, if needed, to revisit specific areas of interest multiple times16.
Applications of the method
The methodology described here is widely applicable to studying the effect of genetic or drug-induced alterations in either HSPCs or bone marrow microenvironment components during the early stages of HSPC transplantation. We have shown that Gαs-null bone marrow cells fail to engraft in transplant recipients because they are unable to exit circulation and localize in the bone marrow space18. Colmone et al.19 studied aberrant homing of human HSPCs injected into recipient mice burdened with leukemia. We describe here how to observe the spatial relationship between DiD-labeled HSPC and EGFP-expressing osteoblasts in col2.3-EGFP transplant recipient mice15; however, virtually any bone marrow cellular or extracellular component can be visualized by confocal/multiphoton intravital microscopy, provided that it is selectively labeled with sufficient intensity.
Comparisons with other methods
Numerous approaches have been developed to study HSPC localization within the bone marrow, starting from bone sectioning and the use of immunofluorescent techniques4,8,20. Confocal/two-photon intravital microscopy is the only one allowing single-cell resolution imaging in live animals16. A number of methodologies have been developed that allow a direct observation of femur bone marrow. Xie et al.21 used ex vivo femur epiphysis cultures and observed HSPC dynamics in the proximity of the bone resection while the tissue was still viable. Solutions allowing in vivo imaging of femur bone marrow include the drilling of the femoral cortex to a thickness penetrable by light22 or the use of endoscopic probes inserted in the knee area and moved toward the head of the femur23. Calvarium bone marrow imaging is minimally invasive, and even though skull and femur develop from distinct embryonic progenitor cells and through different developmental processes, in both bone types, HSPCs are present at similar frequencies and are functionally identical16.
Limitations
The primary limitation of confocal/multiphoton intravital microscopy is the penetration depth, which is currently ~200 µm. Calvarium bone marrow cavities are relatively small; however, we can only visualize approximately the upper half of the bone marrow space. We assume that the deeper half of calvarium bone marrow is structurally and functionally identical to the upper half.
We have proposed that observations made in the calvarium bone marrow might apply to the trabecular area of long bones; however, one has to be aware that the microenvironment of HSPCs located within long bone diaphyses is likely to be very different, for instance, because of the possibility that it may be located at a much greater distance from any endosteal surface. In general, it is impossible to extrapolate observations from calvarium bone marrow structures to other bone marrow areas.
Another limitation of the current imaging methodology is that, despite the calvarial sutures providing spatial reference points, tracking cell positions over long periods of time is inefficient, as the development of scar tissue between imaging sessions impairs further observation of sometimes wide areas. Moreover, little is known about the effects of irradiation, transplantation and even imaging itself on the calvarium bone and stroma, and further studies are necessary to develop highly reliable tracking methodology. Finally, the proliferative characteristics of injected HSPCs dictate the length of time for which they can be observed. Labeled HSPCs injected in non-irradiated recipients do not proliferate and can be detected weeks after transplantation16, whereas the same cells injected in irradiated recipients undergo cell division and dilute the label to undetectable levels within a few days (C.L.C. and C.P.L., unpublished observations).
Experimental design
Mice and conditioning
Before starting an in vivo imaging project, compliance with the relevant local guidelines and regulations for the use of vertebrate species needs to be ensured. Calvarium intravital microscopy experiments are generally classified as generating moderate discomfort and pain to the animals.
Donor and recipient mice need to be syngenic to ensure long-term HSPC engraftment. To observe functional, engrafting HSPCs following transplantation, the recipient mice need to be preconditioned in order to destroy resident hematopoietic cells and make the bone marrow niche permissive to engraftment. In our hands, the most effective and reliable method is a lethal dose of γ-irradiation. We used 9.5 Gy in a single dose or split between two doses of 4.75 Gy 3 h apart for C57/Black6 (C57/B6) mice; however, the most appropriate treatment may vary depending on the mouse strain used and the irradiator available. We therefore recommend titrating the optimal irradiation dose (9–11 Gy) and protocol by monitoring long-term peripheral blood chimerism in a pilot bone marrow transplantation experiment.
If an irradiator is not available, similar engraftment results can be obtained by conditioning the recipient mice with chemotherapeutic agents24.
Cell numbers
In principle, any bone marrow HSPC population can be analyzed with in vivo imaging. We describe here the harvesting and preparation of long-term repopulating HSPCs (LT-HSPCs) such as LKS (that is, Lineagelow, c-Kit+ and Sca-1+) CD34−Flk2− or LKS CD48−CD150+; refs. 17,25). In our experience, transplantation of ~10,000 LT-HSPCs leads to observation of ~10 cells homed to the calvarium bone marrow, and this amount of LT-HSPCs can be obtained from four adult C57/B6 donor mice. In our work, lineage depletion with magnetic columns has been highly reliable, although Dynabeads (Invitrogen) provide a reliable alternative.
Cell sorting
Several combinations of cell surface markers can be used to identify and isolate HSPC sub-populations. In our study, reliable in vivo data for LT-HSPCs were obtained when sorting LKS CD34−Flk2− or LKS CD48−CD150+ populations17,25. In principle, any other hematopoietic population can be used for in vivo imaging experiments and most fluorescence-activated cell sorting (FACS) instruments allow collection of multiple cell populations at the same time (multiway sorting). Similar to the imaging of multiple parameters, the cocktails of antibodies used for sorting need to be conjugated to fluorophores with appropriate excitation and emission spectra, in order to be easily distinguished. Examples of such antibodies and fluorophores that can be used to identify LT-HSPCs are provided in Table 1. Briefly, whole bone marrow cells are first labeled with a cocktail of biotin-conjugated antibodies against differentiation markers (see Table 1 ‘Lineage cocktail’); this is followed by incubation with streptavidin-coated magnetic beads. When flowing through columns placed inside a magnet, differentiated cells (labeled by the antibody cocktail) are retained in the column, whereas undifferentiated or little-differentiated cells flow through. We refer to the eluted cells as lineage-depleted bone marrow. In a second step, lineage-depleted cells are labeled with antibodies, allowing the identification of HSCs (see Table 1 ‘FACS’); together with fluorophore-conjugated streptavidin, this allows the elimination of the remaining Lineage-positive (i.e., differentiated) cells. When labeled cells flow through the FACS instrument, initial gates are drawn on the basis of the size and granularity of the cells to eliminate debris and cell doublets. A subsequent gate containing c-Kit–bright, Lineage-dim cells separates the Lineagelow cell population. Within this population, c-Kit+ Sca1+ cells are the so-called LKS population. Long-term repopulating HSCs are subsequently identified as CD34−Flk2− or CD150+CD48− cells. Further subgates can be used to obtain purer populations; however, in our study, they are unlikely to provide sufficient numbers of cells for in vivo imaging purposes.
Table 1.
Antibodies and fluorophores for HSPC harvesting.
| Antigen | Conjugate | Clone | Vendor | Cat. no. | Dilution |
|---|---|---|---|---|---|
| Lineage cocktail | |||||
| Gr1 | Biotin | RB6-8C5 | BD Biosciences | 553125 | —a |
| Mac1 | Biotin | M1/70 | BD Biosciences | 553309 | —a |
| B220 | Biotin | Ra3-6B2 | BD Biosciences | 553086 | —a |
| Ter119 | Biotin | Ter119 | BD Biosciences | 553672 | —a |
| CD4 | Biotin | GK1.5 | BD Biosciences | 553728 | —a |
| CD8 | Biotin | 53-6.7 | BD Biosciences | 553029 | —a |
| CD3 | Biotin | 145-2C11 | BD Biosciences | 553060 | —a |
| FACS | |||||
| c-Kit | APC | 2B8 | BioLegend | 105812 | 1:100 |
| Sca-1 | Pacific blue (PB) | D7 | BioLegend | 108120 | 1:100 |
| CD34 | FITC | RAM34 | eBioscience | 11-0341-85 | 1:100 |
| Flk2 | Phycoerythrin (PE) | A2F10.1 | BD Biosciences | 553542 | 1:200 |
| CD48 | FITC | HM48-1 | BioLegend | 103404 | 1:1000 |
| CD150 | PE | Tc15-12F12.2 | BioLegend | 115904 | 1:100 |
| Streptavidin | Pacific orange (PO) | Invitrogen | S32365 | 1:500 |
A selected combination of these markers and fluorophores can be used for HSPC sorting to obtain LKS CD34 −Flk2− or LKS CD150+CD48− cell populations. The ideal color assortment and supplier can vary depending on the cell sorter used. Note that streptavidin staining allows gating out the remaining lineage-positive cells during the sorting.
For dilution, see REAGENT SETUP and Step 5 of PROCEDURE.
For each sorting, it is essential to prepare not only the cell suspension stained with the cocktail of chosen antibodies but also a series of controls (compensation controls or single-color controls) necessary to arrange the correct compensation settings. To prepare compensation controls, some total bone marrow cells are aliquotted and stained individually with each fluorophore-conjugated antibody used in the FACS cocktail, and some Lineage-depleted cells are stained with fluorophore-conjugated streptavidin. Lineage-depleted cells are used as a single-color Lineage control because whole bone marrow cells would produce a brighter signal than the sorted sample, leading to unnecessary compensation. One further aliquot is left unstained. The unstained sample is used to set up the sorter voltage settings and each single-color control is run to set up the appropriate compensation settings (mathematical algorithms that eliminate ‘bleed through’ signal of one channel into another, so that a bright signal in one channel is not erroneously collected as a signal in a different channel)26. Further information on setting up the sorter voltage settings, compensations settings and carrying out the cell sorting can be found in refs25–27.
