Abstract
Molecular pathways that control the specification, migration, and number of available smooth muscle progenitor cells play key roles in determining blood vessel size and structure, capacity for tissue repair and remodeling, and progression of age-related disorders. Defects in these pathways will produce malformations of developing blood vessels, depletion of SMC progenitor pools for vessel wall maintenance and repair, and aberrant activation of alternative differentiation pathways in vascular disease. A better understanding of the molecular mechanisms that uniquely specify and maintain vascular SMC precursors is essential if we are to utilize advances in stem and progenitor cell biology and somatic cell reprogramming for applications directed to the vessel wall.
Keywords: specification, stem cell, embryo, adventitia, differentiation
Introduction
Advances in reprogramming of differentiated adult somatic cells have focused renewed attention on molecular pathways that specify lineage- and cell type-specific progenitor cells 1,2,3. Multiple sources of vascular SMC progenitor cells in embryos have been identified by fate mapping studies thereby allowing for experimental analysis of the mechanisms that uniquely specify these progenitors for a smooth muscle fate 4,5. Moreover, recognition that resident SMC progenitor cells are maintained in adult vessel wall raises important new questions about the roles they play in vascular repair, remodeling and disease 6,7,8,9.
Elucidation of molecular pathways that control cell fate decisions within SMC lineages is of fundamental importance at a basic level. It is also important as a basis for development of progenitor cell therapies applied to the vessel wall. Vascular SMC differentiation in embryonic development is recognized by the appearance of cytoskeletal and contractile protein isoforms, including SMα-actin (Acta2), SM-calponin (Cnn1), and SM-myosin heavy chain (Myh11), that confer a functional smooth muscle contractile phenotype on these cells 10,11,12. No such well-characterized markers are available for identification of SMC progenitors per se that, by definition, are specified for a smooth muscle fate, but do not express differentiated SMC marker proteins. In the absence of established markers that would selectively identify SMC progenitor cells, we recognize these cells by their ability to differentiate to SMCs in vitro or in vivo. However, as discussed further below, misuse of SMC markers and particularly the reliance on SMα-actin (Acta2) expression as a sole criterion for differentiation of a progenitor cell into a SMC is problematic and can lead to the false conclusion that the progenitor cell type being studied has the capacity to produce functional SMCs.
Phenotype Plasticity of Vascular Smooth Muscle
Since the earliest ultrastructural studies of smooth muscle tissue, it has been apparent that SMCs exhibit a wide range of phenotypic variation 13,14,15,16,17. In fact, this diversity was considered for many years to be evidence of not one but two different cell types in the tunica media of artery walls, one for contraction and another for synthesis of extracellular matrix (ECM) proteins 18. In 1979, Chamley-Campbell et al 19 proposed that these two principle functions of arterial medial cells in vivo, ECM synthesis and agonist-induced contraction, were embodied in different but interconvertible “synthetic” and “contractile” SMC phenotypes. SMCs adapted to growth in cell culture, as well as SMCs in developing embryonic vessels and proliferating at sites of vascular injury, exhibit a synthetic phenotype, whereas fully differentiated SMCs in mature adult vessels display a contractile phenotype 16,19. This variation was initially referred to as SMC phenotypic modulation 19,20. More recently the term SMC phenotypic switching has come into common use to describe this reversible transition 11. It is important to realize that the two SMC phenotypes share considerable overlap and that contractile cells can replicate and synthetic cells can possess contractile filaments.
Transcriptional Control of Smooth Muscle Differentiation
Analysis of the requirements for transcription of SMC marker genes, including SMα-actin (Acta2), SM22α (Tagln), SM-calponin (Cnn1), and SM-myosin heavy chain (Myh11), in differentiated SMCs revealed the importance of interactions of serum response factor (SRF; srf), a MADS box-containing DNA binding protein, with a DNA sequence (CC(AT)6GG) known as a CArG box 21,22,23,24. The central MADS domain of SRF (Srf) provides a highly conserved molecular platform for protein-protein interactions and signal-responsive transcriptional regulation that is essential for cytoskeletal organization and SMC differentiation 25,26,27,28. The discovery of the myocardin (Myocd) family of SRF-dependent transcriptional coactivators was a critically important step toward understanding how SMC-selective gene transcription is achieved by combinatorial interactions with the more widely expressed DNA binding protein SRF (Srf) 29,30,31,32,33,34,35,36,37. The identification of transcriptional corepressors and chromatin-associated silencers of SRF/myocardin-dependent gene expression is particularly important for characterization of mechanisms by which SMC progenitors are formed and maintained in vivo 38,12,39. The combinatorial nature of SMC selective transcription provides for rapid and versatile control of SMC phenotype in response to a multitude of environmental cues 29,40. An additional component of SMC identity that is not well characterized is cell type-specific alternative splicing. For example, myocardin is alternatively spliced into two forms; a long form (935 amino acids) that is selectively expressed in cardiac muscle and contains a Mef2-interacting domain and a short form (856 amino acids) that lacks a Mef2 binding motif and is SMC-specific 41. Further work shows that the SMC isoform of myocardin synergistically interacts with Nkx3.1 (Nkx3-1, bagpipe) whereas the cardiac isoform of myocardin does not 42. Vascular SMC-selective alternative splicing has also been reported for α-tropomyosin, metavinculin, smoothelin, and Ca(v)1.2 calcium channel α-subunit. SMC progenitors may also be maintained as such by covalent modifications of either SRF or myocardin. For example, phosphorylation of Ser162 in the MADS domain inhibits DNA binding and transcriptional activity of SRF 28. Moreover, sumoylation of myocardin at Arg445 in a PIAS1-dependent manner was shown to be an activating modification for SRF-mediated myogenesis 43. In addition, myocardin proteins contain a B-box domain similar to that found in Elk1. Competitive binding to SRF via these related B-box domains shifts in favor of Elk1 when growth factors stimulate the MAP kinase pathway and toward myocardin as growth factor stimulation diminishes 44.
Origins of Vascular Smooth Muscle Progenitors
Over the last three decades, fate-mapping studies have identified at least 8 independent origins for vascular SMC progenitors 4,5,45,46. Each of these progenitors has a distinctly different lineage history, yet each produces a similar cell type that transcribes a common set of SMC marker genes (SMαActin (Acta2), SM22α (Tagln), SM-calponin (Cnn1), SM-MHC (Myh11) upon differentiation. How progenitor cells from such different developmental origins become specified for a common smooth muscle fate, and how they are maintained as smooth muscle myoblasts during the migration, proliferation and heterotypic cell-cell interactions required to position them for differentiation around nascent blood vessels is not known (Figure 1). Moreover, in perinatal and adult vessels recent evidence suggests that SMC progenitor cells reside in a privileged signaling domain or niche environment within the tunica adventitia 6,8,9 (Figure 2). Their persistence in this perivascular location throughout adult life suggests that cells and matrix components of the adventitial niche provide important signals that maintain the progenitor phenotype and prevent premature SMC differentiation 9,47. Maintenance of SMC progenitors requires transcriptional silencing of SRF-dependent SMC differentiation marker genes within these different environments. Current evidence suggests two types of pathways function coordinately to prevent premature SMC differentiation and thereby allow for the cell migration and cell division events required to position and expand SMC progenitor cell pools during vessel wall growth and repair. One pathway acts at the level of regional chromatin structure to limit access of SRF and its coactivators to critical paired CArG box elements found in most SMC marker genes 12,48,49,50. A second pathway acts at the level of SRF bound to CArG sequences in SMC target genes to silence gene transcription that would otherwise be activated by the occupancy of SRF and one or more SRF coactivators on SMC gene targets 39,44,51,52.
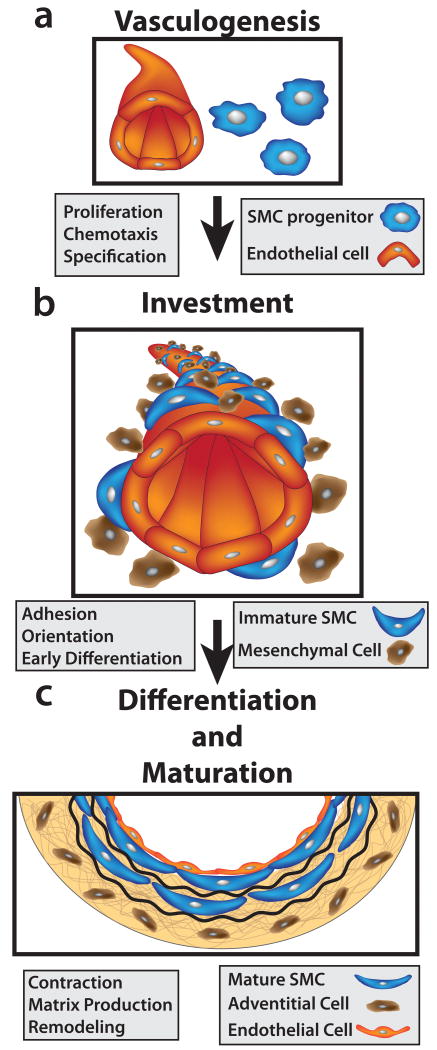
Figure 1. Development of vascular smooth muscle from embryonic progenitors.
(a) Vasculogenesis: Vascular development begins when angioblasts differentiate into endothelial cells (red) that self-assemble into a nascent capillary-like vascular network. (b) Investment: Increasing cardiac output from the developing heart stimulates production of mesenchymal cell chemoattractants by endothelial cells. SMC progenitors (brown) begin to invest the vessel wall around E10.5 in the mouse. Close contact with endothelial cells initiates SMC differentiation (blue cells). The position of the vessel within the embryo determines what type of SMC progenitor will be involved in producing the tunica media. Proliferation of SMC progenitor cells is required to supply numbers of SMCs sufficient for continued vascular development. (c) Differentiation and maturation: As layers of SMCs are added to the developing artery wall, a cross-linked extracellular matrix is formed that defines the structure of the mature tunica media.
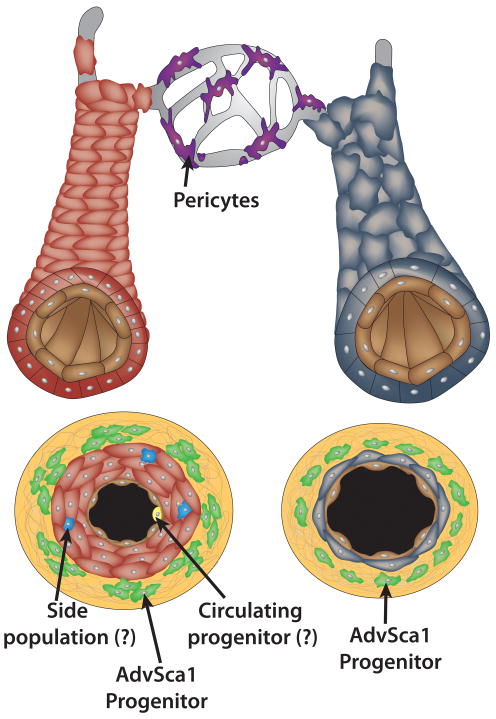
Figure 2. Sources of vascular smooth muscle progenitor cells in adults.
In large arteries (red) and veins (grey), resident SMC progenitors have been identified in the adventitial layer (green cell clusters) 6,7,9 and in the medial layer (scattered blue cells) 180. Some reports suggest that SMCs in atherosclerotic plaques and intimal masses can arise from circulating, bone marrow-derived progenitor cells (yellow) 173,174 while others report finding no evidence to support that possible origin 175,176. In microvessels, pericytes (purple) have been proposed to have multilineage differentiation potentials 162,166, and can act as SMC progenitor cells 159,163.
