Abstract
Chemical synapses are highly specialized cell–cell contacts for communication between neurons in the CNS characterized by complex and dynamic protein networks at both synaptic membranes. The cytomatrix at the active zone (CAZ) organizes the apparatus for the regulated release of transmitters from the presynapse. At the postsynaptic side, the postsynaptic density constitutes the machinery for detection, integration, and transduction of the transmitter signal. Both pre- and postsynaptic protein networks represent the molecular substrates for synaptic plasticity. Their function can be altered both by regulating their composition and by post-translational modification of their components. For a comprehensive understanding of synaptic networks the entire ensemble of synaptic proteins has to be considered. To support this, we established a comprehensive database for synaptic junction proteins (SynProt database) primarily based on proteomics data obtained from biochemical preparations of detergent-resistant synaptic junctions. The database currently contains 2,788 non-redundant entries of rat, mouse, and some human proteins, which mainly have been manually extracted from 12 proteomic studies and annotated for synaptic subcellular localization. Each dataset is completed with manually added information including protein classifiers as well as automatically retrieved and updated information from public databases (UniProt and PubMed). We intend that the database will be used to support modeling of synaptic protein networks and rational experimental design.
Keywords: chemical synapse, postsynaptic density, synaptic junction, cytomatrix at the active zone, proteomics, rat, mouse, human
Introduction
Mammalian brain development culminates in the integration of billions of neurons and astrocytes into a functional network courtesy of trillions of specialized contact sites, the synapses. These highly specialized membrane microdomains – or synaptic junctions – are assumed to be the subcellular substrate for higher order functions such as learning and memory. Sherrington and Foster coined the term “synapse” in 1897 (Gray, 1987), and although physiological evidence on the existence of chemical synapses accumulated in the following decades, it took until the middle of the twentieth century to confirm the presence of such structures by means of electron microscopy (Palay and Palade, 1955; de Robertis, 1965). Among the different kinds of synapses the chemical, glutamatergic synapse between a presynaptic axon and a postsynaptic dendrite is the most predominant type in the mammalian brain. It is a highly asymmetrical structure encompassing a presynaptic terminal and a postsynaptic compartment, and is surrounded by the extracellular matrix and glial cells. The axonal, presynaptic side contains the machineries for neurotransmitter exocytosis and vesicle recycling, and harbors a distinct submembraneous structure, the cytomatrix at the active zone (CAZ) – also termed presynaptic grid, presynaptic dense projection, or presynaptic particle web (for a review and further literature see Schoch and Gundelfinger, 2006) – where neurotransmitter release takes place.
The electron-dense postsynaptic density (PSD) is the most striking structural element on the postsynaptic side, and was first described as the “subsynaptic membrane” by Eccles (1957), “postsynaptic thickening” by Gray (1959) and de Robertis (1959), or as the “subsynaptic web, a web of filaments (or canaliculi)” by de Robertis et al. (1961). The term currently most frequently used, “postsynaptic density”/“PSD” was introduced by Akert et al. (1969). Cotman and Taylor presented in the early 1970s (Cotman and Taylor, 1972, 1974; Cotman et al., 1974) a first protocol for the isolation of the synaptic junctions along with an enrichment of the PSD, and provided a structural description of this biochemical fraction. In combination with electron microscopy, several protocols for biochemical enrichment and detailed purification and extraction of synaptic structures from different species and brain regions were developed through the early 1980s (Gray and Whittaker, 1962; Cotman and Taylor, 1974; Matus and Walters, 1975; Cohen et al., 1977; Matus et al., 1981). An additional procedure was reported recently by Suzuki (2011). In the following years a number of major PSD components were identified from these preparations (e.g., Carlin et al., 1980; Kennedy et al., 1983; Siekevitz, 1985; Cho et al., 1992; Walsh and Kuruc, 1992; Kistner et al., 1993; for review see Kennedy, 1993, 1997; Ziff, 1997). The degree of enrichment and the purity of isolated synaptic junctions varies, depending on the protocol. In contrast to more “crude” preparations such as synaptosomes (Gray and Whittaker, 1962; Cohen et al., 1977; Matus et al., 1981; Suzuki and Tanaka, 1984), synaptodendrosomes (Steward et al., 1988), or synaptoneurosomes (Hollingsworth et al., 1985), which contain a considerable amount of dendritic and presynaptic material, the enrichment for the morphologically distinct, disk-like PSD structure necessitates additional detergent extractions, and ultracentrifugation steps (Gray and Whittaker, 1962; Cohen et al., 1977; Matus et al., 1981; Suzuki and Tanaka, 1984). However, we want to emphasize here that all so-called “PSD preparations” can indeed only enrich for the morphological structure of the PSD and are not pure fractions of this disk-like, 30–50 nm thick postsynaptic membrane and scaffolding entity. As discussed below all of these preparations contain proteins of all compartments contributing to the synaptic junction including the PSD, the presynaptic CAZ, the synaptic extracellular matrix (perisynaptic extracellular matrix and synaptic cleft), and astroglial endfeet contacting synapses. Hence, we refer to the proteins found within this fraction as the detergent-resistant synaptic junction proteins to avoid confusions about either a true, i.e., morphological residence within the morphological structure of the PSD (referred to as “established” PSD proteins, i.e., enriched in the detergent-resistant synaptic junction fraction AND verified by immunogold EM for localization in the PSD) or any physico-chemical or interaction-based presence within the fraction of detergent-resistant synaptic junction proteins. The more or less misleading, historically used term “PSD protein fraction” or “PSD preparation” is not used here. See also, term definitions in the glossary, Table 1.