It is good practice to check the purity of the obtained cell population by occasionally analyzing some of the obtained cells (at least once per each sorter used). This should be done by running some of the obtained cells through the cell sorter to check what percentage of them falls again within the gates used for sorting. If they all do, then sorting purity is 100%. Because of the small numbers of LT-HSCs typically obtained, it may not possible to perform such analysis after every sort.
Fluorescent labeling
Particular care must be taken when choosing the label for imaging cells. Fluorescent signals need to be very intense in order to be detected efficiently through bone. DiD and other similar lipophilic dyes (e.g., Invitrogen Vybrant labeling solutions) are nontoxic to mouse HSPCs and provide uniform bright labeling16. They were initially developed to visualize membrane dynamics; depending on the cell type and the staining protocol used, it has been reported that the membrane distribution of the dyes can vary28. The labeling needs to be optimized for each cell type of interest in order to select the most efficient and least toxic reagent and protocol. Carboxyfluorescein succinimidyl ester (CFSE), PKH and quantum dot-based dyes are valid alternatives to the lipophilic dyes29.
Transgenic reporter strains are extremely valuable to highlight specific bone marrow compartments or hematopoietic lineages30,31. Strong promoters should be chosen to ensure efficient imaging of fluorescent reporters expressed at the highest levels. Lower fluorescence might still permit some detection, however, albeit with a high risk of missing part of the cells or structures of interest (C.L.C., unpublished observation).
We have visualized bone marrow vasculature by injecting Qtracker 800 (nontargeted quantum dots 800, Invitrogen). Depending on each specific experimental setup, other circulating fluorophores (e.g., Qtracker 655, Angiosense (VisEn) or fluorescently labeled dextran)12,16,32 can be valid alternatives.
Our typical setup allows the observation of (i) bone through the SHG signal of collagen14, one of the main components of calcified bone; (ii) osteoblasts by means of high EGFP expression characterizing the col2.3-EGFP reporter strain15; (iii) transplanted cells by brightly labeling them with the lipophilic dye DiD; and (iv) vasculature by injecting nontargeted quantum dots 800 (ref. 16). As all fluorophores are distributed along the electromagnetic spectrum, both for their absorption and emission, a window is left to analyze autofluorescent signal using the 532-nm laser and a 560- to 640-nm emission filter. Autofluorescent cells are present in high numbers in the bone marrow; however, their identity is unknown. They produce signals of identical shape and similar intensity in both the DiD and the ‘autofluorescence’ channel, and sometimes also in the EGFP channel. Because they appear in all channels, whereas labeling fluorophores have a distinctive excitation-emission spectrum, autofluorescent cells are easily identified and excluded from further analysis. We work with ×25 or ×30 water immersion objectives, using the combination of lasers and filters described in Table 2.
Table 2.
Combination of lasers and emission filters used for five-parameter acquisition.
| Observed component |
Excitation laser/ wavelength (nm) |
Emission filter (nm) |
|---|---|---|
| Bone collagen (SHG) | MaiTai titanium sapphire/840 | 400–500 |
| GFP | Argon/491 | 505–590 |
| Autofluorescence | He-Ne/532 | 560–640 |
| DiD | He-Ne/633 | 650–760 |
| Qtracker 800 | He-Ne/532 | > 795 |
Microscopy sessions
In vivo imaging experiments usually require some troubleshooting, and therefore, we recommend imaging no more than one or two mice in each experiment, at least initially. Mouse and microscope setup may require careful and detailed optimization, often for each imaging session, and results might not be comparable if the last mouse is imaged many hours (e.g., >8 h) after the first one. It is possible to delay the injection of cells; however, keeping primary hematopoietic cells ex vivo can be detrimental to their function and viability. We have observed highly reproducible results when injecting the same cell population in experimental replicates on separate days; therefore, we recommend obtaining statistically significant data through smaller repeated experiments rather than larger, less consistent ones.
Image analysis
Several software packages can be used for the final image analysis, from the open source ImageJ, to Adobe Photoshop, to more complex 3D-oriented packages. To date, we have been unable to identify a software package that allows full automation of the process while generating reliable positional data. We recommend working with ImageJ, as it is a fully open-source software platform, therefore ensuring complete control over image processing and avoiding any risk of artifact generation.
Generally, high-quality images are the easiest to analyze; however, it is not always possible to acquire such images as a result of fluctuations in microscope performance and because of the nature of the tissue surrounding the cells of interest. Moreover, better-quality images inevitably require longer exposure of the tissue to the laser, thus increasing the likelihood of damaging the tissue. It is therefore important to establish an efficient imaging routine, compromising between speed of acquisition and image quality, so that the necessary information can be obtained while causing as little damage as possible to the tissue.
The intensity of the signals collected through intravital microscopy depends not only on the brightness of the label or fluorophore used but also on the composition of the surrounding tissue. For this reason, we have not taken into account discrepancies in signal levels and we have limited our analysis to the identification of cells and structures with signal above noise levels.
Controls
Different fluorophores are detected with varying efficiency. If you are injecting multiple cell populations, we recommend performing dye-swap control experiments to ensure that similar numbers of cells are observed with each dye; in particular, ensure that different labeling protocols do not affect cell migration and position. For example, if cell population A is labeled with fluorophore 1 and cell population B is labeled with fluorophore 2, the experiment should be repeated by labeling cell population A with fluorophore 2 and cell population B with fluorophore 1.
It is also ideal to image test and control cells or recipient mice within the same experiment, for example, by purifying test and control cells and injecting them in littermate recipients or by splitting the same cell population between test and control recipient mouse. Even if a large amount of cells can be obtained, we recommend imaging one test and one control condition each day and repeating the experimental setup rather than imaging three test recipient mice on one day and three control mice on a separate day. We have not observed differences in HSPC localization when imaging mice within few hours from transplantation; however, we recommend swapping the order of recipient mice during repeats (e.g., during the first experiment, the test mouse is imaged first and the control mouse second, but during the second experiment, the control mouse is imaged first and the test mouse second).
Finally, for each cell imaged, it is important to validate the signal of the label against autofluorescence in order to avoid further analysis of false-positive events. Imaging one or two mice that have not been injected with any cell or label can be helpful to become familiar with bone and autofluorescent signal, especially if low fluorescence is expected.
MATERIALS
REAGENTS
Donor mice (wild-type C57/B6 mice can be purchased from Jackson Laboratories)
 Please note that all animal husbandry and experimentation must be performed according to all relevant guidelines from the appropriate local regulatory board.
Please note that all animal husbandry and experimentation must be performed according to all relevant guidelines from the appropriate local regulatory board.Recipient mice (if interested in imaging osteoblasts, col2.3-EGFP reporter mice express high levels of EGFP in all osteoblasts and osteocytes15)
Mouse hematopoietic stem and progenitor cells (see PROCEDURE)
PBS (Cellgro, cat. no. 21-031)
FCS (Valley Biomedical, cat. no. BS3032)
Antibodies and streptavidin (see Table 1)
Streptavidin-conjugated magnetic beads (Miltenyi Biotec, cat. no. 130-048-101)
DiD (1,1′-dioctadecyl-3,3,3′,3′-tetramethylindodicarbocyanine perchlorate, Invitrogen, cat. no. V22887)
Qtracker 800 (nontargeted quantum dots; Invitrogen, cat. no. Q21071MP)
Ketamine HCl (Henry Schein, cat. no. 1129300)
 Ketamine is a controlled substance; local regulations on storage and use logging must be followed.
Ketamine is a controlled substance; local regulations on storage and use logging must be followed.Xylazine (Bayer)
 Xylazine is a controlled substance; local regulations on storage and use logging must be followed.
Xylazine is a controlled substance; local regulations on storage and use logging must be followed.Isoflurane (Baxter Anesthetic & Critical Care, cat. no. 2099589)
 Isoflurane is harmful. A scavenging system must be in place to collect any excess gas released.
Isoflurane is harmful. A scavenging system must be in place to collect any excess gas released.  Isoflurane is only needed as an alternative to using a ketamine/xylazine cocktail for anesthesia.
Isoflurane is only needed as an alternative to using a ketamine/xylazine cocktail for anesthesia.Analgesics (e.g., buprenorphine)
 Only needed if performing survival surgery.
Only needed if performing survival surgery.Methocell (VisEn Medical)
 An equivalent aqueous ointment-glycerol solution or sterile saline solution can alternatively be used, depending on the microscope setup.
An equivalent aqueous ointment-glycerol solution or sterile saline solution can alternatively be used, depending on the microscope setup.
EQUIPMENT
LD columns (Miltenyi Biotec, cat. no. 130-042-901)
Steriflip filter (Millipore, cat. no. SCGP00525)
Alcohol swabs (Kendall Healthcare, cat. no. 6818)
Veterinary glue (3M, cat. no. 1469SB) or surgical thread (Ethicon)
 These are only needed if performing survival surgery.