Epigenetic Controls of Smooth Muscle Differentiation
Genomic DNA is organized into repeating units called nucleosomes consisting of histone octamers wrapped twice by stretches of DNA each an average of 146 bp in length. This compacts genomic DNA about sevenfold 53,54. Eukaryotic gene transcription, however, takes place in the context of nucleosomal DNA that is further compacted into higher-ordered structures collectively known as chromatin. Chromatin is highly dynamic and organized into chromosomal domains that adopt specific conformations and positions within the overall three dimensional structure of the nucleus 55. Reversible covalent modifications of histone tail residues by acetylation, methylation, phosphorylation and ubiquitination or histone tail proteolytic cleavage play critical roles in the control of eukaryotic gene expression and cell fate by modifying accessibility of the chromatin-associated DNA template to sequence-specific DNA binding proteins 56,57,58. Collectively, covalent histone tail modifications together with ATP-dependent DNA conformation-modifying enzymes produce an epigenetic landscape that, to a large extent, determines what genes are available to the general transcriptional machinery for gene expression 54. Essentially all genes, including SRF-dependent SMC differentiation marker genes, operate within this complex three-dimensional network of higher-ordered chromatin structure.
With respect to SRF-DNA interactions in SMCs and their progenitors, current evidence suggests that CArG box elements are more accessible for SRF binding in SMCs than the same DNA sequences are in non-SMCs 59. This would help to explain why SMC marker genes are silent in non-SMCs that nonetheless express SRF together with one or more myocardin-related transcription factors (MRTFs) 30 or other known SRF coactivators 60,61,62,63,64. The covalent modifications of specific histone residues (called histone marks) in SMC compared to non-SMCs are complex (reviewed in 49). In principle, histone acetylation on lysine residues is rapid and reversible and correlates with ongoing rates of gene transcription. The addition of negatively charged acetyl groups is thought to disrupt histone protein-DNA interactions effectively relaxing chromatin structure and thereby enabling increased transcription factor access to binding sites on DNA. Counteracting this activating histone modification is a family of histone deacetylases (HDACs) that catalyze the removal of histone acetyl groups and inhibit gene transcription 65. Recruitment of SRF to CArG elements in SMC promoter/enhancer regions closely correlates with acetylation of lysine residues on histones H3 and H4 59,66,67. Histone acetylation at these loci may be catalyzed by the ability of myocardin to bind and recruit histone acetyltransferases, such as P300 (Ep300) 38,68. Further evidence that transcription of SMC marker genes is enhanced by histone acetylation is the finding that treatment of SMCs with trichostatin A, an HDAC inhibitor, increased endogenous SM22α (Tagln) gene expression in vitro 67. Histone lysine residues can also be methylated 69. A well-characterized methylation of histone H3 at lysine 9 (H3K9) is associated with gene silencing. Like histone acetylation, histone methylation is also reversible. A family of jumonji domain-containing histone demethylases that catalyze the removal of methyl groups from specific histone lysine residues has been identified 70,71. Indeed, Lockman et al reported that the jumonji domain-containing H3K9 histone demethylase, Jmjd1a/JHDM2a, is an MRTF-A (Mkl1) interacting protein that stimulates SMC marker gene expression by demethylating H3K9 residues in SMC target genes 72. Therefore, pathways that control the type and extent of covalent histone modifications on intact chromatin surrounding SMC differentiation target genes are clearly important to identify in future studies to better understand how SMC progenitor cells are formed and maintained in vivo.
Of particular interest for the focus of this review are epigenetic changes found in SMC progenitors that are not found when compared to differentiated SMCs or to progenitors of non-SMC lineages. One promising epigenetic modification that correlates with smooth muscle identity was reported by McDonald et al 59 using an A404 clonal cell line derived from the multipotential teratocarcinoma cell line P19. In the presence of retinoic acid, A404 cells rapidly adopt a SMC phenotype including expression of SM-MHC (Myh11) 66. Employing CArG box mutants in the SMα-actin (Acta2) promoter that cannot bind SRF, McDonald et al showed that failure of SRF binding to DNA led to pronounced decreases in H3K9 acetylation and H3K79 dimethylation most likely because these histone modifications are added to SMC chromatin after SRF binding and transcriptional activation 59. By contrast, H3K4 dimethylation (H3K4me2) was unaffected by the presence or absence of SRF bound to SMα-actin (Acta2) promoter CArG box elements 59. Moreover, chromatin immunoprecipitation (ChIP) assays showed that SRF-myocardin complexes that activate transcription physically associate with H3K4me2 whereas transcription silencing SRF-Elk1 complexes do not. Finally, in the presence of PDGF-BB, a stimulus known to downregulate mature SMC marker gene expression, reductions in SRF bound to H3K4me2 were found 59. These results suggest that H3K4me2 is an epigenetic histone mark that may be an important element of a molecular profile that confers SMC identity onto a progenitor cell.
If true, histone methyltransferases responsible for the dimethyl modification of H3K4 would be key targets of one or more pathways that confer SMC specification on a multipotential progenitor cell in vivo. There are at least two well-characterized classes of histone methyltransferases with this activity, the mixed lineage leukemia (MLL) family and SET1 family 73,74. But these histone methyltransferase enzymes are widely expressed and are not smooth muscle lineage specific. Moreover, they do not possess intrinsic specificity for histone residues on smooth muscle marker chromatin as opposed to chromatin anywhere else in the genome. This suggests that there must be more specific guidance cues that recruit histone methyltransferase-containing complexes to smooth muscle gene targets in progenitor cells. Preliminary findings raise the interesting possibility that a previously identified DNA binding homeodomain protein that associates with SRF and recruits histone acetyltransferase activity to SMC target genes, known as Pitx2 75,76, serves to guide MLL-type histone methyltransferases to SMC target genes 77. While clearly an important objective, identification of these molecular guidance cues for placement of critical histone methyl marks on SMC target gene loci in SMC progenitor cells still leaves open the question of how gene targets so marked are maintained in a transcriptionally silent state until an appropriate time during embryonic development or adult homeostasis for SMC differentiation.
In addition to histone modifications, chromatin structure is also controlled by a family of ATPase-dependent chromatin remodeling enzymes that are key components of the SWI/SNF complex 78. In vertebrates, the two major ATPase subunits of the SWI/SNF complex are Brahma (Brm1) and Brahma-related gene 1 (Brg1). Zhang et al reported that the myocardin family member, MRTFA (Mkl1, Mal), recruits Brg1 to SRF-dependent SMC marker gene targets to promote gene transcription 50. The SW13 cell line, deficient in both Brg1 and Brm1, was unable to support MRTFA-mediated increases in SRF-dependent SMC marker gene activity 50. Chromatin immunoprecipitation (ChIP) assays showed that dominant negative forms of Brg1 strongly inhibited the MRTFA-enhanced SRF binding to promoter regions of SMC marker genes without affecting SRF binding to CArG elements in immediate early genes such as c-fos. Similar results were reported for effects of Brg1 interaction with myocardin (Myocd) on SMC target gene expression 79. Therefore mechanisms to limit the expression or function of ATPase-dependent chromatin remodeling complexes at SMC differentiation marker genes may be important elements in the maintenance of transcriptional silencing in SMC progenitor cells.
Silencing of SMC Differentiation Marker Genes in SMC Progenitors
When considering how SMC progenitors are formed and maintained, it is important to realize that SRF and CArG box chromatin interacts not only with potent coactivators like myocardin but also with potent transcriptional silencer complexes as well 39,80. For example, the muscle segment homeobox proteins (Msx1 and Msx2) form ternary complexes with SRF (Srf) and myocardin (Myocd) that block DNA binding of the SRF-myocardin complex to CArG box motifs in SMC marker genes 80. Another example is KLF4 (Klf4), a zinc finger protein 81 that binds to conserved GC-rich elements located near paired CArG boxes in the SMα-actin (Acat2) promoter and other SMC marker genes 82. Klf4 physically binds to SRF, recruits HDAC2 (Hdac2) and HDAC5 (Hdac5) to SMC marker genes, and blocks SRF association with CArG box sequences in intact chromatin 52. The forkhead transcription factor FoxO4 (Foxo4) physically interacts with myocardin and inhibits its SRF-coactivator function thus acting as a repressor for SMC differentiation 51. In response to signals that activate the PI3-kinase/Akt pathway, Foxo4 is exported from the nucleus, and SMC differentiation target genes become transcriptionally active. The co-repressors Msx1, Klf4 and Foxo4 were found to be coexpressed with SRF and myocardin in SMC progenitor cells obtained from neonatal aortic adventitia 9. More recently, the HMG box-containing protein HMG2L1 was shown to physically interact with myocardin and abolish the binding of myocardin-SRF complexes to CArG box elements in SMC promoters. Overexpression of HMG2L1 in SMCs repressed SMC marker gene expression whereas depletion of endogenous HMG2L1 increased expression of the same SMC marker genes 83.
PRISM (PR domain in smooth muscle) (Prdm6) is a PR and SET domain-containing protein with transcriptional repressor activity that is selectively expressed in smooth muscle tissues 84. It is expressed as early as E11.5 in the aortic wall corresponding to initial events in tunica media formation 84. It is also expressed in developing airway, tracheal and bladder smooth muscle. PRISM is capable of recruiting histone methyltransferases and other chromatin remodeling enzymes including class I HDACs through interactions of these factors with its PR domain and thereby mediate transcriptional repression in SMC progenitors. Among the gene targets whose expression in SMCs is repressed by PRISM are Gata6 and myocardin (Myocd). Indeed, siRNA-mediated knockdown of PRISM produced a concomitant induction of SMC differentiation marker genes, including SM-MHC, suggesting some level of constitutive repressor activity is maintained by PRISM in differentiated SMCs. Other members of the PR/SET domain family are reported to play important roles in the specification of neural crest progenitors 85 and slow-twitch skeletal muscle fibers 86.
Control of cell differentiation by transcriptional corepressors plays important roles in progenitor cell maintenance in skeletal muscle and other progenitor cell types 87. For example, skeletal muscle myoblasts are maintained, in part, by a MEF2-dependent corepressor complex that normally silences muscle-specific gene expression 88. This complex consists of MEF2 (Mef2), MITR (Mef2-interacting transcriptional repressor), HP1 (heterochromatin interacting protein-1), Hdac4 and Hdac5 89. Activation of calcium/calmodulin-dependent protein kinase leads to phosphorylation of Hdac4 and Hdac5 resulting in nuclear export of these transcriptional repressors 90. The Mef2 released from phosphorylated HDACs associates with MyoD (Myod1) and E-protein heterodimers resulting in transcriptional activation of skeletal muscle structural genes. Mef2, like Srf, is a MADS box-containing DNA binding protein that provides a platform for protein-protein interactions in skeletal myoblasts. Another important class of negative transcriptional regulators is the Groucho/TLE gene family, which consists of four unlinked genes named TLE1-4 in humans and Grg1-4 in mouse 91. Grg/TLE proteins interact with engrailed homology-1 (EH1) domain-containing DNA binding proteins and recruit class II HDACs to chromatin via a glycine and proline-rich domain in the N-terminal half of Grg/TLE proteins 92. EH1 domain proteins important in vascular development include the forkhead box transcription factors of the FoxA, FoxC and FoxD families, T-box proteins, and muscle-segment homeobox proteins of the Msx family 93.
