Abstract
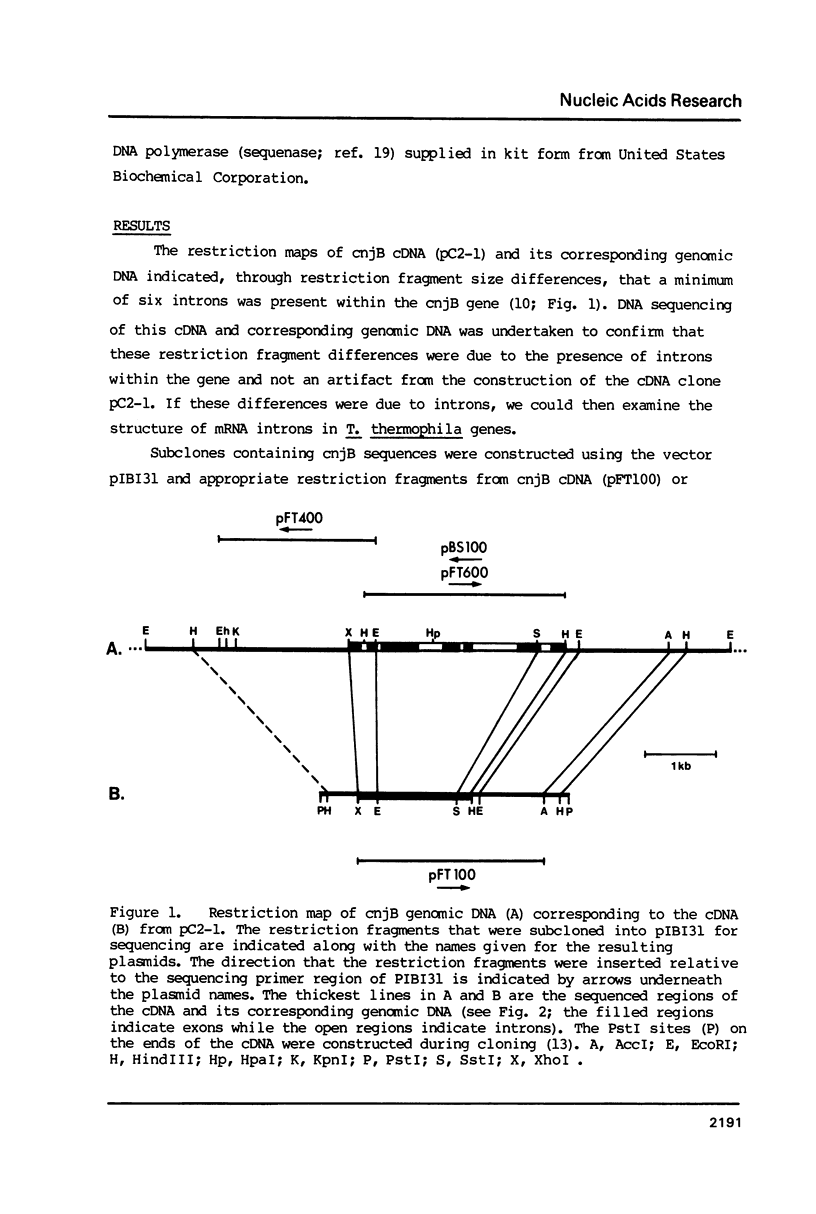
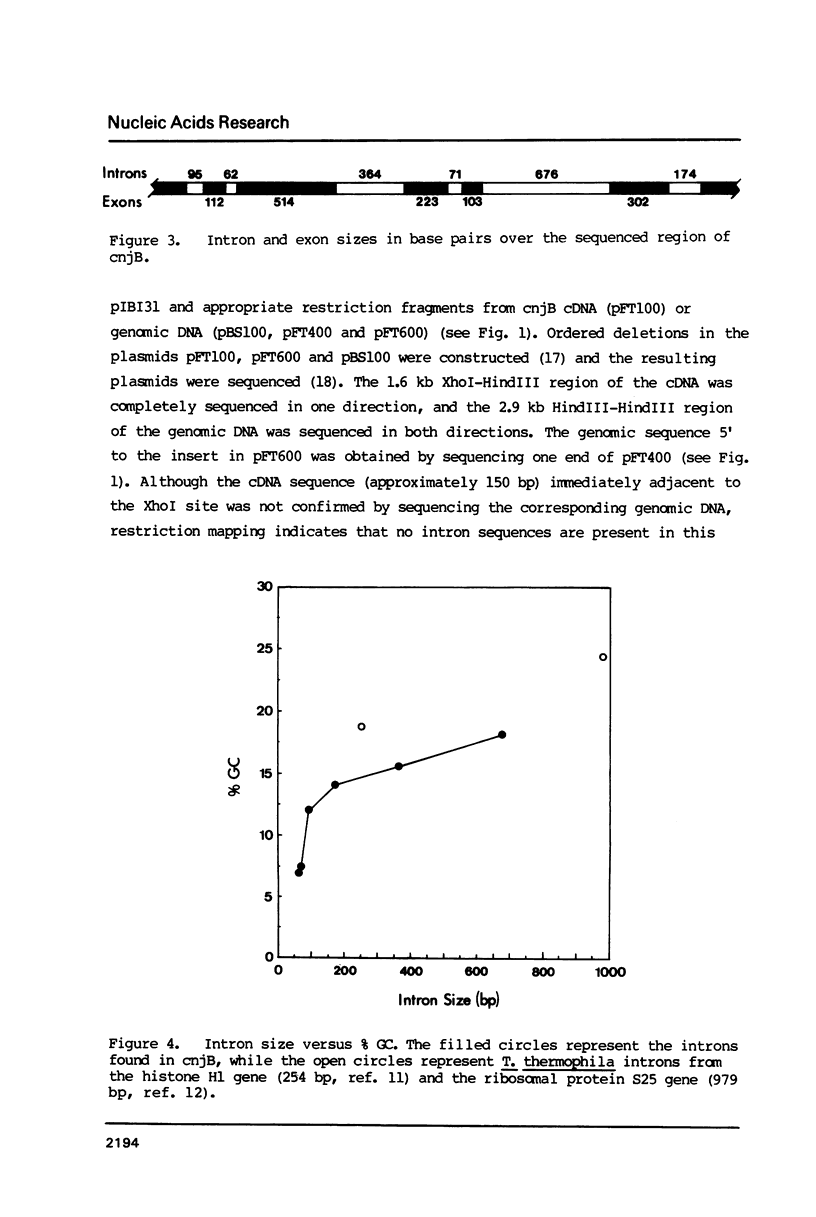
Multiple introns have been found in a gene from a ciliated protozoan. This Tetrahymena thermophila gene (cnjB) is large (7.5 kb mRNA) and active only during conjugation, the organism's sexual cycle. Six introns ranging in size from 62 bp to 676 bp were found when we sequenced a 3.1 kb segment of the cnjB gene together with its corresponding cDNA. We estimate, by extrapolation of our current data, a total of approximately 30 introns in this gene with a total gene size (introns plus exons) of 15 kb or more. The number of introns is surprising given the scarcity of introns in ciliate genes examined to date. Our findings constitute the first example of multiple introns in a ciliate gene. Having the sequence of several introns has allowed us to construct consensus sequences for T. thermophila mRNA introns. The 5' and 3' intron junctions resemble those of general nuclear mRNA (GT/AG rule is followed) but differences are seen. In particular, stretches of 10 or more adenines and thymines are found adjacent to the conserved GT and AGs at the junctions. Unusual aspects of the coding region of this gene are discussed.
Full text
PDF










Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baba M. L., Darga L. L., Goodman M., Czelusniak J. Evolution of cytochrome C investigated by the maximum parsimony method. J Mol Evol. 1981;17(4):197–213. doi: 10.1007/BF01732758. [DOI] [PubMed] [Google Scholar]
- Bannon G. A., Bowen J. K., Yao M. C., Gorovsky M. A. Tetrahymena H4 genes: structure, evolution and organization in macro- and micronuclei. Nucleic Acids Res. 1984 Feb 24;12(4):1961–1975. doi: 10.1093/nar/12.4.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calzone F. J., Stathopoulos V. A., Grass D., Gorovsky M. A., Angerer R. C. Regulation of protein synthesis in Tetrahymena. RNA sequence sets of growing and starved cells. J Biol Chem. 1983 Jun 10;258(11):6899–6905. [PubMed] [Google Scholar]
- Caron F., Meyer E. Does Paramecium primaurelia use a different genetic code in its macronucleus? Nature. 1985 Mar 14;314(6007):185–188. doi: 10.1038/314185a0. [DOI] [PubMed] [Google Scholar]
- Cupples C. G., Pearlman R. E. Isolation and characterization of the actin gene from Tetrahymena thermophila. Proc Natl Acad Sci U S A. 1986 Jul;83(14):5160–5164. doi: 10.1073/pnas.83.14.5160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink G. R. Pseudogenes in yeast? Cell. 1987 Apr 10;49(1):5–6. doi: 10.1016/0092-8674(87)90746-x. [DOI] [PubMed] [Google Scholar]