These are only needed if performing survival surgery.γ-Irradiator with mouse holder (most commonly containing a Cesium-137 source33)
Dissecting/surgery tools (scissors, forceps, scalpel; Fine Science Tools, cat. nos. 14084-08, 11000-12 and Fisher Scientific, cat. no. 14-840-01)
Mortar and pestle (Fisher/CoorsTek, cat. nos. 12-961AA, 12-961-5AA)
Falcon tubes (50 and 15 ml; BD Bioscience, cat. nos. 352070, 352096)
Eppendorf tubes with conical bottom (1.5 ml; Eppendorf, cat. no. 022600028)
Cell strainers (40-µm pores; BD Bioscience, cat. no 352340)
Magnets (e.g., QuadroMACS, Miltenyi, cat. no. 130-042-302)
Cell sorter (e.g., BD FACS ARIA, BD)
Insulin syringes (Fisher Scientific, cat. no. 14-829-1B)
Two-photon/confocal microscope with in vivo imaging setup (custom made16,17,34 or commercially available (e.g., Zeiss, Leica); see EQUIPMENT SETUP for details)
Mouse warmer (e.g., electric heated pad, Petsavers)
Image analysis software (we recommend ImageJ; http://rsbweb.nih.gov/ij/index.html)
Incubator
REAGENT SETUP
 All reagents need to be sterile.
All reagents need to be sterile.
Harvest medium
Harvest medium is composed of PBS with 2% (vol/vol) FCS. Prepare at least 500 ml. It can be prepared in advance and stored at 4 °C for a few days.
Sorting collection medium
Sorting collection medium is made up of PBS with 10% (vol/vol) FCS. Prepare 1–2 ml. It can be prepared in advance and stored at 4 °C for a few days.
Lineage antibody cocktail
The cocktail is composed of biotin-conjugated Ter119, Gr1, B220, Mac1, CD3, CD4 and CD8 antibodies (see Table 1 for details). Mix these in a 1:1:1:1:1:1:1 ratio. It can be prepared in advance and stored in aliquots of 0.5–1 ml at 4 °C until needed.
Degassed PBS
Prepare ~40 ml of PBS in a 50-ml Falcon tube, connect it to a Steriflip filter and keep it under vacuum for a few minutes without filtering it.
 This reagent is best prepared fresh just before use. Eliminating gas from all solutions applied to the columns increases the efficiency of the purification step.
This reagent is best prepared fresh just before use. Eliminating gas from all solutions applied to the columns increases the efficiency of the purification step.
Ketamine-xylazine cocktail
To a bottle containing 10 ml ketamine HCl (50 mg ml−1; 500 mg), add 750 µl xylazine (100 mg ml−1; 75 mg). Shake well. Store at room temperature (20–25 °C) in the dark for up to 3 months.
EQUIPMENT SETUP
Cell sorter
Several cell sorters are available, and it is important to optimize the antibody staining and fluorophore combination according to the advice of the available FACS instrument. For each sort, unstained and single-color controls must be prepared for compensation purposes, using the same cell type (e.g., bone marrow cells) but only a single antibody (at the same concentration used in the cocktail staining) in each tube (see Experimental design).
Microscope
A schematic representation of our custom-made mouse imaging setup is presented in Figure 1. The in vivo microscope needs to be equipped with a heated mouse holder, either custom made or commercially available (e.g., Stoelting homeothermic blanket system, cat. no. 50300V). A 3D electronically controlled stage is helpful for tracking x-y positions and for z-stack acquisition. Immersion objectives (for example ×25 or ×30) with 0.9 numerical aperture are ideal for imaging through physiological solution or aqueous medium. We use water immersion objectives and image through a cover slip separating the glycerol solution from the water. Alternatively, water dipping objectives with similar magnification and numerical aperture can be used without cover slips and directly positioned on top of the saline-covered calvarium. Non-descanned detectors maximize the efficiency of two-photon microscopy signal acquisition and improve collection of low signals.
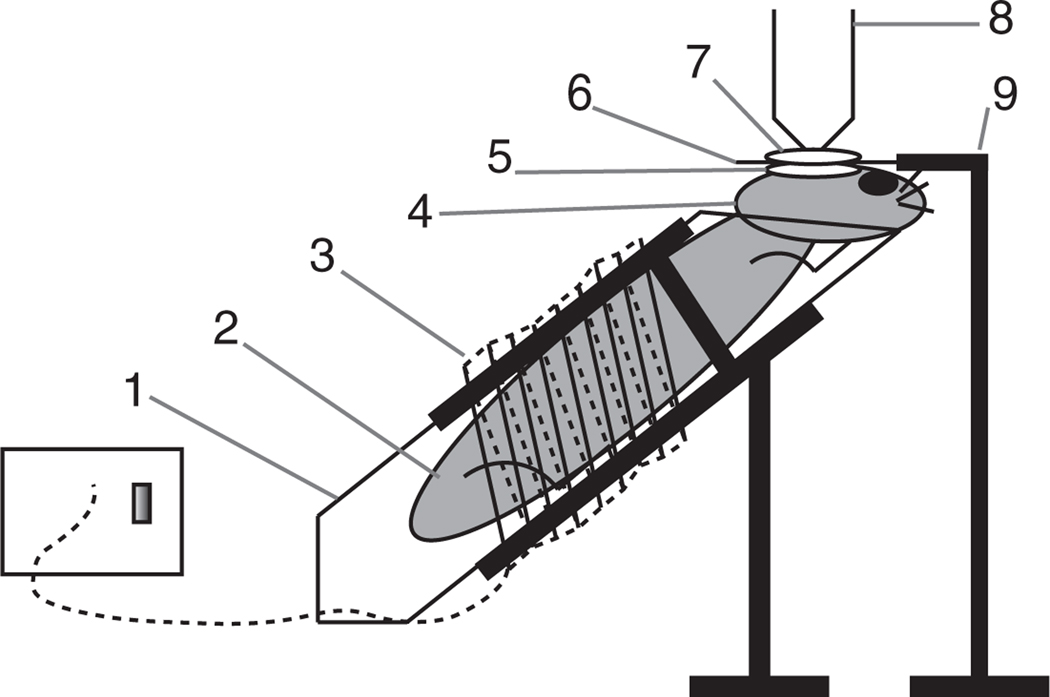
Figure 1.
Diagram representing a custom-made mouse imaging setup. (1) Mouse holder, obtained by modifying a 50-ml Falcon tube and placing it into a clamp/holder. (2) The mouse is positioned comfortably within the tube. (3) Heating module ensures that the mouse body temperature is kept at 37 °C. (4) The mouse head is positioned so that the imaged area is as horizontal as possible by resting the chin on the tube rim. (5) A drop of aqueous ointment or physiological saline solution is applied over the calvarium. (6) Cover slip. (7) A drop of water is placed over the cover slip. (8) Water immersion objective (alternatively, water dipping objectives can be used without the cover slip and can be placed directly onto the mouse calvarium). (9) The cover slip holder keeps the cover slip perfectly horizontal. Note: the drawing is not to scale. An isoflurane cone and scavenging system can be placed in proximity of the nose of the animal.
Table 2 summarizes our preferred combination of lasers and filters. Lasers and filter sets can differ from the ones listed and should be chosen on the basis of the fluorophores used for each experiment. It is important to minimize overlap between the collected signals. A pulsed femtosecond laser is crucial to detect collagen by SHG signal.
PROCEDURE
Conditioning of recipient mice  15 min to 3.5 h
15 min to 3.5 h
-
1
Administer the appropriate dose of γ-irradiation to the recipient mice (see Experimental design for further details). Arrange timing so that the recipient mice will be injected within 24 h of the irradiation.
HSPC harvest  4–6 h
4–6 h
-
2
Euthanize four adult donor mice (8–16 weeks old) and harvest bone marrow from hind limbs and spine by crushing the dissected bones in harvest medium with a mortar and pestle and filtering the cell suspension using a cell strainer. Collect whole bone marrow cells from each mouse in one 50-ml Falcon tube. For further details see refs. 33,35.
-
3
Fill each of the four tubes from Step 2 with harvest medium, spin at 300–350g for 5 min at 4–25 °C.
-
4
Aspirate the supernatant, loosen the pellets by scraping the tubes on the tube rack and resuspend the cells in each tube in 1 ml harvest medium. Set aside 50 µl of cell suspension from any of the tubes to use later (Step 14) for unstained and compensation controls during FACS (see Experimental design for further details).
-
5
To allow isolation of Lineage-negative cells, add 50 µl of Lineage antibody cocktail (see REAGENT SETUP and Table 1) to each tube, mix briefly and incubate at 4 °C for 15 min.
-
6
During this incubation, prepare 40 ml of degassed PBS.
-
7
Fill each tube from Step 5 with harvest medium, spin at 300–350g for 5 min at 4–25 °C.
-
8
Aspirate supernatant from each tube, loosen each pellet by scraping the tubes on the tube rack and resuspend each pellet in 1 ml of degassed PBS. Remove potential clumps by pushing them to the side of the tube and aspirating them in the pipette tip (change tip afterward).
-
9
Add 30 µl of streptavidin-conjugated magnetic beads to each tube, mix briefly and incubate at 4 °C for 15 min.
-
10
During the incubation, position four LD columns on the magnets and load each of them with 3 ml of degassed PBS. After the PBS has run through the columns, position a 15-ml collection tube under each column.
-
11
Add 2 ml of degassed PBS to each tube from Step 9 and load the resulting 3 ml of cell suspension onto a column and through a cell strainer. Repeat for all tubes. The same cell strainer can be used for all columns. Wait until the cell suspension has entirely run through the column.
-
12
Collect any remaining cells in each tube from Step 9 with 3 ml of degassed PBS and load them onto each column, again using the same cell strainer.
-
13
Combine the four eluates in two 15-ml tubes and spin them at 500g for 4 min at 4–25 °C. Aspirate the supernatant form each tube and loosen each pellet by scraping the tubes against their rack.