Roles for Noncoding RNA in Formation and Fate of SMC Progenitors
A growing number of studies now indicate the important roles played by noncoding RNAs 94,95 in development and differentiation of vascular smooth muscle 96. For example, maintenance of fully differentiated phenotypes of vascular SMCs is dependent on expression of microRNA-143 (miR-143) and miR-145 97,98,99,100,101. miRNAs are a class of ∼ 22-nucleotide noncoding small RNAs that play essential roles in regulating gene expression by posttranscriptional mechanisms including arrest of translation and degradation of mRNAs 95. Loss of function for miR-143/145 results in arteries with thinner walls and SMCs with a noticeable lack of differentiated features. Systemic blood pressure and smooth muscle contractile activity are reduced in miR-143/145-deficient mice 100, as is the ability to migrate due to disarray of actin stress fibers 99. Expression of miR-145 alone was sufficient to convert multipotent neural crest cells into vascular SMCs suggesting that miR-145 targets are important for maintaining the SMC progenitor phenotype in the neural crest lineage 97. Likewise, overexpression of miR-145 was able to substantially rescue the vascular SMC defects resulting from the loss of dicer, an endonuclease required for miRNA synthesis 95, in SMCs 102. Identified targets of miR-143/145 include two gene products that are known repressors of SRF-myocardin-induced transcription, namely Elk1 and Klf4, as well as the potent SRF coactivator MRTF-B (Mkl2) 97,99. In addition, miR-145 has been shown to downregulate expression of Klf5, a factor associated with repression of the mature differentiated SMC phenotype 99,101. Conversely, miR-221 and miR-222 down-regulate factors that promote expression of the differentiated SMC phenotype thus leading to dedifferentiation of SMCs 103,104. These two miRNAs are induced by PDGF-BB and target myocardin (Myocd) transcripts for down-regulation 104.
Recent studies suggest the paradoxical idea that gene silencing requires transcription initiation 105. Thus, current evidence suggests that long noncoding RNA molecules (lncRNAs) recruit transcriptional repressors to specific sites in the genome to silence gene expression. For example, mammalian X chromosome-inactivation requires noncoding Xist RNA that forms a hairpin structure that recruits polycomb group repressor complexes (PRC2) to the X inactivation center to silence gene transcription 106,107. Likewise, a noncoding RNA transcript from the INK4b/ARF locus directs repression of that locus by recruiting PRC2 silencer complexes 108. Most promoters are now known to produce short transcripts in both 5′ and 3′ orientations that encompass CpG islands frequently found in promoter regions 109. Short hairpin structures produced by these non-coding transcripts recruit PRC2 repressor complexes that spread across a local region of DNA, catalyze trimethylation of histone H3K27 (H3K27me3) and silence gene expression. Activation of transcription from this repressed locus requires demethylation of H3K27me3 by histone demethylase activity 110. As discussed above, expression of a jumonji domain-containing H3K9 histone demethylase, Jmjd1a/JHDM2a, in SMCs is associated with transcriptional upregulation of smooth muscle differentiation marker gene expression 72. More recently a positive role for lncRNAs acting as transcriptional enhancers was reported suggesting that regulatory functions for this important class of noncoding RNAs will likely turn out to be complex and interesting 111. A role for lncRNAs in control of cell fate in general, and SMC fate in particular, is an important area for future work.
Signal-Responsive Differentiation of SMC Progenitors
As discussed above, SMC progenitor cells are poised to differentiate to SMCs but are restrained from doing so by redundantly acting repressor/silencer mechanisms. Reductions in the activity of SRF-dependent corepressor complexes or increases in the activity of SRF-dependent coactivator complexes are sufficient to trigger SMC differentiation in these cells. A principle pathway for formation of SRF-coactivator complexes involves extracellular signal-dependent activation of RhoA-GTPase (Rhoa) and its downstream effectors Rho kinase (Rock1), LIM kinase (Limk1), protein kinase N (Pkn1), and mammalian diaphanous (mDia) proteins. Activation of Rock1 and Limk1-dependent signaling leads to actin polymerization, stress fiber formation and cytoskeletal reorganization 112,113,114. Activated Rock1 is reported to translocate to the nucleus, phosphorylate P300 and promote SMC gene transcription 115,116,117. Of particular importance was the finding by Treisman's group that Rhoa- GTPase-mediated actin treadmilling, specifically the polymerization of G-actin to F-actin, results in the mobilization of potent SRF coactivators MAL/MRTFs/MKLs from inhibitory binding sites on G actin followed by their translocation to the nucleus 32,118,119. Accumulation of MRTFs in the nucleus and their partnering with SRF and other coactivators 60,61,62,63,64 leads to transcriptional activation of SMC target genes and SMC differentiation by mechanisms discussed above. It is reasonable to assume that in actively migrating SMC progenitors, repetitive actin filament turnover during extension of lamellipodial and filopodial projections constantly regenerates cytoplasmic G actin and maintains nuclear MRTF activities at low levels. Once these cells reach their destination and take up positions in developing tunica media, F-actin predominates and MRTFs are found in their active nuclear forms. This mechanism would couple morphogenesis of blood vessel walls with differentiation of a major constituent cell type (SMC) present within those walls. For example, conditional deletion of integrin-linked kinase (Ilk) produces overactive Rhoa and Rock1-mediated signaling in SMC progenitors resulting in dermal arterioles with smaller diameters and loss of circumferential SMC alignment consistent with premature differentiation, excessive contraction and lack of proliferative expansion of SMC progenitor pools 120. These findings reinforce the idea that SMC progenitor pool sizes are critical determinants of blood vessel size, length, and function. The relative state of cytoskeletal actin polymerization, and thus levels of active MRTFs, depends upon Rhoa-GTPase-mediated signaling pathways that respond to factors present in the local environment thus emphasizing the importance of niche-dependent signaling for the maintenance of a SMC progenitor phenotype 9,47.
Smooth Muscle Progenitors and the Embryonic Vascular System
Genetic approaches to developmental fate mapping reveal the intricate diversity of vascular SMC lineages in development, but they leave us wondering how progenitor cells of such diverse embryonic origins and developmental histories can differentiate into a common cell type 4,5,46. For example, the dorsal surface of the neural tube contains progenitor cells that migrate into the pharyngeal arch complex between E8.5 and E9.5 and produce SMCs that form the walls of the great arteries 45,121,122,123. Clonal analysis ex vivo shows that individual neural crest progenitor cells are multipotent 124,125. Thus factors in the environment through which these cells migrate play essential roles in specifying their fate 126,127,128. SMCs in the descending aorta originate from progenitors in epithelial somites that express Pax3 and FoxC2 5,129,130. SMC fate specification occurs when expression levels of FoxC2 exceed those of the promyogenic factor Pax3 130 and since FoxC2 represses transcription of Pax3, a SMC fate is stabilized over the alternative skeletal muscle fate. The abundance ratio of FoxC2 and Pax3 is controlled by signals in the somite microenvironment 130 and thus resembles SMC fate specification in cardiac neural crest progenitors during delamination and migration from the neural tube. A likely scenario based on current data (Figure 1) is that SMC progenitor cells thus specified make contact with endothelial cells 131, engage notch-dependent signaling pathways 132,133,134,135,136, mobilize MRTF-B (Mkl2), and initiate SMC differentiation 137,138. It is important to point out that the requirement for MRTF-B (Mkl2) is restricted to cardiac neural crest-derived SMCs indicating that SMC progenitors originating from other sources in the embryo must have different molecular requirements for SMC differentiation even though the same SMC differentiation marker genes are activated in all cases 137,138. Indeed, analysis of MRTF-A (Mkl1, Mal)-deficient mice revealed that neural crest-derived vascular SMCs differentiated normally whereas myoepithelial cells of mammary gland ductal tissues failed to develop a contractile phenotype 139.
SMCs in the coronary vasculature arise from a separate population of embryonic progenitors. The majority of coronary SMCs (CoSMCs) can be traced back to origins in the proepicardium (PE), a transient collection of mesothelial cells that appears at the sinoatrial junction at about E8.5 in the mouse 140,141,142. Specification of lateral plate mesoderm for a proepicardial fate is mediated by an antagonistic interplay between BMP and FGF signaling 143,144. In avian embryos, soluble factors produced by the developing liver bud, but not lung bud, direct multipotent mesoderm progenitors to adopt a proepicardial identity 145. In zebrafish embryos, an early role for Tbx5 in specifying PE progenitor cells was identified that results in competence to respond to BMP4 (Bmp4) released by the developing myocardium 146. PE cells thus specified reach the heart around E9.5 in the mouse and grow out over the surface of the myocardium to form a single layer of epicardial cells. Then around E13.5 to E14.5 some epicardial cells undergo an epithelial to mesenchymal transition (EMT), loose their epicardial phenotype and adopt a pre-SMC phenotype in the subepicardium in vivo 142,147,148,149. Epicardial cell EMT is associated with a Rhoa-GTPase and Rock1-mediated cytoskeletal actin reorganization that is required for CoSMC differentiation 115,150. Epicardial cells from adult hearts appear to retain their specification to differentiate into CoSMCs, at least in vitro 151. Maintenance of a CoSMC progenitor phenotype in the epicardium may be due, in part, to high levels of epicardial expression of EH1 domain-containing T-box proteins including Tbx5 and Tbx18 152,153,154.
In addition to cardiac neural crest and proepicardium, a third distinct source of SMC progenitor cells in the early embryo is found in an Islet-1 (Isl1)-positive cell population that contributes multiple lineages to the developing heart 155. Isl1 is a LIM-homeobox transcription factor that marks the second heart field of cardiac progenitors and is required for formation of the atria, right ventricle, and cardiac outflow tract 156. Genetic fate mapping studies using Isl1-cre to mark early cardiac progenitor cells in the mouse showed that SMCs in the walls of the aortic root, pulmonary trunk and coronary stems are produced from Isl1-positive progenitors that are distinct from progenitors in the cardiac neural crest. Also labeled by Isl1-cre were endothelial cells, endocardial cells, and myocardial cells of the right ventricle suggesting that the progenitor population is multipotential 155. Clonal analysis of Isl1+ progenitor cells verified their unique multilineage potential 155,157. Similar results were reported by Kattman et al who isolated VEGF-R2/Flk1-positive progenitor cells from head-fold stage mouse embryos and showed that single progenitor cells generated colonies that contained cardiac myocytes, endothelial cells and vascular SMCs thus confirming their multilineage potential 158.
Smooth Muscle Progenitors and the Adult Vascular System
Pericytes are smooth muscle-like microvascular mural cells with progenitor-like properties (Figure 2). Proper investment of microvessel walls with pericytes is a required step in vascular development and angiogenesis 159,160. However, little is known about the developmental origins of pericytes as very few lineage-mapping studies have addressed whether or not pericytes share common origins in the embryo with vascular SMCs. One exception to this is in the brain where pericytes and cerebral vascular SMCs have been shown to originate from cephalic neural crest progenitor cells 161. Many pericytes exhibit the potential for multilineage mesenchymal cell differentiation 162,163. Pericytes are classically defined as cells embedded underneath the basal lamina of microvascular endothelial cells. This position is strikingly similar to that of muscle stem cells known as satellite cells that reside under the basal lamina of skeletal muscle myofibers. In both cases, the position of resident pericytes and satellite cells under a basal lamina may be important to efficiently receive signals released from endothelial cells or myofibers respectively that maintain their progenitor phenotypes. Cossu and coworkers have shown that pericytes isolated from injured skeletal muscle tissue and injected intra-arterially have the remarkable ability to home to injured skeletal muscle, reconstitute satellite cell pools, form skeletal muscle myofibers, and promote muscle regeneration in vivo 164,165,166. This property may reflect an origin of some pericytes from mesoangioblasts, a population of multipotential mesenchymal progenitor cells that appear early in vascular development as VEGFR2/Flk1-positive cells associated with the abluminal side of the dorsal aorta 167.