- Gilbert W., Marchionni M., McKnight G. On the antiquity of introns. Cell. 1986 Jul 18;46(2):151–153. doi: 10.1016/0092-8674(86)90730-0. [DOI] [PubMed] [Google Scholar]
- Godiska R. Structure and sequence of the H surface protein gene of Paramecium and comparison with related genes. Mol Gen Genet. 1987 Jul;208(3):529–536. doi: 10.1007/BF00328151. [DOI] [PubMed] [Google Scholar]
- Hanyu N., Kuchino Y., Nishimura S., Beier H. Dramatic events in ciliate evolution: alteration of UAA and UAG termination codons to glutamine codons due to anticodon mutations in two Tetrahymena tRNAs. EMBO J. 1986 Jun;5(6):1307–1311. doi: 10.1002/j.1460-2075.1986.tb04360.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattori M., Sakaki Y. Dideoxy sequencing method using denatured plasmid templates. Anal Biochem. 1986 Feb 1;152(2):232–238. doi: 10.1016/0003-2697(86)90403-3. [DOI] [PubMed] [Google Scholar]
- Helftenbein E. Nucleotide sequence of a macronuclear DNA molecule coding for alpha-tubulin from the ciliate Stylonychia lemnae. Special codon usage: TAA is not a translation termination codon. Nucleic Acids Res. 1985 Jan 25;13(2):415–433. doi: 10.1093/nar/13.2.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S. Unidirectional digestion with exonuclease III creates targeted breakpoints for DNA sequencing. Gene. 1984 Jun;28(3):351–359. doi: 10.1016/0378-1119(84)90153-7. [DOI] [PubMed] [Google Scholar]
- Horowitz S., Bowen J. K., Bannon G. A., Gorovsky M. A. Unusual features of transcribed and translated regions of the histone H4 gene family of Tetrahymena thermophila. Nucleic Acids Res. 1987 Jan 12;15(1):141–160. doi: 10.1093/nar/15.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horowitz S., Gorovsky M. A. An unusual genetic code in nuclear genes of Tetrahymena. Proc Natl Acad Sci U S A. 1985 Apr;82(8):2452–2455. doi: 10.1073/pnas.82.8.2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaine B. P., Spear B. B. Nucleotide sequence of a macronuclear gene for actin in Oxytricha fallax. Nature. 1982 Feb 4;295(5848):430–432. doi: 10.1038/295430a0. [DOI] [PubMed] [Google Scholar]