-
14
Resuspend each pellet in 0.5 ml of harvest medium and combine the two cell suspensions in one of the two tubes (1 ml total). Set aside 10 µl of cell suspension as Lineage compensation control (see Experimental design for details). Add fluorescent-labeled streptavidin and antibodies to label the HSPC population of interest for FACS (each antibody is added and diluted as detailed in Table 1)12,26. For compensation purposes, prepare unstained and single-color controls for each antibody used by aliquotting and staining the total bone marrow cell suspension set aside in Step 4 (as described in Table 3). We recommend using Lineage-depleted cells for the Lineage compensation control, as they provide the same signal intensity as the population to be sorted. Incubate all tubes at 4 °C for 20 min, spin at 500g for 5 min (at 4–25 °C), aspirate the supernatant and resuspend each pellet in 200–800 µl of harvest medium.
-
15
Use unstained and single-color control samples to set up FACS voltages and compensation parameters. Then, sort the desired HSPC subpopulation into a 1.5-ml microcentrifuge tube with a conical bottom, prefilled with 1 ml of sorting collection medium (see Experimental design and EQUIPMENT SETUP). For LT-HSPCs, we recommend sorting either Lineagelow c-Kit+ Sca1+ CD34−Flk2− cells or Lineagelow c-Kit+ Sca1+ CD48−CD150+ cells25.
Table 3.
An example series of compensation controls.
| Cells and amount | Amount of harvest medium | Antibody |
|---|---|---|
| Bone marrow cells from Step 4, 10 µl | 190 µl | None, unstained control |
| Bone marrow cells from Step 4, 10 µl | 190 µl | c-Kit, APC (2 µl) |
| Bone marrow cells from Step 4, 10 µl | 190 µl | Sca-1, PB (2 µl) |
| Bone marrow cells from Step 4, 10 µl | 190 µl | CD34, FITC (2 µl) |
| Bone marrow cells from Step 4, 10 µl | 190 µl | Flk2, PE (2 µl) |
| Lineage-depleted cells from Step 14, 5 µl | 245 µl | Streptavidin, PO (0.5 µl) |
Unstained and single-color controls are necessary in order to set up appropriate voltage and compensation settings for cell sorting. The same antibodies used for the cell mixture sorting are used, at the same dilutions. Lineage-depleted cells are already labeled by the Lineage cocktail and therefore only require streptavidin staining.
HSPC labeling and injection  30 min
30 min
-
16
Spin the sorted cells at 500g for 5 min at 4–25 °C. After this, it should be possible to distinguish a small white pellet for samples of ~7,000 cells and more.
-
17
Remove the supernatant using a P1000 or P200 pipette, making sure to avoid collecting the pellet.
 If the pellet is hard to see, position the pipette tip on the opposite side of the tube relative to where the pellet should be and do not aspirate the entire contents. The goal is to remove as much serum as possible without losing any cells.
If the pellet is hard to see, position the pipette tip on the opposite side of the tube relative to where the pellet should be and do not aspirate the entire contents. The goal is to remove as much serum as possible without losing any cells. -
18
Resuspend the cells in PBS. It is crucial not to add serum in this step, as it will decrease the efficiency of the subsequent staining. HSPCs and, especially, LT-HSPCs are rare; therefore, the counting is performed by the sorter itself while harvesting the cells. We recommend resuspending up to 100,000 cells in 100 µl PBS, and a concentration of 106 cells per ml for higher cell numbers. We recommend transferring volumes >500 µl to a 15- or 50-ml tube.
-
19
Add 0.5 µl of DiD (or the necessary amount to reach a final concentration of 5 µM) and vortex immediately.
 Vortex the mixture immediately in order to prevent the lipophilic dye from forming aggregates on the surface of the solution.
Vortex the mixture immediately in order to prevent the lipophilic dye from forming aggregates on the surface of the solution. -
20
Incubate at 37 °C for 10 min. A tissue culture incubator works fine for this purpose.
-
21
Fill the tube with PBS and spin at 500g for 5 min at 4–25 °C. After this, it should be possible to see a blue pellet. If the pellet is still completely white, repeat the staining procedure (Steps 19–21).
-
22
Remove the supernatant in the same way as in Step 17 and resuspend the cells in PBS or saline solution for intravenous injection. We recommend a volume of 300 µl to avoid losing too many cells in case of a problematic injection. An insulin syringe, with no ‘dead space’ adjacent to the needle, should be used to avoid cell loss.
-
23
Intravenous injection of the cells can be performed by tail vein injection (option A) or retro-orbital injection (option B).
-
Tail vein injection
 5 min
5 minWarm the tail of the recipient mouse to dilate the veins by placing the mouse under a heating lamp or using a tail warmer.
Inject the labeled cells. We recommend injecting the cells as soon as possible after staining. If possible, the mouse should be warmed while the cells are being washed.
-
Retro-orbital injection
 5 min
5 min-
Induce anesthesia using isoflurane inhalation. Force the mouse to inhale isoflurane by placing it in an induction chamber and slowly raising the amount of isoflurane mixed with oxygen to 1–4% (vol/vol).
 Isoflurane is harmful and, therefore, an appropriate scavenging system has to be in place to collect any excess gas released.
Isoflurane is harmful and, therefore, an appropriate scavenging system has to be in place to collect any excess gas released. The mouse should reach deep anesthesia within a few minutes. Ensure that this is the case using one of the approved methods according to institutional and/or governmental regulations (e.g., reaction to toe pinch).
The mouse should reach deep anesthesia within a few minutes. Ensure that this is the case using one of the approved methods according to institutional and/or governmental regulations (e.g., reaction to toe pinch). Inject the cells retro-orbitally by inserting the needle behind the eye bulb, on the side closer to the nose (with this method there is no need to keep the mouse warm, as the total anesthesia time is very short).
-
Allow the mouse to recover from anesthesia.
 Depending on how the experiment is planned, the mouse can be prepared for imaging immediately after injection or intravital microscopy can be performed hours or days later. To analyze homing patterns, we recommend performing the imaging within a few hours of the injection12.
Depending on how the experiment is planned, the mouse can be prepared for imaging immediately after injection or intravital microscopy can be performed hours or days later. To analyze homing patterns, we recommend performing the imaging within a few hours of the injection12.
-
-
Intravital microscopy  1–3 h for each mouse
1–3 h for each mouse
-
24
Anesthetize the mouse using either option A for ketamine-xylazine injection or option B for isoflurane inhalation.
-
Ketamine-xylazine injection
Inject appropriate dose of ketamine-xylazine cocktail (80 mg kg−1 initial dose and 12 mg kg−1 final dose, respectively) intraperitoneally.
-
Isoflurane inhalation
Induce anesthesia using isoflurane inhalation as described in Step 23B(i).
-
-
25
If you are interested in visualizing vasculature, inject nontargeted quantum dots intravenously. A volume of 30 µl of 2 µM solution of Qtracker 655 or 800 should produce a sufficiently bright signal. Higher amounts of shorter-wavelength Qtracker might be required. Dilute the Qtracker in PBS or saline to a final volume of 100–150 µl and use an insulin syringe to minimize reagent waste.
-
26
Clip the scalp hair using scissors or a small veterinary hair clipper and remove the hair fragments using an alcohol swab.

-
27
When deep anesthesia is achieved (see Step 24B(i)), make an incision through the scalp using either option A for a single longitudinal incision or option B for a C-shaped incision (Fig. 2). The incision can be made with a sharp scalpel or scissors.
 Follow aseptic surgical technique to maintain sterility.
Follow aseptic surgical technique to maintain sterility.-
Single longitudinal incision
 < 1 min
< 1 min-
Make a single incision, starting between the ears and following the head midline until 1–2 mm from the nose area.
 This is the fastest incision and is perfect for single imaging sessions. It is also the fastest incision to resuture, and it is therefore recommended for mice that (i) are going to be imaged only once and (ii) will be kept alive for a long follow-up after imaging.
This is the fastest incision and is perfect for single imaging sessions. It is also the fastest incision to resuture, and it is therefore recommended for mice that (i) are going to be imaged only once and (ii) will be kept alive for a long follow-up after imaging.
-
-
C-shaped incision
 1 min
1 minMake a first incision starting near one ear and ending near the other ear.
Continue at a ~60° angle, pointing toward and reaching 1–2 mm from one side of the nose.
-
Continue at a ~90° angle and make a smaller incision in the vicinity of the nose.
 This incision takes longer to prepare and to suture; however, it minimizes the formation of scar tissue above the area to be imaged, and it is therefore recommended for mice that are going to be imaged two or more times.
This incision takes longer to prepare and to suture; however, it minimizes the formation of scar tissue above the area to be imaged, and it is therefore recommended for mice that are going to be imaged two or more times.
-
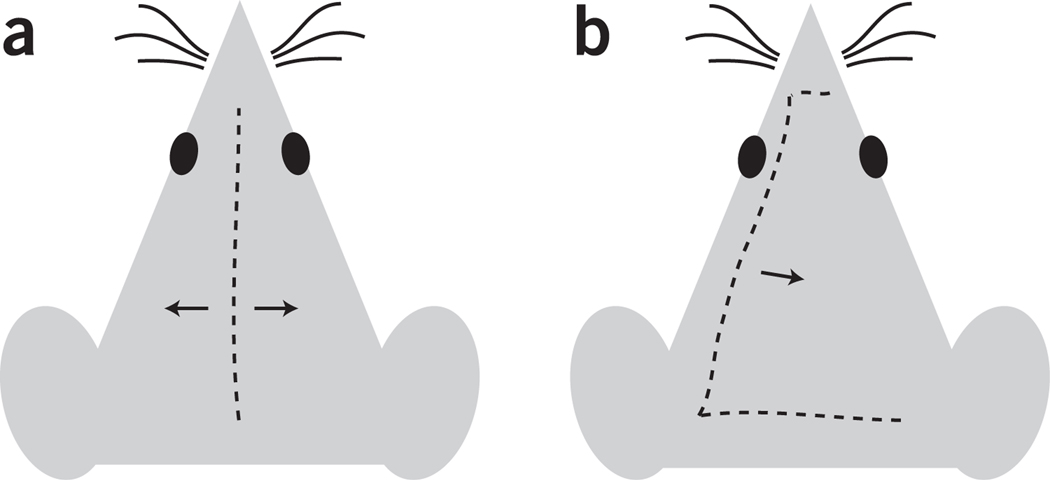
Figure 2.