The possibility that vascular SMC progenitor cells reside in the outer layer of artery wall, the adventitia and associated perivascular tissues, has been suggested several times over the years (Figure 2) 168,169. For most investigators, these suggestions were regarded as anecdotal and data consistent with a role for the adventitia as harboring vascular progenitor cells was largely ignored. However, in a survey of adult vessels in ApoE-/- mice, Hu et al reported finding SMC marker-negative, stem cell antigen-1 (Sca1; Ly6a)pos progenitor cells in the adventitia of large arteries 6. When isolated from genetically-marked animals and put back on the adventitial side of vein grafts, adventitia-derived Sca1pos cells were found in the media at 2 weeks and in the neointima at 4 weeks after transplantation where they no longer expressed Sca1 and became immunopositive for SMC differentiation markers 6. Similarly, a population of CD34pos/PECAM1neg cells was found in human internal thoracic artery with a capacity to form capillary-like microvessels in ex vivo aortic ring assays 7. The concentration of these cells in the inner adventitia led Zengin et al to refer to this location as a “vasculogenic zone” in the artery wall 7. More recently, a novel domain of sonic hedgehog (Shh) signaling was described that is restricted almost entirely to the adventitia of large and medium-sized arteries and veins, and colocalizes with the vasculogenic zone described by Zengin et al 9. Within this Shh signaling domain a population of Sca1pos/CD34pos progenitor cells was found with the capacity to differentiate into mural cells (pericytes and SMCs) in vitro and promote angiogenesis in matrigel implants in vivo 9. Another factor implicated in an adventitial microenvironment for SMC progenitors is stromal cell-derived factor 1-alpha (SDF-1α; Cxcl12) 170. SMC-specific deletion of PTEN (Pten), a dual-specificity lipid and protein phosphatase, increased production of SDF-1α by Pten-deficient SMCs and resulted in accumulation of CXCR4-positive circulating progenitor cells in a perivascular, adventitial location 170. These reports are particularly interesting in light of past studies that suggest roles for adventitial cells in artery wall thickening in pulmonary hypertension 8 and in neointimal formation after vascular injury 168. For example, animals exposed to chronic hypoxia develop markedly thickened pulmonary artery walls (reviewed in 8). This form of hypertensive remodeling is characterized by additional layers of smooth muscle forming on the adventitial side of the pulmonary artery wall and may well consist of SMCs that originate from local progenitors resident in the adventitia 8. Likewise, mice that are haploinsufficient for tropoelastin (Eln) exhibit additional layers of SMCs and elastin that form on the adventitial side of the aorta during late stages of embryogenesis around E16.5 to postnatal day 3 171,172. Whether SMC progenitor cells in the adventitia can detect mechanical stretch of the artery wall, or changes in relative hypoxia, or respond to soluble signals associated with wall remodeling stimuli are important questions for future studies.
Experiments to determine if circulating progenitor cells of bone marrow origin contribute intimal SMCs to atherosclerotic plaques were initially interpreted as supporting such an origin 173,174. In those early studies, up to 50% of intimal SMCs within mouse atherosclerotic plaques were reported to be derived from progenitors of bone marrow origin 174. However, as work progressed on this possibility, an origin for intimal SMCs from marrow-derived progenitors seemed less likely 175,176. A large influx of marrow-derived inflammatory cells, mostly monocytes and macrophages, occurs early in lesion formation. High-resolution confocal imaging is required to determine if the marker used to detect cells of bone marrow origin is actually coexpressed with markers used to identify intimal SMCs 175. This is not trivial as intimal SMCs and inflammatory cells are often in very close proximity, if not actual physical contact, during these early time points. In addition, common methods for marrow-derived mononuclear cell isolation lead to their contamination with platelet membrane fragments (microparticles, MPs) carrying markers used to identify other cell types (e.g, CD31, endothelial cells)177. By extrapolation, inflammatory cell MPs may be shed and incorporated into nearby SMCs leading to false conclusions about their origins. Upon extending the time course of experiments out to 16 weeks after wire injury to femoral artery, Daniel et al clearly showed that bone marrow-derived cells of any type were dramatically reduced in numbers while intimal cells expressing smooth muscle markers continued to increase 176. By 16 weeks after injury there were very few, if any, cells that coexpressed the bone marrow lineage marker and SMC specific proteins. Taken together, the current data suggests that inflammatory cells contribute an early paracrine activity that diminishes greatly with time, and that there is little, if any, long-term contribution of marrow-derived progenitor cells to the vascular SMC population in these vessels.
Myofibroblasts are derived from resident tissue fibroblasts and are found in abundance in a number of different reactive and pathogenic conditions 178. Myofibroblasts are present in granulation tissue during wound healing, in connective tissue stroma surrounding solid tumors, and are abundant in fibrotic tissue. Because myofibroblasts express some of the commonly used SMC markers, such as SMα-actin (Acta2) and SM22α (Tagln) it is frequently assumed that these cells are SMC progenitors that can go on to complete a differentiation sequence and become SMCs. In fact, although SMCs and myofibroblasts both express the SMα-actin (Acta2) gene, they use distinct molecular mechanisms to do so 77. For example, analysis of MCAT element mutations in the SMα-actin promoter/enhancer in transgenic mice showed that myofibroblasts within granulation tissue of skin wounds required intact MCAT elements for reporter gene expression whereas vascular SMCs as well as SMCs of the stomach, bladder and intestine did not 77. Moreover, different TEF-1 (Tead1) family members associate with MCAT elements in myofibroblasts (RTEF-1; Tead4) compared to SMCs (TEF-1; Tead1) 77. In addition, myofibroblasts differ from SMCs in an absence of expression of smoothelins, relatively late stage differentiation markers for SMCs 178,179. Thus activation of SMC markers in myofibroblasts occurs by distinct molecular pathways compared to SMCs, arguing that resident tissue fibroblasts or myofibroblasts are not SMC progenitors under most conditions.
Summary
The number of different smooth muscle types far exceeds that of skeletal or cardiac muscle. This diversity reflects a multiplicity of different kinds of SMC progenitors found in embryonic and adult tissues. It also reflects the versatility of a common molecular mediator for SMC differentiation, a complex of SRF and a myocardin family member occupying one or more CArG box cis-elements. Through interactions with a large number of co-activators and co-repressors that recruit epigenetic regulators of regional chromatin structure, this core transcription module is made responsive to a wide variety of extracellular signaling cues that specify and direct cell fate. Maintenance of SMC progenitor pools requires signals that allow for proliferative expansion during tissue growth or repair together with activation of redundant transcriptional silencing mechanisms to maintain the SMC progenitor phenotype and prevent SMC differentiation. A more complete understanding of the identity of these signals and the molecular mechanisms that are utilized to form and maintain vascular SMC progenitor cells will enable advances in somatic cell reprogramming and stem cell biology to be more effectively applied to disorders of the vessel wall.
Acknowledgments
We acknowledge helpful discussions with our colleagues DaZhi Wang, Joseph Miano, and Robert J. Schwartz.
Sources of Funding: This work was supported by National Institutes of Health Grants HL-93594 and HL-19242 (to MWM), by American Heart Association Fellowships 09PRE2240020 (to JNR) and 09PRE2060165 (to VJH), the Curriculum in Genetics & Molecular Biology Graduate Program, University of North Carolina, Chapel Hill, NC, and the Seattle Children's Research Institute, Seattle, WA.
Non-standard Abbreviations and Acronyms
- ACLP
aortic carboxypeptidase-like protein
- APEG
aorta preferentially-expressed gene
- BMP
bone morphogenic protein
- CArG
(CC(AT)6GG)
- ChIP
chromatin immunoprecipitation
- CoSMC
coronary smooth muscle cell
- En
embryonic day n
- ECM
extracellular matrix
- Flk1
fetal liver kinase-1
- H3K4me2
histone H3 lysine-4 dimethyl modified
- HAT
histone acetyltransferase
- HDAC
histone deacetylase
- HP1
heterochromatin interacting protein-1
- MADS
mcm-agamous-deficiens-srf
- miRNA
micro-RNA
- MKL
megakaryoblastic leukemia
- MLL
mixed lineage leukemia
- MRTF
myocardin-related transcription factor
- PIAS1
protein inhibitor of activated STAT-1
- PRC2
polycomb group repressor complex 2
- Rock1
rho kinase-1
- SCA1
stem cell antigen-1
- siRNA
small interfering RNA
- SMC
smooth muscle cell
- SRF
serum response factor
- VEGF-R2
vascular endothelial cell growth factor receptor-2
Footnotes
Disclosures: None.
References
- 1.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 2.Zhou Q, Brown J, Kanarek A, Rajagopal J, Melton D. In vivo reprogramming of adult pancreatic exocrine cells to beta-cells. Nature. 2008;455:627–632. doi: 10.1038/nature07314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ieda M, Fu J, Delgado-Olguin P, Vedantham V, Hayashi Y, Bruneau B, Srivastava D. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell. 2010;142:375–386. doi: 10.1016/j.cell.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Majesky M. Developmental basis of vascular smooth muscle diversity. Arterioscl Thromb Vasc Biol. 2007;27:1248–1258. doi: 10.1161/ATVBAHA.107.141069. [DOI] [PubMed] [Google Scholar]
- 5.Wasteson P, Johansson B, Jukkola T, Breuer S, Akyurek L, Partanen J, Lindahl P. Developmental origin of smooth muscle cells in the descending aorta in mice. Development. 2008;135:1823–1832. doi: 10.1242/dev.020958. [DOI] [PubMed] [Google Scholar]
- 6.Hu Y, Zhang Z, Torsney E, Afzal A, Davison F, Metzler B, Xu Q. Abundant progenitor cells in the adventitia contribute to atherosclerosis of vein grafts in ApoE-deficient mice. J Clin Invest. 2004;113:1258–1265. doi: 10.1172/JCI19628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zengin E, Chalajour F, Gehling U, Ito W, Treede H, Lauke H, Weil J, Reichenspurner H, Kilic N, Ergun S. Vascular wall resident progenitor cells: a source for postnatal vasculogenesis. Development. 2006;133:1543–1551. doi: 10.1242/dev.02315. [DOI] [PubMed] [Google Scholar]