- Kimmel A. R., Firtel R. A. Intervening sequences in a Dictyostelium gene that encodes a low abundance class mRNA. Nucleic Acids Res. 1980 Dec 11;8(23):5599–5610. doi: 10.1093/nar/8.23.5599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martindale D. W., Bruns P. J. Cloning of abundant mRNA species present during conjugation of Tetrahymena thermophila: identification of mRNA species present exclusively during meiosis. Mol Cell Biol. 1983 Oct;3(10):1857–1865. doi: 10.1128/mcb.3.10.1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martindale D. W., Martindale H. M., Bruns P. J. Tetrahymena conjugation-induced genes: structure and organization in macro- and micronuclei. Nucleic Acids Res. 1986 Feb 11;14(3):1341–1354. doi: 10.1093/nar/14.3.1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen H., Andreasen P. H., Dreisig H., Kristiansen K., Engberg J. An intron in a ribosomal protein gene from Tetrahymena. EMBO J. 1986 Oct;5(10):2711–2717. doi: 10.1002/j.1460-2075.1986.tb04555.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomoto M., Imai N., Saiga H., Matsui T., Mita T. Characterization of two types of histone H2B genes from macronuclei of Tetrahymena thermophila. Nucleic Acids Res. 1987 Jul 24;15(14):5681–5697. doi: 10.1093/nar/15.14.5681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preer J. R., Jr, Preer L. B., Rudman B. M., Barnett A. J. Deviation from the universal code shown by the gene for surface protein 51A in Paramecium. Nature. 1985 Mar 14;314(6007):188–190. doi: 10.1038/314188a0. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sogin M. L., Elwood H. J., Gunderson J. H. Evolutionary diversity of eukaryotic small-subunit rRNA genes. Proc Natl Acad Sci U S A. 1986 Mar;83(5):1383–1387. doi: 10.1073/pnas.83.5.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabor S., Richardson C. C. DNA sequence analysis with a modified bacteriophage T7 DNA polymerase. Proc Natl Acad Sci U S A. 1987 Jul;84(14):4767–4771. doi: 10.1073/pnas.84.14.4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waring R. B., Davies R. W. Assessment of a model for intron RNA secondary structure relevant to RNA self-splicing--a review. Gene. 1984 Jun;28(3):277–291. doi: 10.1016/0378-1119(84)90145-8. [DOI] [PubMed] [Google Scholar]
- Wu M., Allis C. D., Richman R., Cook R. G., Gorovsky M. A. An intervening sequence in an unusual histone H1 gene of Tetrahymena thermophila. Proc Natl Acad Sci U S A. 1986 Nov;83(22):8674–8678. doi: 10.1073/pnas.83.22.8674. [DOI] [PMC free article] [PubMed] [Google Scholar]