Diagram representing two options for scalp incision. (a) Longitudinal incision, recommended for single or last imaging session. (b) C-shaped incision, recommended for repeated imaging sessions.
-
28
Separate the skin flaps by pulling toward the sides with forceps.
-
29
Thoroughly clean the skull surface by flushing with sterile PBS and wiping with sterile gauze until all the residual hairs are eliminated.

-
30
Apply a drop of warm glycerol-based gel or physiological saline solution on the skull to avoid drying. Methocell is a commercially available solution; however, glycerol or many aqueous eye ointments can be used.
 Do not allow the skull surface to dry (when this happens, translucency is lost and replaced by a whiter, thicker appearance), as this reduces image clarity. It is important to use a sufficient amount of gel or saline to cover the area; however, excessive amounts of gel tend to overflow and may cause suffocation if they cover the nostrils.
Do not allow the skull surface to dry (when this happens, translucency is lost and replaced by a whiter, thicker appearance), as this reduces image clarity. It is important to use a sufficient amount of gel or saline to cover the area; however, excessive amounts of gel tend to overflow and may cause suffocation if they cover the nostrils. -
31
Place the mouse on the microscope stage. This step varies depending on each specific situation, the microscope model, the stage and so on. In general, place the mouse in the most comfortable position (this reduces gasping), while keeping the imaged area as horizontal as possible (see Fig. 1 for our custom-made imaging setup).
-
32
Observe SHG signal (see Experimental design, EQUIPMENT SETUP and Table 2) to identify a few crucial reference locations, such as the sagittal suture, the intersection between the central and coronal sutures, the bifurcation of the central suture where closer to the nose region (Fig. 3).
-
33
Look for injected cells by observing DiD and autofluorescence signals (see Experimental design, EQUIPMENT SETUP and Table 2).
 A greater number of components can be visualized simultaneously while scanning; however, prolonged exposure to the laser leads to increased tissue damage and photobleaching. The most efficient method for finding all transplanted cells that homed to the cavities is to systematically scan the whole region using overlapping fields of view and a zigzag motion to ensure that no area is left unchecked.
A greater number of components can be visualized simultaneously while scanning; however, prolonged exposure to the laser leads to increased tissue damage and photobleaching. The most efficient method for finding all transplanted cells that homed to the cavities is to systematically scan the whole region using overlapping fields of view and a zigzag motion to ensure that no area is left unchecked.
-
34
Whenever a potential DiD-labeled cell is found, acquire an image of the best focal plane containing both the DiD and autofluorescence signal from the cell of interest. To confirm that the observed signal is from a DiD-labeled cell, immediately check that the DiD signal is at least twofold more intense than the autofluorescence signal from the same cell of interest (non-DiD-labeled, autofluorescent cells will present similar signal intensity in both channels and all autofluorescent cells within the field will have the same signal intensity ratio between DiD and autofluorescent channels; DiD-labeled cells, however, have weak or no autofluorescent signal compared with DiD signal).
 Use settings that allow the brightest signal acquisition, but that avoid oversaturation of the image (most microscopy acquisition software can show a pseudocolored image of the field of view with oversaturated areas highlighted in color). Oversaturated images distort cell size and distance between objects of interest.
Use settings that allow the brightest signal acquisition, but that avoid oversaturation of the image (most microscopy acquisition software can show a pseudocolored image of the field of view with oversaturated areas highlighted in color). Oversaturated images distort cell size and distance between objects of interest. -
35
Having confirmed that the observed signal is from a DiD-labeled cell, you should acquire further images containing details about other cellular and extracellular components surrounding the cell of interest (Fig. 4). For example, acquire a z-stack with slices at 1–10-µm steps, from the focal plane of the cell up to the endosteal surface and the bone covering it. It is advisable to place the cell of interest in the middle of the field of view to maximize the information about the surrounding area in all directions. It is important to compromise between the quality of the z-stack and the amount of information acquired during the whole imaging session. Z-stacks with many slices (e.g., 1–2-µm steps) and high definition are easier to analyze further; however, they require a long acquisition time and expose the tissue to a higher degree of laser-induced damage than z-stacks with fewer slices (e.g., 10-µm steps) and reduced resolution. The lowest resolution allowing the discrimination of the contour of the cells and structures of interest should be used to minimize the time required for each stack, as well as to maximize the number of stacks acquired and cells analyzed in each experiment. When imaging col2.3-EGFP osteoblasts (see Experimental design, EQUIPMENT SETUP and Table 2) or other brightly labeled structures, it can be hard to avoid oversaturation, as sometimes a very bright area is located somewhere in the field, but dimmer cells/structures are in focus near the cell of interest. If both brighter and dimmer areas are of interest, it is helpful to acquire multiple images of the same field of view using multiple appropriate laser power and detection settings.
 Establish an appropriate strategy for naming the images. For example, each image series, corresponding to a separate cell, could have an increasing number, and various letter/number combinations could be used to distinguish images of different structures or at different depths within the series. This will help during the analysis process.
Establish an appropriate strategy for naming the images. For example, each image series, corresponding to a separate cell, could have an increasing number, and various letter/number combinations could be used to distinguish images of different structures or at different depths within the series. This will help during the analysis process.
-
36
End the imaging experiment using option A or maintain the experimental animal for further analysis using option B.
-
End imaging experiment
 minutes
minutesEuthanize the mouse according to institutionally approved methods.
-
Further analysis
 up to months
up to monthsTo keep the mouse alive for further follow-up analysis, first remove the glycerol solution from the skull by washing with sterile saline solution.
-
Suture the incision using either veterinary glue or stitching. Veterinary glues guarantee the quickest closure of the wound and are especially well suited if the effect of ketamine-xylazine is about to run out or if the mouse will not be imaged again. However, glues might cause irritation and scar formation, and, whenever time allows (especially when you are planning to image the mouse again), it is advisable to perform surgical suturing. This can be done with a hypoallergenic suturing thread and the most convenient type of needle and stitching technique. Running sutures can be used along the incisions, and square knot stitches are recommended for the corners. Good suturing technique minimizes scar formation.
 Even though some degree of scar formation is inevitable and scarring tends to build up over time, a second imaging session can be delayed for a few days, depending on the proliferation rate of the injected cells (DiD labeling becomes undetectable after the first few cell divisions due to dilution between daughter cells). The acquired images can be processed and analyzed for any length of time after the imaging. Delaying the final processing and analysis until all imaging sessions within a series have been completed increases consistency within the experiments.
Even though some degree of scar formation is inevitable and scarring tends to build up over time, a second imaging session can be delayed for a few days, depending on the proliferation rate of the injected cells (DiD labeling becomes undetectable after the first few cell divisions due to dilution between daughter cells). The acquired images can be processed and analyzed for any length of time after the imaging. Delaying the final processing and analysis until all imaging sessions within a series have been completed increases consistency within the experiments.
-
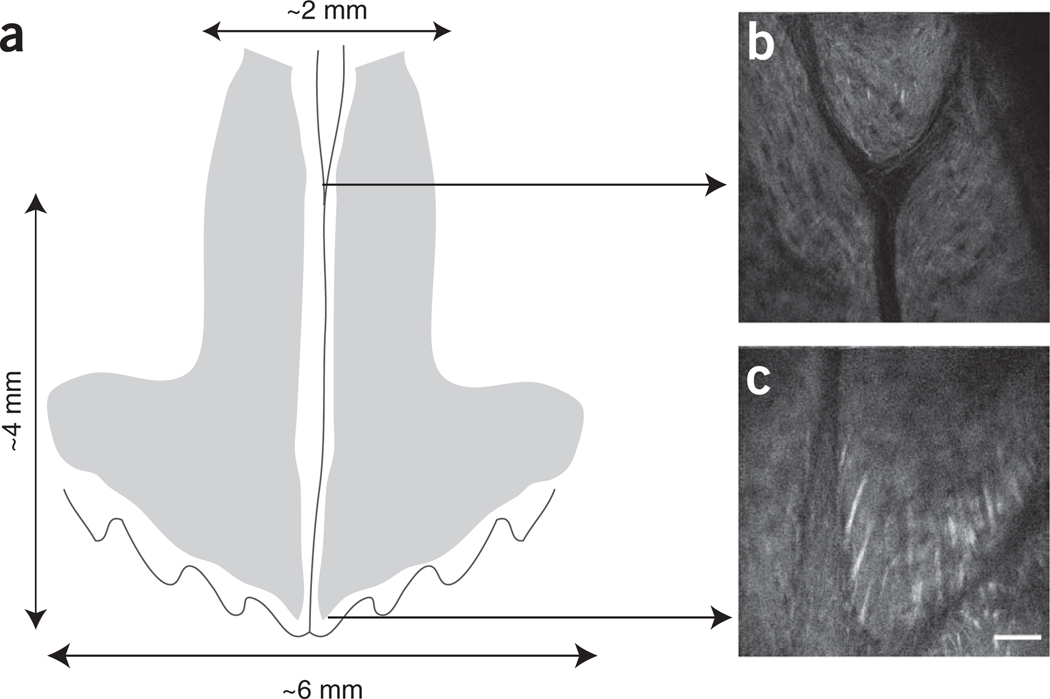
Figure 3.