- 8.Stenmark K, Davie N, Frid M, Gerasimovskaya E, Das M. Role of the adventitia in pulmonary vascular remodeling. Physiology. 2006;21:134–145. doi: 10.1152/physiol.00053.2005. [DOI] [PubMed] [Google Scholar]
- 9.Passman J, Dong X, Wu S, Maguire C, Hogan K, Bautch V, Majesky M. A sonic hedgehog signaling domain in the arterial adventitia supports resident Sca1+ smooth muscle progenitor cells. Proc Natl Acad Sci U S A. 2008;105:9349–9354. doi: 10.1073/pnas.0711382105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miano J, Cserjesi P, Ligon K, Periasamy M, Olson E. Smooth muscle myosin heavy chain exclusively marks the smooth muscle lineage during mouse embryogenesis. Circ Res. 1994;75:803–812. doi: 10.1161/01.res.75.5.803. [DOI] [PubMed] [Google Scholar]
- 11.Owens G, Kumar M, Wamhoff B. Molecular regulation of vascular smooth muscle cell differentiation in development and disease. Physiol Rev. 2004;84:767–801. doi: 10.1152/physrev.00041.2003. [DOI] [PubMed] [Google Scholar]
- 12.Miano J, Long X, Fujiwara K. Serum response factor: master regulator of the actin cytoskeleton and contractile apparatus. Am J Physiol Cell Physiol. 2007;292:C70–C81. doi: 10.1152/ajpcell.00386.2006. [DOI] [PubMed] [Google Scholar]
- 13.Pease D, Paule W. Electron microscopy of elastic arteries: The thoracic aorta of the rat. J Ultrastruct Res. 1960;3:469–483. doi: 10.1016/s0022-5320(60)90023-x. [DOI] [PubMed] [Google Scholar]
- 14.Fritz K, Jarmolych J, Daoud A. Association of DNA synthesis and apparent dedifferentiation of aortic smooth muscle cells in vitro. Exp Mol Pathol. 1970;12:354–362. doi: 10.1016/0014-4800(70)90066-3. [DOI] [PubMed] [Google Scholar]
- 15.Poole J, Cromwell S, Benditt E. Behavior of smooth muscle cells and formation of extracellular structures in the reaction of arterial walls to injury. Am J Pathol. 1971;62:391–414. [PMC free article] [PubMed] [Google Scholar]
- 16.Chamley J, Campbell G, Burnstock G. Dedifferentiation, redifferentiation and bundle formation of smooth muscle cells in tissue culture: the influence of cell number and nerve fibres. J Embryol Exp Morph. 1974;32:297–323. [PubMed] [Google Scholar]
- 17.Gerrity R, Cliff W. The aortic tunica media of the developing rat: I. Quantitative stereologic and biochemical analysis. Lab Invest. 1975;32:585–600. [PubMed] [Google Scholar]
- 18.Wissler R. The arterial medial cell, smooth muscle or multifunctional mesenchyme? J Atheroscl Res. 1968;8:201–213. doi: 10.1016/s0368-1319(68)80056-0. [DOI] [PubMed] [Google Scholar]
- 19.Chamley-Campbell J, Campbell G, Ross R. The smooth muscle cell in culture. Physiol Rev. 1979;59:1–61. doi: 10.1152/physrev.1979.59.1.1. [DOI] [PubMed] [Google Scholar]
- 20.Gabbiani G, Schmid E, Winter S, Chaponnier C, De Chastonay C, Vandekerckhove J, Weber K, Franke W. Vascular smooth muscle cells differ from other smooth muscle cells: predominance of vimentin filaments and a specific type of actin. Proc Natl Acad Sci U S A. 1981;78:298–300. doi: 10.1073/pnas.78.1.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Treisman R. Identification of a protein-binding site that mediates transcriptional response of the c-fos gene to serum factors. Cell. 1986;46:567–574. doi: 10.1016/0092-8674(86)90882-2. [DOI] [PubMed] [Google Scholar]
- 22.Norman C, Runswick M, Pollock R, Triesman R. Isolation and properties of cDNA clones encoding SRF, a transcription factor that binds to the c-fos serum response element. Cell. 1988;55:989–1003. doi: 10.1016/0092-8674(88)90244-9. [DOI] [PubMed] [Google Scholar]
- 23.Belaguli N, Schildmeyer L, Schwartz R. Organization and myogenic restricted expression of the murine serum response factor gene: a role for autoregulation. J Biol Chem. 1997;272:18222–18231. doi: 10.1074/jbc.272.29.18222. [DOI] [PubMed] [Google Scholar]
- 24.Miano J. Serum response factor: toggling between disparate programs of gene expression. J Mol Cell Cardiol. 2003;35:577–593. doi: 10.1016/s0022-2828(03)00110-x. [DOI] [PubMed] [Google Scholar]
- 25.Pellegrini L, Tan S, Richmond T. Structure of serum response factor core bound to DNA. Nature. 1995;376:490–498. doi: 10.1038/376490a0. [DOI] [PubMed] [Google Scholar]
- 26.Hassler M, Richmond T. The B-box dominates SAP-1-SRF interactions in the structure of the ternary complex. EMBO J. 2001;20:3018–3028. doi: 10.1093/emboj/20.12.3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schratt G, Philippar U, Berger J, Schwarz H, Heidenreich O, Nordheim A. Serum response factor is crucial for actin cytoskeletal organization and focal adhesion assembly in embryonic stem cells. J Cell Biol. 2002;156:737–750. doi: 10.1083/jcb.200106008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Iyer D, Chang D, Marx J, Wei L, Olson E, Parmacek M, Balasubramanyam A, Schwartz R. Serum response factor MADS box serine-162 phosphorylation switches proliferation and myogenic gene programs. Proc Natl Acad Sci U S A. 2006;103:4516–4521. doi: 10.1073/pnas.0505338103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang D, Chang P, Wang Z, Sutherland L, Richardson J, Small E, Krieg P, Olson E. Activation of cardiac gene expression by myocardin, a transcriptional cofactor for serum response factor. Cell. 2001;105:851–862. doi: 10.1016/s0092-8674(01)00404-4. [DOI] [PubMed] [Google Scholar]
- 30.Wang D, Li S, Hockemeyer D, Sutherland L, Wang Z, Schratt G, Richardson J, Nordheim A, Olson E. Potentiation of serum response factor activity by a family of myocardin-related transcription factors. Proc Natl Acad Sci USA. 2002;99:14855–14860. doi: 10.1073/pnas.222561499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen J, Kitchen C, Streb J, Miano J. Myocardin: A component of a molecular switch for smooth muscle differentiation. J Mol Cell Cardiol. 2002;34:1345–1356. doi: 10.1006/jmcc.2002.2086. [DOI] [PubMed] [Google Scholar]
- 32.Miralles F, Posern G, Zaromytidou A, Treisman R. Actin dynamics control SRF activity by regulation of its coactivator MAL. Cell. 2003;113:329–342. doi: 10.1016/s0092-8674(03)00278-2. [DOI] [PubMed] [Google Scholar]
- 33.Selvaraj A, Prywes R. Megakaryoblastic leukemia-1/2, a transcriptional co-activator of serum response factor, is required for skeletal myogenic differentiation. J Biol Chem. 2003;278:41977–41987. doi: 10.1074/jbc.M305679200. [DOI] [PubMed] [Google Scholar]
- 34.Du K, Ip H, Li J, Chen M, Dandre F, Yu W, Lu M, Owens G, Parmacek M. Myocardin is a critical serum response factor cofactor in the transcriptional program regulating smooth muscle cell differentiation. Mol Cell Biol. 2003;23:2425–2437. doi: 10.1128/MCB.23.7.2425-2437.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yoshida T, Sinha S, Dandre F, Wamhoff B, Hoofnagle M, Kremer B, Wang D, Olson E, Owens G. Myocardin is a key regulator of CArG-dependent transcription of multiple smooth muscle marker genes. Circ Res. 2003;92:856–864. doi: 10.1161/01.RES.0000068405.49081.09. [DOI] [PubMed] [Google Scholar]
- 36.Teg Pipes G, Creemers E, Olson E. The myocardin family of transcriptional coactivators: versatile regulators of cell growth, migration, and myogenesis. Genes Dev. 2006;20:1545–1556. doi: 10.1101/gad.1428006. [DOI] [PubMed] [Google Scholar]
- 37.Parmacek M. Myocardin-related transcription factors: critical coactivators regulating cardiovascular development and adaptation. Circ Res. 2007;100:633–644. doi: 10.1161/01.RES.0000259563.61091.e8. [DOI] [PubMed] [Google Scholar]
- 38.Cao D, Wang Z, Zhang C, Oh J, Xing W, Li S, Richardson J, Wang D, Olson E. Modulation of smooth muscle gene expression by association of histone acetyltransferases and deacetylases with myocardin. Mol Cell Biol. 2005;25:364–376. doi: 10.1128/MCB.25.1.364-376.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kawai-Kowase K, Owens G. Multiple repressor pathways contribute to phenotypic switching of vascular smooth muscle cells. Am J Physiol Cell Physiol. 2007;292:C59–C69. doi: 10.1152/ajpcell.00394.2006. [DOI] [PubMed] [Google Scholar]
- 40.Wang D, Olson E. Control of smooth muscle development by the myocardin family of transcriptional coactivators. Curr Opin Genet Dev. 2004;14:558–566. doi: 10.1016/j.gde.2004.08.003. [DOI] [PubMed] [Google Scholar]
- 41.Creemers E, Sutherland L, Oh J, Barbosa A, Olson E. Coactivation of MEF2 by the SAP domain proteins myocardin and MASTR. Mol Cell. 2006;23:83–96. doi: 10.1016/j.molcel.2006.05.026. [DOI] [PubMed] [Google Scholar]
- 42.Imamura M, Long X, Nanda V, Miano J. Expression and functional activity of four myocardin isoforms. Gene. 2010;464:1–10. doi: 10.1016/j.gene.2010.03.012. [DOI] [PubMed] [Google Scholar]
- 43.Wang J, Li A, Wang Z, Feng X, Olson E, Schwartz R. Myocardin sumoylation transactivates cardiogenic genes in pluripotent 10T1/2 fibroblasts. Mol Cell Biol. 2007;27:622–632. doi: 10.1128/MCB.01160-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang Z, Wang D, Hockemeyer D, McAnally J, Nordheim A, Olson E. Myocardin and ternary complex factors compete for SRF to control smooth muscle gene expression. Nature. 2004;428:185–198. doi: 10.1038/nature02382. [DOI] [PubMed] [Google Scholar]
- 45.LeLievre C, Le Douarin N. Mesenchymal derivatives of the neural crest: Analysis of chimeric quail and chick embryos. J Embryol Exp Morphol. 1975;134:125–154. [PubMed] [Google Scholar]
- 46.Gittenberger-de Groot A, DeRuiter M, Bergwerff M, Poelmann R. Smooth muscle cell origin and its relation to heterogeneity in development and disease. Arterioscler Thromb Vasc Biol. 1999;19:1589–1594. doi: 10.1161/01.atv.19.7.1589. [DOI] [PubMed] [Google Scholar]
- 47.Tilki D, Hohn H, Ergün B, Rafii S, Ergün S. Emerging biology of vascular wall progenitor cells in health and disease. Trends Mol Med. 2009;15:501–509. doi: 10.1016/j.molmed.2009.09.004. [DOI] [PubMed] [Google Scholar]
- 48.Chow K, Hogan M, Schwartz R. Phased cis-acting promoter elements interact at short distances to direct avian skeletal alpha-actin gene transcription. Proc Natl Acad Sci USA. 1991;88:1301–1305. doi: 10.1073/pnas.88.4.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McDonald O, Owens G. Programming smooth muscle plasticity with chromatin dynamics. Circ Res. 2007;100:1428–1441. doi: 10.1161/01.RES.0000266448.30370.a0. [DOI] [PubMed] [Google Scholar]
- 50.Zhang M, Fang H, Zhou J, Herring B. A novel role of Brg1 in the regulation of SRF/MRTFA-dependent smooth muscle-specific gene expression. J Biol Chem. 2007;282:25708–25716. doi: 10.1074/jbc.M701925200. [DOI] [PubMed] [Google Scholar]
- 51.Liu Z, Wang Z, Yanagisawa H, Olson E. Phenotypic modulation of smooth muscle cells through interaction of FoxO4 and myocardin. Dev Cell. 2005;9:261–270. doi: 10.1016/j.devcel.2005.05.017. [DOI] [PubMed] [Google Scholar]
- 52.Liu Y, Sinha S, McDonald O, Shang Y, Hoofnagle M, Owens G. Kruppel-like factor 4 abrogates myocardin-induced activation of smooth muscle gene expression. J Biol Chem. 2005;280:9719–9727. doi: 10.1074/jbc.M412862200. [DOI] [PubMed] [Google Scholar]
- 53.Luger K, Mader A, Richmond R, Sargent D, Richmond T. Crystal structure of the nucleosome core particle at 2.8A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 54.Ho L, Crabtree G. Chromatin remodeling during development. Nature. 2010;463:474–484. doi: 10.1038/nature08911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Duan Z, Andronescu M, Schutz K, McIlwain S, Kim Y, Lee C, Shendure J, Fields S, Blau C, Noble W. A three-dimensional model of the yeast genome. Nature. 2010;465:363–367. doi: 10.1038/nature08973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jenuwein T, Allis C. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 57.Ruthenburg A, Li H, Patel D, Allis C. Multivalent engagement of chromatin modifications by linked binding modules. Nat Rev Mol Cell Biol. 2007;8:983–994. doi: 10.1038/nrm2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chi P, Allis C, Wang G. Covalent histone modifications--miswritten, misinterpreted and mis-erased in human cancers. Nat Rev Cancer. 2010;10:457–469. doi: 10.1038/nrc2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.McDonald O, Wamhoff B, Hoofnagle M, Owens G. Control of SRF binding to CArG box chromatin regulates smooth muscle gene expression in vivo. J Clin Invest. 2006;116:36–48. doi: 10.1172/JCI26505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Carson J, Fillmore R, Schwartz R, Zimmer W. The smooth muscle gamma-actin gene promoter is a molecular target for the mouse bagpipe homologue, mNkx3-1, and serum response factor. Keywords. 2000;275:39061–39072. doi: 10.1074/jbc.M006532200. [DOI] [PubMed] [Google Scholar]
- 61.Herring B, Kriegel A, Hoggatt A. Identification of Barx2B, a serum response factor-associated homeodomain protein. Keywords. 2001;276:14482–14489. doi: 10.1074/jbc.M011585200. [DOI] [PubMed] [Google Scholar]
- 62.Chang D, Belaguli N, Iyer D, Roberts W, Wu S, Dong X, Marx J, Moore M, Beckerle M, Majesky M, Schwartz R. Cysteine-rich LIM-only proteins CRP1 and CRP2 are potent smooth muscle differentiation cofactors. Dev Cell. 2003;4:107–118. doi: 10.1016/s1534-5807(02)00396-9. [DOI] [PubMed] [Google Scholar]
- 63.Sundberg-Smith L, DiMichele L, Sayers R, Mack C, Taylor J. The LIM protein leupaxin is enriched in smooth muscle and functions as an serum response factor cofactor to induce smooth muscle cell gene transcription. Circ Res. 2008;102:1502–1511. doi: 10.1161/CIRCRESAHA.107.170357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jin L, Gan Q, Zieba B, Goicoechea S, Owens G, Otey C, Somlyo A. The actin associated protein palladin is important for the early smooth muscle cell differentiation. PLoS One. 2010;22:e12823. doi: 10.1371/journal.pone.0012823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Haberland M, Montgomery R, Olson E. The many roles of histone deacetylases in development and physiology: implications for disease and therapy. Nat Rev Genet. 2009;10:32–42. doi: 10.1038/nrg2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Manabe I, Owens G. Recruitment of serum response factor and hyperacetylation of histones at smooth muscle-specific regulatory regions during differentiation of a novel P19-derived in vitro smooth muscle differentiation system. Circ Res. 2001;88:1127–1134. doi: 10.1161/hh1101.091339. [DOI] [PubMed] [Google Scholar]
- 67.Qiu P, Li L. Histone acetylation and recruitment of serum response factor and CREB-binding protein onto SM22 promoter during SM22 gene expression. Circ Res. 2002;90:858–865. doi: 10.1161/01.res.0000016504.08608.b9. [DOI] [PubMed] [Google Scholar]
- 68.Qiu P, Ritchie R, Gong X, Hamamori Y, Li L. Dynamic changes in chromatin acetylation and the expression of histone acetyltransferase and histone deacetylases regulate the SM22alpha transcription in response to Smad3-mediated TGFbeta1 signaling. Biochem Biophys Res Commun. 2006;348:351–358. doi: 10.1016/j.bbrc.2006.07.009. [DOI] [PubMed] [Google Scholar]
- 69.Suzuki M, Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nature Rev Genet. 2008;9:465–476. doi: 10.1038/nrg2341. [DOI] [PubMed] [Google Scholar]
- 70.Tsukada Y, Fang J, Erdjument-Bromage H, Warren M, Borchers C, Tempst P, Zhang Y. Histone demethylation by a family of JmjC domain-containing proteins. Nature. 2006;439:811–816. doi: 10.1038/nature04433. [DOI] [PubMed] [Google Scholar]
- 71.Klose R, Yamane K, Bae Y, Zhang D, Erdjument-Bromage H, Tempst P, Wong J, Zhang Y. The transcriptional repressor JHDM3A demethylates trimethyl histone H3 lysine 9 and lysine 36. Nature. 2006;442:312–316. doi: 10.1038/nature04853. [DOI] [PubMed] [Google Scholar]
- 72.Lockman K, Taylor J, Mack C. The histone demethylase, jmjd1a, interacts with the myocardin factors to regulate SMC differentiation marker gene expression. Circ Res. 2008;101:e115–e123. doi: 10.1161/CIRCRESAHA.107.164178. [DOI] [PubMed] [Google Scholar]
- 73.Shi Y, Whetstine J. Dynamic regulation of histone lysine methylation by demethylases. Mol Cell. 2007;25:1–14. doi: 10.1016/j.molcel.2006.12.010. [DOI] [PubMed] [Google Scholar]
- 74.Ruthenburg A, Allis C, Wysocka J. Methylation of lysine 4 on histone H3: intricacy of writing and reading a single epigenetic mark. Mol Cell. 2007;25:15–30. doi: 10.1016/j.molcel.2006.12.014. [DOI] [PubMed] [Google Scholar]
- 75.Lu M, Pressman C, Dyer R, Johnson R, Martin J. Function of Rieger syndrome gene in left-right asymmetry and craniofacial development. Nature. 1999;401:276–278. doi: 10.1038/45797. [DOI] [PubMed] [Google Scholar]
- 76.Shang Y, Yoshida T, Amendt B, Martin J, Owens G. Pitx2 is functionally important in the early stages of vascular smooth muscle cell differentiation. J Cell Biol. 2008;181:461–473. doi: 10.1083/jcb.200711145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gan Q, Yoshida T, Li J, Owens G. Smooth muscle cells and myofibroblasts use distinct transcriptional mechanisms for smooth muscle α-actin expression. Circ Res. 2007;101:883–892. doi: 10.1161/CIRCRESAHA.107.154831. [DOI] [PubMed] [Google Scholar]
- 78.D'Alessio J, Wright K, Tjian R. Shifting players and paradigms in cell-specific transcription. Mol Cell. 2009;36:924–931. doi: 10.1016/j.molcel.2009.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhou J, Zhang M, Fang H, El-Mounayri O, Rodenberg J, Imbalzano A, Herring B. The SWI/SNF chromatin remodeling complex regulates myocardin-induced smooth muscle-specific gene expression. Arterioscl Thromb Vasc Biol. 2009;29:921–928. doi: 10.1161/ATVBAHA.109.187229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hayashi N, Nakamura S, Nishida W, Sobue K. Bone morphogenetic protein-induced Msx1 and Msx2 inhibit myocardin-dependent smooth muscle gene transcription. Mol Cell Biol. 2006;26:9456–9470. doi: 10.1128/MCB.00759-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Haldar S, Ibrahim O, Jain M. Kruppel-like Factors (KLFs) in muscle biology. J Mol Cell Cardiol. 2007;43:1–10. doi: 10.1016/j.yjmcc.2007.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Adam P, Regan C, Hautmann M, Owens G. Positive- and negative-acting Kruppel-like transcription factors bind a transforming growth factor beta control element required for expression of the smooth muscle cell differentiation marker SM22alpha in vivo. J Biol Chem. 2000;275:37798–37806. doi: 10.1074/jbc.M006323200. [DOI] [PubMed] [Google Scholar]
- 83.Zhou J, Hu G, Wang X. Repression of smooth muscle differentiation by a novel high mobility group box-containing protein, HMG2L1. J Biol Chem. 2010;285:23177–23185. doi: 10.1074/jbc.M110.109868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Davis C, Haberland M, Arnold M, Sutherland L, McDonald O, Richardson J, Childs G, Harris S, Owens G, Olson E. PRISM/PRDM6, a transcriptional repressor that promotes the proliferative gene program in smooth muscle cells. Mol Cell Biol. 2006;26:2626–2636. doi: 10.1128/MCB.26.7.2626-2636.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Roy S, Ng T. Blimp-1 specifies neural crest and sensory neuron progenitors in the zebrafish embryo. Curr Biol. 2004;14:1772–1777. doi: 10.1016/j.cub.2004.09.046. [DOI] [PubMed] [Google Scholar]
- 86.Baxendale S, Davison C, Muxworthy C, Wolff C, Ingham P, Roy S. The B-cell maturation factor Blimp-1 specifies vertebrate slow-twitch muscle fiber identity in response to Hedgehog signaling. Nat Genet. 2004;36:88–93. doi: 10.1038/ng1280. [DOI] [PubMed] [Google Scholar]
- 87.Schuettengruber B, Cavalli G. Recruitment of polycomb group complexes and their role in the dynamic regulation of cell fate choice. Development. 2009;136:3531–3542. doi: 10.1242/dev.033902. [DOI] [PubMed] [Google Scholar]
- 88.McKinsey T, Zhang C, Olson E. MEF2: a calcium-dependent regulator of cell division, differentiation and death. Trends Biochem Sci. 2002;27:40–47. doi: 10.1016/s0968-0004(01)02031-x. [DOI] [PubMed] [Google Scholar]
- 89.Zhang C, McKinsey T, Olson E. Association of class II histone deacetylases with heterochromatin protein 1: potential role for histone methylation in control of muscle differentiation. Mol Cell Biol. 2002;22:7302–7312. doi: 10.1128/MCB.22.20.7302-7312.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.McKinsey T, Zhang C, Olson E. Signal-dependent nuclear export of a histone deacetylase regulates muscle differentiation. Nature. 2000;408:106–111. doi: 10.1038/35040593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sekiya T, Zaret K. Repression by Groucho/TLE/Grg proteins: genomic site recruitment generates compacted chromatin in vitro and impairs activator binding in vivo. Mol Cell. 2007;28:291–303. doi: 10.1016/j.molcel.2007.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Cinnamon E, Paroush Z. Context-dependent regulation of Groucho/TLE-mediated repression. Curr Opin Genet Dev. 2008;18:435–440. doi: 10.1016/j.gde.2008.07.010. [DOI] [PubMed] [Google Scholar]