Imaged area and specific spatial reference points. (a) Diagram representing reciprocal positioning of calvarium sutures (vertical line: sagittal suture; undulated line: coronal suture) and bone marrow cavities (gray areas). Note: the drawing is not to scale. The orientation of the head is as shown in Figure 2. (b,c) Examples of SHG signal from two reference points: (b) sagittal suture bifurcation and (c) intersection of sagittal and coronal sutures (right side). Scale bar, 50 µm (×30 objective mounted on our custom-made microscope).
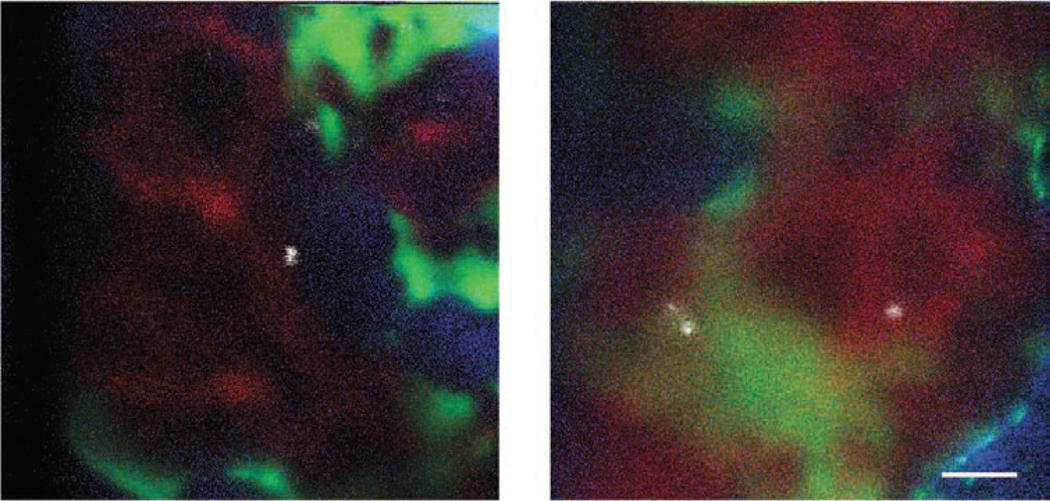
Figure 4.
Examples of bone marrow calvarium intravital microscopy images obtained with lasers and filters listed in Table 2. Two fields of view containing DiD-labeled cells (white: DiD signal), vasculature (red: quantum dot 800 signal), osteoblasts (green: EGFP signal) and bone collagen (blue: SHG signal) are presented. Note that the vasculature is more clearly demarcated in nonirradiated (left) than in irradiated (right) recipient mice. Scale bar, 50 µm.
Image analysis  at least 3–6 h
at least 3–6 h
-
37
Confirm that the imaged cells are indeed DiD-labeled and not autofluorescent cells. If DiD and autofluorescence signals are acquired separately, merge the two images. Make manual adjustments to ensure that all signals are taken into consideration. The autofluorescent signal is often very dim, whereas DiD-labeled cells can be very bright. Generally, it should be possible to adjust levels to confirm that all autofluorescence signals have identical patterns in both channels. Some autofluorescent cells are dimmer and others brighter, but such variations are identical in both channels. On the basis of our experience, we advise to count as a ‘real’ DiD signal any signal that is at least 1.7-fold brighter in the DiD channel compared with the autofluorescence channel, relative to the background (Fig. 5). This type of measurement can be obtained, for example, in ImageJ, by selecting DiD-labeled and autofluorescent cells as regions of interest and measuring the average signal intensity in each channel.
-
38
Eliminate all images acquired for cells that do not meet the selection criteria described above (Step 37).
-
39
Generate the clearest image of each cell. To do this, eliminate background and autofluorescence noise by adjusting DiD signal output levels or thresholding functions (in any image analysis software).
 A clearer, more contrasted image of the cell of interest will help with measuring later, as it will make it easier to judge cell boundaries. However, this adjustment has to be performed carefully to avoid altering the size of the cell of interest (see Fig. 5).
A clearer, more contrasted image of the cell of interest will help with measuring later, as it will make it easier to judge cell boundaries. However, this adjustment has to be performed carefully to avoid altering the size of the cell of interest (see Fig. 5). -
40
Generate an image series containing the adjusted, best focal plane signal from the cell of interest (from Step 39) and EGFP or SHG (respectively, osteoblasts or bone) signal from the same focal plane and the planes above. Generate a three-color image series containing the cell of interest in red, osteoblasts in green and bone signal in blue. Depending on how the images are acquired, this step may involve eliminating the DiD channel signal from all slices above the cell of interest and replacing it with the one generated in Step 39. The red signal will be identical throughout the series, whereas blue and green signals will change according to depth (Fig. 6).
-
41
Use the autolevel function to optimize the signal in the blue and green channels in each image of the series (e.g., in ImageJ, click ‘Image’ ‘Adjust’ ‘Window/Level’ ‘Auto’). Eliminate background noise whenever possible by further adjusting the minimum output intensity (Imin). To select the appropriate Imin value, choose an acquired image in which no light reached the sample, then check the intensity curve of all pixels and select the Imax of that curve as Imin for the adjusted images. For example, if a system generates a background image with intensity of 0–7, then input 7 as Imin. Adjusting the Imin ensures that areas with no signal (e.g., areas containing nonfluorescent cells) are black in the final image, thus enhancing the contrast and facilitating the measurement step (Step 42).
 The most efficient way to perform this step is by batch-processing all the images simultaneously. Batch processing increases consistency and decreases the time required to deal with a large number of images.
The most efficient way to perform this step is by batch-processing all the images simultaneously. Batch processing increases consistency and decreases the time required to deal with a large number of images. -
42
Measure cell position by recording the length of a line drawn from the edge of the cell to the edge of the structure of interest (in our case, bone or osteoblast) throughout the image series; subsequently, apply Pythagoras’ theorem when measuring images that are above the focal plane containing the cell of interest (Fig. 6). Ensure that correct cell boundaries are taken into consideration, not only for the transplanted cells but also for osteoblasts and bone. See the TROUBLESHOOTING table for additional details. If the measurements are taken in pixels, convert the pixels to µm. From each z-stack, select the shortest 3D distance from the cell to the structure of interest.

Troubleshooting advice can be found in Table 4.
Figure 5.
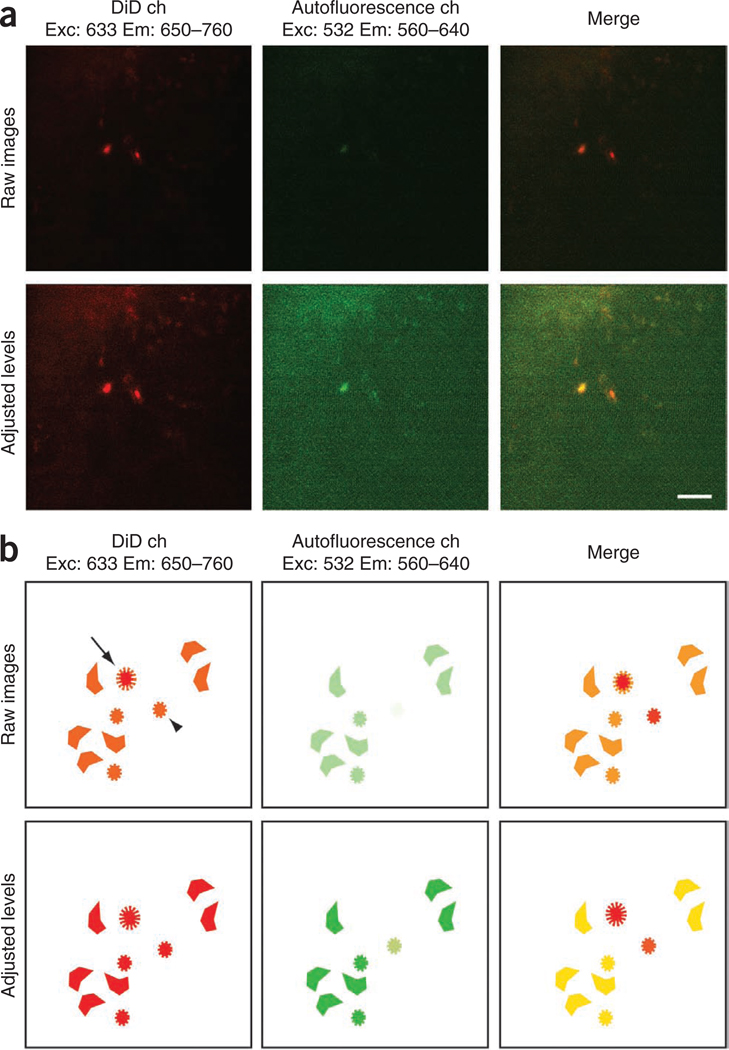
Validation of DiD signal through comparison with autofluorescence signal (measuring DiD:autofluorescence signal ratio). (a) Examples of raw and adjusted images containing a bright DiD-labeled (right) cell and a bright autofluorescent (left) cell. (b) Schematic diagram of bright and dim DiD-labeled cells (arrow and arrowhead), together with autofluorescent cells in the same field of view. Top row in both a and b, only minor color differences between DiD-labeled and autofluorescent cells are seen in the initial merged image (top right). Bottom row in both a and b, level adjustments highlight autofluorescent cells. Note that adjusting output levels increases the size of all cells, including the DiD-positive cells. In the adjusted, merged image (bottom right), autofluorescent cells have similar intensities in both channels, whereas the DiD-labeled cell present a more intense red color. Adjusted images can be used to measure the DiD/autofluorescence signal intensity ratio for autofluorescent cells and for the cell of interest (1 versus 1.7 and above). ch, channel. Scale bar, 50 µm.