- 93.Copley R. The EHI motif in metazoan transcription factors. BMC Genomics. 2006;6:169–177. doi: 10.1186/1471-2164-6-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 95.Bartel D. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Liu N, Olson E. MicroRNA regulatory networks in cardiovascular development. Dev Cell. 2010;18:510–525. doi: 10.1016/j.devcel.2010.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Cordes K, Sheehy N, White M, Berry E, Morton S, Muth A, Lee T, Miano J, Ivey K, Srivastava D. miR-145 and miR-143 regulate smooth muscle cell fate and plasticity. Nature. 2009;460:705–710. doi: 10.1038/nature08195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Elia L, Quintavalle M, Zhang J, Contu R, Cossu L, Latronico M, Peterson K, Indolfi C, Catalucci D, Chen J, Courtneidge S, Condorelli G. The knockout of miR-143 and -145 alters smooth muscle cell maintenance and vascular homeostasis in mice: correlates with human disease. Cell Death Differ. 2009;16:1590–1598. doi: 10.1038/cdd.2009.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Xin M, Small E, Sutherland L, Qi X, McAnally J, Plato C, Richardson J, Bassel-Duby R, Olson E. MicroRNAs miR-143 and miR-145 modulate cytoskeletal dynamics and responsiveness of smooth muscle cells to injury. Genes Dev. 2009;23:2166–2178. doi: 10.1101/gad.1842409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Boettger T, Beetz N, Kostin S, Schneider J, Kruger M, Hein L, Braun T. Acquisition of the contractile phenotype of arterial smooth muscle cells depends on the miR-143/145 gene cluster. J Clin Invest. 2009;119:2634–2647. doi: 10.1172/JCI38864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Cheng Y, Liu X, Yang J, Lin Y, Xu D, Lu Q, Deitch E, Huo Y, Delphin E, Zhang C. MicroRNA-145, a novel smooth muscle cell phenotypic marker and modulator, controls vascular neointimal lesion formation. Circ Res. 2009;105:158–166. doi: 10.1161/CIRCRESAHA.109.197517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Albinsson S, Suarez Y, Skoura A, Offermanns S, Miano J, Sessa W. MicroRNAs are necessary for vascular smooth muscle growth, differentiation, and function. Arterioscler Thromb Vasc Biol. 2010;30:1118–1126. doi: 10.1161/ATVBAHA.109.200873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Liu X, Cheng Y, Zhang S, Lin Y, Yang J, Zhang C. A necessary role of miR-221 and miR-222 in vascular smooth muscle cell proliferation and neointimal hyperplasia. Circ Res. 2009;104:476–487. doi: 10.1161/CIRCRESAHA.108.185363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Davis B, Hilyard A, Nguyen P, Lagna G, Hata A. Induction of microRNA-221 by platelet-derived growth factor signaling is critical for modulation of vascular smooth muscle phenotype. J Biol Chem. 2009;284:3728–3738. doi: 10.1074/jbc.M808788200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Guenther M, Young R. Repressive transcription. Science. 2010;329:150–151. doi: 10.1126/science.1193995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zhao J, Sun B, Erwin J, Song J, Lee J. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science. 2008;322:750–756. doi: 10.1126/science.1163045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lee J. Lessons from X-chromosome inactivation: long ncRNA as guides and tethers to the epigenome. Genes Dev. 2009;23:1831–1842. doi: 10.1101/gad.1811209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Yap K, Li S, Munoz-Cabello A, Raguz S, Zeng L, Mujtaba S, Gil J, Walsh M, Zhou M. Molecular interplay of the nonoconding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol Cell. 2010;38:662–674. doi: 10.1016/j.molcel.2010.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Selia A, Calabrese J, Levine S, Yeo G, Rahl P, Flynn R, Young R, Sharp P. Divergent transcription from active promoters. Science. 2008;322:1849–1851. doi: 10.1126/science.1162253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Cloos P, Christensen J, Agger K, Helin K. Erasing the methyl mark: histone demethylases at the center of cellular differentiation and disease. Genes Dev. 2008;22:1115–1140. doi: 10.1101/gad.1652908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Orom U, Derrien T, Beringer, Gumireddy K, Gardini A, Bussotti G, Lai F, Zytnicki M, Notredame C, Huang Q, Guigo R, Shiekhattar R. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143:46–58. doi: 10.1016/j.cell.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Fukata M, Makagawa M, Kaibuchi K. Roles of Rho-family GTPases in cell polarisation and directional migration. Curr Opin Cell Biol. 2003;15:590–597. doi: 10.1016/s0955-0674(03)00097-8. [DOI] [PubMed] [Google Scholar]
- 113.Somogyi K, Rørth P. Evidence for tension-based regulation of Drosophila MAL and SRF during invasive cell migration. Dev Cell. 2004;7:85–93. doi: 10.1016/j.devcel.2004.05.020. [DOI] [PubMed] [Google Scholar]
- 114.Posern G, Treisman R. Actin' together: serum response factor, its cofactors and the link to signal transduction. Trends Cell Biol. 2006;16:588–596. doi: 10.1016/j.tcb.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 115.Lu J, Landerholm T, Wei J, Dong X, Wu S, Liu X, Nagata K, Inagaki M, Majesky M. Coronary smooth muscle differentiation from proepicardial cells requires rhoA-mediated actin reorganization and p160 rho-kinase activity. Dev Biol. 2001;240:404–418. doi: 10.1006/dbio.2001.0403. [DOI] [PubMed] [Google Scholar]
- 116.Mack C, Somlyo A, Hautmann M, Somlyo A, Owens G. Smooth muscle differentiation marker gene expression is regulated by rhoA-mediated actin polymerization. J Biol Chem. 2001;276:341–347. doi: 10.1074/jbc.M005505200. [DOI] [PubMed] [Google Scholar]
- 117.Tanaka T, Nishimura D, Wu R, Amano M, Iso T, Kedes L, Nishida H, Kaibuchi K, Hamamori Y. Nuclear Rho kinase, ROCK2, targets p300 acetyltransferase. J Biol Chem. 2006;281:15320–15329. doi: 10.1074/jbc.M510954200. [DOI] [PubMed] [Google Scholar]
- 118.Sotiropoulos A, Gineitis D, Copeland J, Triesman R. Signal-regulated activation of serum response factor is mediated by changes in actin dynamics. Cell. 1999;98:159–169. doi: 10.1016/s0092-8674(00)81011-9. [DOI] [PubMed] [Google Scholar]
- 119.Cen B, Selvaraj A, Prywes R. Myocardin/MKL family of SRF coactivators: key regulators of immediate early and muscle specific gene expression. J Cell Biochem. 2004;93:74–82. doi: 10.1002/jcb.20199. [DOI] [PubMed] [Google Scholar]
- 120.Kogata N, Tribe R, Fassler R, Way M, Adams R. Integrin-linked kinase controls vascular wall formation by negatively regulating Rho/ROCK-mediated vascular smooth muscle cell contraction. Genes Dev. 2009;23:2278–2283. doi: 10.1101/gad.535409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Kirby M, Waldo K. Neural crest and cardiovascular patterning. Circ Res. 1995;77:211–215. doi: 10.1161/01.res.77.2.211. [DOI] [PubMed] [Google Scholar]
- 122.Jiang X, Rowitch D, Soriano P, McMahon A, Sucov H. Fate of the mammalian cardiac neural crest. Development. 2000;127:1607–1616. doi: 10.1242/dev.127.8.1607. [DOI] [PubMed] [Google Scholar]
- 123.Epstein J, Li J, Lang D, Chen F, Brown C, Jin F, Lu M, Thomas M, Liu E, Wessels A, Lo C. Migration of cardiac neural crest cells in Splotch embryos. Development. 2000;127:1869–1878. doi: 10.1242/dev.127.9.1869. [DOI] [PubMed] [Google Scholar]
- 124.Rao M, Anderson D. Immortalization and controlled in vitro differentiation of murine multipotent neural crest stem cells. J Neurobiol. 1997;32:722–746. doi: 10.1002/(sici)1097-4695(19970620)32:7<722::aid-neu7>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 125.Wilson Y, Richards K, Ford-Perriss M, Panthier J, Murphy M. Neural crest cell lineage segregation in the mouse neural tube. Development. 2004;131:6153–6162. doi: 10.1242/dev.01533. [DOI] [PubMed] [Google Scholar]
- 126.Vitelli F, Taddei I, Morishima M, Meyers E, Lindsay E, Baldini A. A genetic link between Tbx1 and fibroblast growth factor signaling. Development. 2002;129:4605–4611. doi: 10.1242/dev.129.19.4605. [DOI] [PubMed] [Google Scholar]
- 127.Chen S, Lechleider R. Transforming growth factor-beta-induced differentiation of smooth muscle from a neural crest stem cell line. Circ Res. 2004;94:1195–1202. doi: 10.1161/01.RES.0000126897.41658.81. [DOI] [PubMed] [Google Scholar]
- 128.Mancini M, Verdi J, Conley B, Nicola T, Spicer D, Oxburgh L, Vary C. Endoglin is required for myogenic differentiation potential of neural crest stem cells. Dev Biol. 2007;308:520–533. doi: 10.1016/j.ydbio.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Esner M, Meihac S, Relaix F, Nicolas J, Cossu G, Buckingham M. Smooth muscle of the dorsal aorta shares a common clonal origin with skeletal muscle of the myotome. Development. 2006;133:737–749. doi: 10.1242/dev.02226. [DOI] [PubMed] [Google Scholar]
- 130.Lagha M, Brunelli S, Messina G, Cumano A, Kume T, Relaix F, Buckingham M. Pax3:FoxC2 reciprocal repression in the somite modulates muscular versus vascular cell fate choice in multipotent progenitors. Dev Cell. 2009;17:892–899. doi: 10.1016/j.devcel.2009.10.021. [DOI] [PubMed] [Google Scholar]
- 131.Paik J, Skoura A, Chae S, Cowan A, Han D, Proia R, Hla T. Sphingosine 1-phosphate receptor regulation of N-cadherin mediates vascular stabilization. Genes Dev. 2004;18:2392–2403. doi: 10.1101/gad.1227804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Doi H, Iso T, Yamazaki M, Akiyama H, Kanai H, Sato H, Kawai-Kowase K, Tanaka T, Maeno T, Okamoto E, Arai M, Kedes L, Kurabayashi M. HERP1 Inhibits myocardin-induced vascular smooth muscle cell differentiation by interfering with SRF binding to CArG box. Arterioscler Thromb Vasc Biol. 2005;25:2328–2334. doi: 10.1161/01.ATV.0000185829.47163.32. [DOI] [PubMed] [Google Scholar]
- 133.High F, Zhang M, Proweller A, Tu L, Parmacek M, Pear W, Epstein J. An essential role for Notch in neural crest during cardiovascular development and smooth muscle differentiation. J Clin Invest. 2007;117:353–363. doi: 10.1172/JCI30070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Regan J, Majesky M. Building a vessel wall with notch signaling. Circ Res. 2009;104:419–421. doi: 10.1161/CIRCRESAHA.109.194233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Liu H, Zhang W, Kennard S, Caldwell R, Lilly B. NOTCH3 is critical for proper angiogenesis and mural cell investment. Circ Res. 2010;107:860–870. doi: 10.1161/CIRCRESAHA.110.218271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Tang Y, Urs S, Boucher J, Bernaiche T, Venkatesh D, Spicer D, Vary C, Liaw L. Notch and transforming growth factor-beta (TGFβ) signaling pathways cooperatively regulate vascular smooth muscle cell differentiation. J Biol Chem. 2010;285:17556–17563. doi: 10.1074/jbc.M109.076414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Li J, Zhu X, Chen M, Cheng L, Zhou D, Lu M, Du K, Epstein J, Parmacek M. Myocardin-related transcription factor B is required in cardiac neural crest for smooth muscle differentiation and cardiovascular development. Proc Natl Acad Sci USA. 2005;102:8916–8921. doi: 10.1073/pnas.0503741102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Oh J, Richardson J, Olson E. Requirement of myocardin-related transcription factor-B for remodeling of branchial arch arteries and smooth muscle differentiation. Proc Natl Acad Sci USA. 2005;102:15122–15127. doi: 10.1073/pnas.0507346102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Li S, Chang S, Qi X, Richardson J, Olson E. Requirement of a myocardin-related transcription factor for development of mammary myoepithelial cells. Mol Cell Biol. 2006;26:5797–5808. doi: 10.1128/MCB.00211-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Mikawa T, Fischman D. Retroviral analysis of cardiac morphogenesis: Discontinuous formation of coronary vessels. Proc Natl Acad Sci USA. 1992;89:9504–9508. doi: 10.1073/pnas.89.20.9504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Manner J, Perez-Pomares J, Macias D, Munoz-Chapuli R. The origin, formation and developmental significance of the epicardium: A review. Cells Tissues Organs. 2001;169:89–103. doi: 10.1159/000047867. [DOI] [PubMed] [Google Scholar]
- 142.Majesky M. Development of coronary vessels. Curr Top Dev Biol. 2004;62:225–259. doi: 10.1016/S0070-2153(04)62008-4. [DOI] [PubMed] [Google Scholar]
- 143.Kruithof B, van Wijk B, Somi S, Kruithof-de Juliob M, Pérez Pomares J, Weesie F, Wessels A, Moorman A, van den Hoff M. BMP and FGF regulate the differentiation of multipotential pericardial mesoderm into the myocardial or epicardial lineage. Dev Biol. 2006;295:507–522. doi: 10.1016/j.ydbio.2006.03.033. [DOI] [PubMed] [Google Scholar]
- 144.Olivey H, Svensson E. Epicardial-myocardial signaling directing coronary vasculogenesis. Circ Res. 2010;106:818–832. doi: 10.1161/CIRCRESAHA.109.209197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Ishii Y, Langberg J, Hurtado R, Lee S, Mikawa T. Induction of proepicardial marker gene expression by the liver bud. Development. 2007;134:3627–3637. doi: 10.1242/dev.005280. [DOI] [PubMed] [Google Scholar]
- 146.Liu J, Stainier D. Tbx5 and Bmp signaling are essential for proepicardium specification in zebrafish. Circ Res. 2010;106:1818–1828. doi: 10.1161/CIRCRESAHA.110.217950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Dettman R, Denetclaw W, Ordahl C, Bristow J. Common epicardial origin of coronary vascular smooth muscle, perivascular fibroblasts, and intermyocardial fibroblasts in the avian heart. Dev Biol. 1998;193:169–181. doi: 10.1006/dbio.1997.8801. [DOI] [PubMed] [Google Scholar]
- 148.Landerholm T, Dong XR, Lu J, Belaguli N, Schwartz R, Majesky M. A role for serum response factor in coronary smooth muscle differentiation from proepicardial cells. Development. 1999;126:2053–2062. doi: 10.1242/dev.126.10.2053. [DOI] [PubMed] [Google Scholar]
- 149.Mellgren A, Smith C, Olsen G, Eskiocak B, Zhou B, Kazi M, Ruiz F, Pu W, Tallquist M. Platelet-derived growth factor receptor β signaling is required for efficient epicardial cell migration and development of two distinct coronary vascular smooth muscle cell populations. Circ Res. 2008;103:1393–1401. doi: 10.1161/CIRCRESAHA.108.176768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Compton L, Potash D, Mundell N, Barnett J. Transforming growth factor-β induces loss of epithelial character and smooth muscle cell differentiation in epicardial cells. Dev Dyn. 2006;235:82–93. doi: 10.1002/dvdy.20629. [DOI] [PubMed] [Google Scholar]
- 151.Smart N, Risebro C, Melville A, Moses K, Schwartz R, Chien K, Riley P. Thymosin beta-4 induced adult epicardial progenitor mobilization and neovascularization. Nature. 2007;445:177–182. doi: 10.1038/nature05383. [DOI] [PubMed] [Google Scholar]
- 152.Kraus F, Haenig B, Kispert A. Cloning and expression analysis of the mouse T-box gene Tbx18. Mech Dev. 2001;100:83–86. doi: 10.1016/s0925-4773(00)00494-9. [DOI] [PubMed] [Google Scholar]
- 153.Hatcher C, Diman N, Kim M, Pennisi D, Song Y, Goldstein M, Mikawa T, Basson C. A role for Tbx5 in proepicardial cell migration during cardiogenesis. Physiol Genomics. 2004;18:129–140. doi: 10.1152/physiolgenomics.00060.2004. [DOI] [PubMed] [Google Scholar]
- 154.Farin H, Bussen M, Schmidt M, Singh M, Schuster-Gossler K, Kispert A. Transcriptional repression by the T-box proteins TBX18 and TBX15 depends on groucho corepressors. J Biol Chem. 2007;282:25748–25759. doi: 10.1074/jbc.M703724200. [DOI] [PubMed] [Google Scholar]
- 155.Moretti A, Caron L, Nakano A, Lam J, Bernshausen A, Chen Y, Qyang Y, Bu L, Sasaki M, Martin-Plug S, Sun Y, Evans S, Laugwitz K, Chien K. Multipotent embryonic Isl1+ progenitor cells lead to cardiac, smooth muscle and endothelial cell diversification. Cell. 2006;127:1151–1165. doi: 10.1016/j.cell.2006.10.029. [DOI] [PubMed] [Google Scholar]
- 156.Cai C, Liang X, Shi Y, Chu P, Pfaff S, Chen J, Evans S. Isl1 identifies a cardiac progenitor population that proliferates prior to differentiation and contributes a majority of cells to the heart. Dev Cell. 2003;5:877–889. doi: 10.1016/s1534-5807(03)00363-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Bu L, Jiang X, Martin-Puig S, Caron L, Zhu S, Shao Y, Roberts D, Huang P, Domain I, Chien K. Human ISL1 heart progenitors generate diverse multipotent cardiovascular cell lineages. Nature. 2009;460:113–117. doi: 10.1038/nature08191. [DOI] [PubMed] [Google Scholar]
- 158.Kattman S, Huber T, Keller G. Multipotent flk1+ cardiovascular progenitor cells give rise to the cardiomyocyte, endothelial and vascular smooth muscle cell lineages. Dev Cell. 2006;11:723–732. doi: 10.1016/j.devcel.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 159.Armulik A, Abramsson A, Betsholtz C. Endothelial/pericyte interactions. Circ Res. 2005;97:512–523. doi: 10.1161/01.RES.0000182903.16652.d7. [DOI] [PubMed] [Google Scholar]
- 160.Gaengel K, Genove G, Armulik A, Betsholtz C. Endothelial-mural cell signaling in vascular development and angiogenesis. Arterioscler Thromb Vasc Biol. 2009;29:630–638. doi: 10.1161/ATVBAHA.107.161521. [DOI] [PubMed] [Google Scholar]
- 161.Etchevers H, Vincent C, Le Douarin N, Couly G. The cephalic neural crest provides pericytes and smooth muscle cells to all blood vessels of the face and forebrain. Development. 2001;128:1059–1068. doi: 10.1242/dev.128.7.1059. [DOI] [PubMed] [Google Scholar]
- 162.Crisan M, Yap S, Casteilla L, Chen C, Corselli M, Park T, Andriolo G, Sun B, Zheng B, Zhang L, Norotte C, Teng P, Traas J, Schugar R, Deasy B, Badylak S, Buhring H, Giacobino J, Lazzari L, Huard J, Péault B. A perivascular origin for mesenchymal stem cells in multiple human organs. Cell Stem Cell. 2008;3:301–313. doi: 10.1016/j.stem.2008.07.003. [DOI] [PubMed] [Google Scholar]
- 163.Corselli M, Chen C, Crisan M, Lazzari L, Péault B. Perivascular ancestors of adult multipotent stem cells. Arterioscler Thromb Vasc Biol. 2010;30:1104–1109. doi: 10.1161/ATVBAHA.109.191643. [DOI] [PubMed] [Google Scholar]
- 164.Sampaolesi M, Blot S, D'Antona G, Granger N, Tonlorenzi R, Innocenzi A, Mognol P, Thibaud J, Galvez B, Barthelemy I, Perani L, Mantero S, Guttinger M, Pansarasa O, Rinaldi C, De Angelis M, Torrente Y, Bordignon C, Bottinelli R, Cossu G. Mesoangioblast stem cells ameliorate muscle function in dystrophic dogs. Nature. 2006;444:574–579. doi: 10.1038/nature05282. [DOI] [PubMed] [Google Scholar]
- 165.Morgan J, Muntoni F. Mural cells paint a new picture of muscle stem cells. Nat Cell Biol. 2007;9:249–251. doi: 10.1038/ncb0307-249. [DOI] [PubMed] [Google Scholar]
- 166.Dellavalle A, Sampaolesi M, Tonlorenzi R, Tagliafico E, Sacchetti B, Perani L, Innocenzi A, Galvez B, Messina G, Morosetti R, Li S, Belicci M, Peretti G, Chamberlain J, Wright W, Torrente Y, Ferrari S, Bianco P, Cossu G. Pericytes of human skeletal muscle are myogenic precursors distinct from satellite cells. Nature Cell Biol. 2007;9:255–267. doi: 10.1038/ncb1542. [DOI] [PubMed] [Google Scholar]
- 167.De Angelis L, Berghella L, Coletta M, Lattanzi L, Zanchi M, Cusella-de Angelis M, Ponzetto C, Cossu G. Skeletal myogenic progenitors originating from embryonic dorsal aorta coexpress endothelial and myogenic markers and contribute to postnatal muscle growth and regeneration. J Cell Biol. 1999;147:869–877. doi: 10.1083/jcb.147.4.869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Scott N, Cipolla G, Ross C, Dunn B, Martin F, Simonet L, Wilcox J. Identification of a potential role for the adventitia in vascular lesion formation after balloon overstretch injury of porcine coronary arteries. Circulation. 1996;93:2178–2187. doi: 10.1161/01.cir.93.12.2178. [DOI] [PubMed] [Google Scholar]
- 169.Sartore S, Chiavegato A, Faggin E, Franch R, Puato M, Ausoni S, Pauletto P. Contribution of adventitial fibroblasts to neointima formation and vascular remodeling: from innocent bystander to active participant. Circ Res. 2001;89:1111–1121. doi: 10.1161/hh2401.100844. [DOI] [PubMed] [Google Scholar]
- 170.Nemenoff R, Simpson P, Furgeson S, Kaplan-Albuquerque N, Crossno J, Garl P, Cooper J, Weiser-Evans M. Targeted deletion of PTEN in smooth muscle cells results in vascular remodeling and recruitment of progenitor cells through induction of stromal cell-derived factor-1alpha. Circ Res. 2008;102:1036–1045. doi: 10.1161/CIRCRESAHA.107.169896. [DOI] [PubMed] [Google Scholar]
- 171.Li D, Faury G, Taylor D, Davis E, Boyle W, Mecham R, Stenzel P, Boak B, Keating M. Novel arterial pathology in mice and humans hemizygous for elastin. J Clin Invest. 1998;102:1783–1787. doi: 10.1172/JCI4487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Faury G, P M, Knutsen R, Boyle W, Heximer S, McLean S, Minkes R, Blumer K, Kovacs A, Kelly D, Li D, Starcher B, Mecham R. Developmental adaptation of the mouse cardiovascular system to elastin haploinsufficiency. J Clin Invest. 2003;112:1419–1428. doi: 10.1172/JCI19028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Saiura A, Sata M, Hirata Y, Nagai R, Makuuchi M. Circulating smooth muscle progenitor cells contribute to atherosclerosis. Nat Med. 2001;7:382–383. doi: 10.1038/86394. [DOI] [PubMed] [Google Scholar]
- 174.Sata M, Saiura A, Kunisato A, Tojo A, Okada S, Tokuhisa T, Hirai H, Makuuchi M, Hirata Y, Nagai R. Hematopoietic stem cells differentiate into vascular cells that participate in the pathogenesis of atherosclerosis. Nat Med. 2002;8:403–409. doi: 10.1038/nm0402-403. [DOI] [PubMed] [Google Scholar]
- 175.Bentzon J, Weile C, Sondergaard C, Hindkjaer J, Kassem M, Falk E. Smooth muscle cells in atherosclerosis originate from the local vessel wall and not circulating progenitor cells in ApoE knockout mice. Arterioscler Thromb Vasc Biol. 2006;26:2696–2702. doi: 10.1161/01.ATV.0000247243.48542.9d. [DOI] [PubMed] [Google Scholar]
- 176.Daniel J, Bielenberg W, Stieger P, Weinert S, Tillmanns H, Sedding D. Time-course analysis on the differentiation of bone marrow-derived progenitor cells into smooth muscle cells during neointima formation. Arterioscler Thromb Vasc Biol. 2010;30:1890–1896. doi: 10.1161/ATVBAHA.110.209692. [DOI] [PubMed] [Google Scholar]
- 177.Prokopi M, Pula G, Mayr U, Devue C, Gallagher J, Xiao Q, Boulanger C, Westwood N, Urbich C, Willeit J, Steiner M, Breuss J, Xu Q, Kiechi S, Mayr M. Proteomic analysis reveals presence of platelet microparticles in endothelial progenitor cell cultures. Blood. 2009;114:723–732. doi: 10.1182/blood-2009-02-205930. [DOI] [PubMed] [Google Scholar]
- 178.Hinz B, Phan S, Thannickal V, Galli A, Bochaton-Piallat M, Gabbiani G. The myofibroblast: one function, multiple origins. Am J Pathol. 2007;170:1807–1816. doi: 10.2353/ajpath.2007.070112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Niessen P, Clement S, Fonato L, Chaponnier C, Teunissen B, Rensen S, van Eyes G, Gabbiani G. Biochemical evidence for the interaction between smoothelin and filamentous actin. Exp Cell Res. 2004;292:170–178. doi: 10.1016/j.yexcr.2003.09.005. [DOI] [PubMed] [Google Scholar]
- 180.Sainz J, Al Haj Zen A, Caligiuri G, Demerens C, Urbain D, Lemitre M, Lafont A. Isolation of “side population” progenitor cells from healthy arteries of adult mice. Arterioscler Thromb Vasc Biol. 2006;26:281–286. doi: 10.1161/01.ATV.0000197793.83391.91. [DOI] [PubMed] [Google Scholar]