Figure 6.
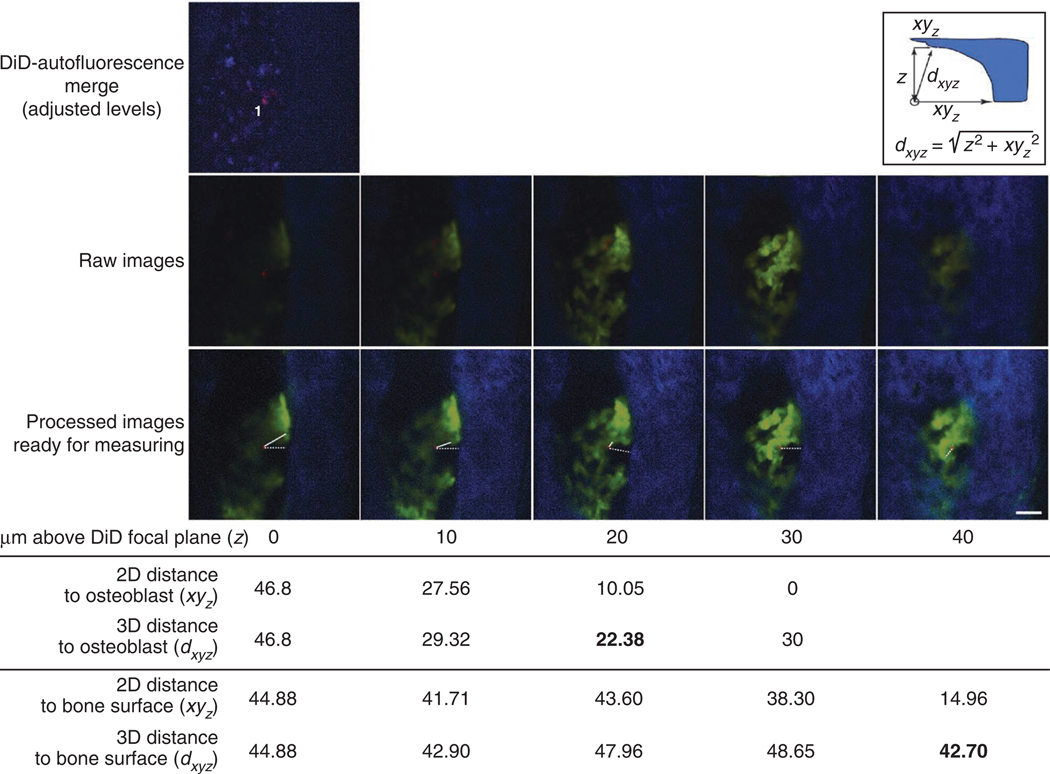
Image analysis procedure. Top image: DiD-autofluorescence merge is checked to eliminate false-positive cells. In the presented case, one DiD-labeled cell is in focus (1). The red signal on the top right of the measured cell is from out-of-focus DiD cells located deeper and not presented in this example. Middle row: raw images of z-stacks containing DiD (red, indicating transplanted cells), EGFP (green, osteoblasts) and SHG (blue, bone cartilage) signals (step: 10 µm; each z-slice is acquired 10 µm above the previous one). Bottom row: Left, osteoblast (green), bone (blue) and DiD (red) signals are shown after optimization and signal enhancement. The same image of the DiD-labeled cell is then superimposed to the images of bone and osteoblasts closer to the top bone surface. Lines are drawn and measured from the edge of the DiD-labeled cell to the closest osteoblast (continuous lines) and bone surface (dashed lines) in focus. Insert: Pythagoras’ theorem is used to calculate the 3D distance from the DiD-labeled cell to the bone and osteoblasts. The DiD-labeled cell is the white circle at the bottom left and the bone is presented here in blue. For clarity, only the formula to calculate the 3D distance to the bone surface is shown. Calculated 2D and 3D distances at each z position are shown in the table below the images. No measurement is calculated at z = 40 because xy30 = 0, and therefore dxy40 cannot be shorter than dxy30. The shortest 3D distances between the DiD-labeled cell and the closest osteoblasts (22.38 µm) and bone surface (42.70 µm) are highlighted in bold. Scale bar, 50 µm.
Table 4.
Troubleshooting table.
| Step | Problem | Possible reason | Solution |
|---|---|---|---|
| 26 and 29 | Hair shadows | Hair fragments lying on | Thoroughly clean the skull surface before imaging, as even the smallest hair will cast a shadow of at least 20–30 µm wide. It can be helpful to clip the scalp hair before the surgery; however, this generates extra hair fragments, which can eventually be deposited on the scalp. Hair clipping can be replaced by thorough cleaning after the incision. Alternatively, depilatory creams can be used; however, these require prolonged anesthesia, which can be problematic with ketamine-xylazine. Regardless, it is crucial to thoroughly clean the skull surface by flushing it with PBS and wiping it with sterile gauze. White hair is particularly difficult to see by eye and therefore special care must be taken when working with white mice |
| 33 | No cells detected | One of the following problems may have occurred:
|
|
| 35 | Insufficient length of anesthesia | Logistical setup and excessively lengthy image acquisition | Perform the surgery near the microscope or microscopy facility. Develop a series of checkpoints to ensure appropriate positioning of the mouse and an optimized imaging routine. Maintain the same imaging routine to ensure efficiency and reproducibility between experiments. If necessary, image only half of the calvarium (e.g., the right half), assuming that the other half would contain a similar number of HSPCs in similar positions (this reduces the collected data and the sample size (n) for future statistical analysis). Alternatively, try a slightly higher dose of ketamine-xylazine cocktail or substitute the cocktail with isoflurane anesthesia |
| Out-of-focus signal | Signal spillage from very bright osteoblasts or difficulty in balancing signals in a z-stack Resolution in the optical plane (xy) is higher than the axial resolution (z) and therefore only vertical bone marrow cavity walls appear clearly demarcated (as opposed to a gradual slice-to-slice shift) | Compare the signal of each structure of interest throughout the image series and make a consistent judgment about what is in focus and what is not as well as where bone and osteoblasts are located within the area of interest (e.g., only consider signal in focus or only consider signal of a certain relative intensity). In many cases, blurred or dim signal in one slice will be strong and clearly in focus in others. Clear and consistent standards for each analysis minimize person-toperson variability. It is expected that measurements repeated by the same or different investigators may differ by few µm | |
| Unexpected mouse death |
|
|
|
| 36B(ii) | Cell tracking difficulties | Shifts in the mouse position between imaging sessions and potential alterations within the bone marrow microenvironment | Carefully reposition the mouse and calculate the angle of positional shift in all three dimensions. Acquiring z-stacks for multiple signals can facilitate recognizing the same area through the spatial relationship of multiple components. Little is known about the effects of irradiation and of the imaging itself on the bone marrow microenvironment. Occasionally, we have observed increased numbers of osteoblasts during the second imaging session (C.L.C. and C.P.L., unpublished observations). If you are interested in measuring HSPC-osteoblast distance a few days after transplantation, we recommend not imaging the mouse until the time point of interest |
Step 1, Recipients conditioning: 15 min to 3.5 h
Steps 2–15, Recipient conditioning and HSPC harvest: 4–6 h
Steps 16–23, Cell labeling and injection: 30 min
Steps 24–36, Single microscopy session: 1–3 h
Steps 37–42, Image analysis: 3–6 h or as long as required
ANTICIPATED RESULTS
When repeatedly injecting similar numbers of HSPCs, similar numbers of cells should be detected in the calvarium bone marrow. We typically observe 10–15 events when injecting ~10,000 LT-HSPCs (LKS CD34−Flk2− or LKS CD48−CD150+), with a detection limit set at ~5,000 LT-HSPCs injected. These numbers vary for different cell populations, possibly because of their different ability to localize in the bone marrow space.
Although some variability is observed between independent recipient mice, we observed consistent trends throughout our experiments, and usually no statistically significant differences are observed when the same HSPC populations are injected in equivalent recipient mice. As differences in bone marrow localization of distinct HSPC populations can be subtle, data from cells observed in multiple recipients can be pooled in order to statistically compare different cell types. In any case, the average distance from any HSPC population to osteoblasts or bone has a very wide standard deviation. Moreover, each measurement is subject to errors. Accordingly, we recommend not to focus on the exact distance (in µm) between specific cell types; rather, we suggest careful analysis of the shapes and shifts of the distributions. Even though specific localization of LT-HSPCs relative to osteoblasts may vary between experiments, the facts remain that most LT-HSPCs are most likely to be located in the proximity of osteoblasts, that the likelihood of identifying LT-HSPCs rapidly decreases when moving further from osteoblasts and that only a few LT-HSPCs are directly adjacent to osteoblasts.
ACKNOWLEDGMENTS
We thank F. Ferraro, S. Lane, S. Lymperi, E. Ozcivici, A. Sanchez-Aguilera and M. Spitaler for critical input on the manuscript. C.L.C. was funded by the European Molecular Biology Organization and Human Frontier Science Program and is currently funded by Imperial College London, Kay Kendall Leukaemia Foundation and Cancer Research UK. C.P.L. and D.T.S. are funded by multiple NIH grants.
Footnotes
AUTHOR CONTRIBUTIONS C.P.L. developed the confocal/multiphoton imaging system for calvarium bone marrow intravital microscopy. C.L.C. developed, performed and undertook troubleshooting of the described protocol. D.T.S. provided guidance and critical input throughout the development of the methodology.
COMPETING FINANCIAL INTERESTS The authors declare competing financial interests (see the HTML version of this article for details).
References
- 1.Calvi LM, et al. Osteoblastic cells regulate the haematopoietic stem cell niche. Nature. 2003;425:841–846. doi: 10.1038/nature02040. [DOI] [PubMed] [Google Scholar]
- 2.Lord BI, Testa NG, Hendry JH. The relative spatial distributions of CFUs and CFUc in the normal mouse femur. Blood. 1975;46:65–72. [PubMed] [Google Scholar]
- 3.Visnjic D, et al. Hematopoiesis is severely altered in mice with an induced osteoblast deficiency. Blood. 2004;103:3258–3264. doi: 10.1182/blood-2003-11-4011. [DOI] [PubMed] [Google Scholar]
- 4.Zhang J, et al. Identification of the haematopoietic stem cell niche and control of the niche size. Nature. 2003;425:836–841. doi: 10.1038/nature02041. [DOI] [PubMed] [Google Scholar]
- 5.Chan CK, et al. Endochondral ossification is required for haematopoietic stem-cell niche formation. Nature. 2009;457:490–494. doi: 10.1038/nature07547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hooper AT, et al. Engraftment and reconstitution of hematopoiesis is dependent on VEGFR2-mediated regeneration of sinusoidal endothelial cells. Cell Stem Cell. 2009;4:263–274. doi: 10.1016/j.stem.2009.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Katayama Y, et al. Signals from the sympathetic nervous system regulate hematopoietic stem cell egress from bone marrow. Cell. 2006;124:407–421. doi: 10.1016/j.cell.2005.10.041. [DOI] [PubMed] [Google Scholar]
- 8.Kiel MJ, et al. SLAM family receptors distinguish hematopoietic stem and progenitor cells and reveal endothelial niches for stem cells. Cell. 2005;121:1109–1121. doi: 10.1016/j.cell.2005.05.026. [DOI] [PubMed] [Google Scholar]
- 9.Mendez-Ferrer S, Lucas D, Battista M, Frenette PS. Haematopoietic stem cell release is regulated by circadian oscillations. Nature. 2008;452:442–447. doi: 10.1038/nature06685. [DOI] [PubMed] [Google Scholar]
- 10.Naveiras O, et al. Bone-marrow adipocytes as negative regulators of the haematopoietic microenvironment. Nature. 2009;460:259–263. doi: 10.1038/nature08099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sugiyama T, Kohara H, Noda M, Nagasawa T. Maintenance of the hematopoietic stem cell pool by CXCL12-CXCR4 chemokine signaling in bone marrow stromal cell niches. Immunity. 2006;25:977–988. doi: 10.1016/j.immuni.2006.10.016. [DOI] [PubMed] [Google Scholar]
- 12.Mazo IB, et al. Hematopoietic progenitor cell rolling in bone marrow microvessels: parallel contributions by endothelial selectins and vascular cell adhesion molecule 1. J. Exp. Med. 1998;188:465–474. doi: 10.1084/jem.188.3.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sipkins DA, et al. In vivo imaging of specialized bone marrow endothelial microdomains for tumour engraftment. Nature. 2005;435:969–973. doi: 10.1038/nature03703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zipfel WR, Williams RM, Webb WW. Nonlinear magic: multiphoton microscopy in the biosciences. Nat. Biotechnol. 2003;21:1369–1377. doi: 10.1038/nbt899. [DOI] [PubMed] [Google Scholar]
- 15.Kalajzic Z, et al. Directing the expression of a green fluorescent protein transgene in differentiated osteoblasts: comparison between rat type I collagen and rat osteocalcin promoters. Bone. 2002;31:654–660. doi: 10.1016/s8756-3282(02)00912-2. [DOI] [PubMed] [Google Scholar]
- 16.Lo Celso C, et al. Live-animal tracking of individual haematopoietic stem/progenitor cells in their niche. Nature. 2009;457:92–96. doi: 10.1038/nature07434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lo Celso C, Wu JW, Lin CP. In vivo imaging of hematopoietic stem cells and their microenvironment. J. Biophotonics. 2009;2:619–631. doi: 10.1002/jbio.200910072. [DOI] [PubMed] [Google Scholar]
- 18.Adams GB, et al. Haematopoietic stem cells depend on Galpha(s)-mediated signalling to engraft bone marrow. Nature. 2009;459:103–107. doi: 10.1038/nature07859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Colmone A, et al. Leukemic cells create bone marrow niches that disrupt the behavior of normal hematopoietic progenitor cells. Science. 2008;32:1861–1865. doi: 10.1126/science.1164390. [DOI] [PubMed] [Google Scholar]
- 20.Nilsson SK, Johnston HM, Coverdale JA. Spatial localization of transplanted hemopoietic stem cells: inferences for the localization of stem cell niches. Blood. 2001;97:2293–2299. doi: 10.1182/blood.v97.8.2293. [DOI] [PubMed] [Google Scholar]
- 21.Xie Y, et al. Detection of functional haematopoietic stem cell niche using real-time imaging. Nature. 2009;457:97–101. doi: 10.1038/nature07639. [DOI] [PubMed] [Google Scholar]
- 22.Kohler A, et al. Altered cellular dynamics and endosteal location of aged early hematopoietic progenitor cells revealed by time-lapse intravital imaging in long bones. Blood. 2009;114:290–298. doi: 10.1182/blood-2008-12-195644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lewandowski D, et al. In vivo cellular imaging pinpoints the role of reactive oxygen species in the early steps of adult hematopoietic reconstitution. Blood. 2010;115:443–452. doi: 10.1182/blood-2009-05-222711. [DOI] [PubMed] [Google Scholar]
- 24.Down JD, Ploemacher RE. Transient and permanent engraftment potential of murine hematopoietic stem cell subsets: differential effects of host conditioning with gamma radiation and cytotoxic drugs. Exp. Hematol. 1993;21:913–921. [PubMed] [Google Scholar]
- 25.Bryder D, Rossi DJ, Weissman IL. Hematopoietic stem cells: the paradigmatic tissue-specific stem cell. Am. J. Pathol. 2006;169:338–346. doi: 10.2353/ajpath.2006.060312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roederer M. Compensation in flow cytometry. Curr. Protoc. Cytom. 2002;(Supplement 22):1.14.1–1.14.20. doi: 10.1002/0471142956.cy0114s22. [DOI] [PubMed] [Google Scholar]
- 27.Fleming HE, et al. Wnt signaling in the niche enforces hematopoietic stem cell quiescence and is necessary to preserveself-renewal in vivo. Cell Stem Cell. 2008;2:274–283. doi: 10.1016/j.stem.2008.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lassailly F, Greissinger E, Bonnet D. “Microenvironmental contaminations” induced by fluorescent lipophilic dyes used for noninvasive in vitro and in vivo cell tracking. Blood. 2010;115:5347–5354. doi: 10.1182/blood-2009-05-224030. [DOI] [PubMed] [Google Scholar]
- 29.Wallace PK, Muirhead KA. Cell tracking 2007: a proliferation of probes and applications. Immunol. Invest. 2007;36:527–561. doi: 10.1080/08820130701812584. [DOI] [PubMed] [Google Scholar]
- 30.Faust N, Varas F, Kelly LM, Heck S, Graf T. Insertion of enhanced green fluorescent protein into the lysozyme gene creates mice with green fluorescent granulocytes and macrophages. Blood. 2000;96:719–726. [PubMed] [Google Scholar]
- 31.Zhang J, et al. CD41-YFP mice allow in vivo labeling of megakaryocytic cells and reveal a subset of platelets hyperreactive to thrombin stimulation. Exp. Hematol. 2007;35:490–499. doi: 10.1016/j.exphem.2006.11.011. [DOI] [PubMed] [Google Scholar]
- 32.Wunder A, Tung CH, Muller-Ladner U, Weissleder R, Mahmood U. In vivo imaging of protease activity in arthritis: a novel approach for monitoring treatment response. Arthritis. Rheum. 2004;50:2459–2465. doi: 10.1002/art.20379. [DOI] [PubMed] [Google Scholar]
- 33.Lo Celso C, Klein RJ, Scadden DT. Hematopoietic stem cells niche. Curr. Protoc. 2007;3:2A.5.1–2A.5.2. doi: 10.1002/9780470151808.sc02a05s3. [DOI] [PubMed] [Google Scholar]
- 34.Veilleux I, Spencer JA, Biss DP, Cote D, Lin CP. In vivo cell tracking with video rate multimodality laser scanning microscopy. IEEE J. Sel. Top. Quantum Electron. 2008;14:10–18. [Google Scholar]
- 35.Lo Celso C, Scadden D. Isolation and transplantation of hematopoietic stem cells (HSCs) J. Vis. Exp. 2007;2:157. doi: 10.3791/157. [DOI] [PMC free article] [PubMed] [Google Scholar]