Table 1.
Glossary of subsynaptic structures and protein fractions.
| Synaptic structure or preparation | Description | Selected literature |
|---|---|---|
| Postsynaptic density (PSD) | The PSD was originally defined at the ultrastructural level as the electron-dense material associated with the postsynaptic membrane. The PSD is particularly prominent at excitatory asymmetric (type I) synapses. Core PSD proteins and their associated partners are major components of ⇒ synaptic junctional protein preparations | Gray (1959), Peters et al. (1991), Ziff (1997) |
| Presynaptic cytomatrix at the active zone (CAZ) | The cytomatrix assembled at the presynaptic active zone of neurotransmitter release – also named presynaptic grid, presynaptic dense projection, or presynaptic particle web is the electron-dense counterpart of the PSD-associated with the cytoplasmic face of the presynaptic membrane. Essential scaffolding components of the CAZ co-purify with ⇒ synaptic junctional protein preparations | Peters et al. (1991), Garner et al. (2000), Phillips et al. (2001) |
| Glial endfeet | Endfeet of astrocytes are also very tightly associated with synaptic junctions leading to the concept of the tripartite synapse. Consequently, astrocytic proteins are also found in ⇒ synaptic junctional protein preparations | Araque et al. (1999), Faissner et al. (2010) |
| Synaptic extracellular matrix (ECM) | ECM components tightly associated with the synapse also do co-fractionate with ⇒ synaptic junctional protein preparations. Components of the perisynaptic ECM and the ECM within the synaptic cleft are supposed to vary | Zuber et al. (2005), Faissner et al. (2010), Dityatev et al. (2010) |
| Synaptic junctional protein fraction | Detergent-resistant biochemical protein preparation enriched for PSD components. Historically this fraction was called “PSD preparation” or “PSD protein fraction.” However, as discussed in this article it contains in addition to PSD core and PSD-associated proteins a variety of proteins of the ⇒ CAZ, of the ⇒ synaptic ECM, of ⇒ glial endfeet as well as protein components of which is unclear whether they are tightly associated with synaptic junctions or co-partition with this protein fraction because of their similar biochemical characteristics or the “stickiness” that causes them to associate with synaptic junctions after tissue homogenization | Carlin et al. (1980), Kennedy (1997), Li et al. (2004) |
| Established PSD proteins | Proteins that have been localized to the postsynaptic density in situ using specific antibodies and immunogold localization in the electron microscope | cf. Table 4 |
The identification of the large number of molecular PSD constituents started to boom with the advent of modern biochemistry and molecular biology technologies at the end of the 1980s. The Kennedy laboratory combined state-of-the-art protein sequencing with a subsequent molecular cloning strategy to identify components of the detergent-resistant synaptic junction fraction. Among those candidates was one of the most prominent proteins of the PSD, PSD-95 (Cho et al., 1992; Hunt et al., 1996). At the same time Garner, Gundelfinger and colleagues isolated over 200 cDNAs for specific synaptic proteins by expression screening of a rat cDNA library with the help of antisera raised against a rat brain synaptic protein preparation (Kistner et al., 1993; Müller et al., 1995; Seidenbecher et al., 1995; Langnaese et al., 1996). Among these cDNA clones were PSD-95/SAP90 (Cho et al., 1992; Kistner et al., 1993), ProSAP1/Shank2 (Boeckers et al., 1999; Naisbitt et al., 1999), the Calcium-binding protein Caldendrin (Seidenbecher et al., 1998), but also the presynaptic proteins Bassoon and Piccolo (Cases-Langhoff et al., 1996; tom Dieck et al., 1998), and the extracellular matrix protein Brevican (Seidenbecher et al., 1995). In the following years, interaction studies revealed that the PSD hosts a tightly organized and controlled protein network for signal transduction. The laboratory of Kennedy and Seeburg for instance showed the association of PSD-95 with the C-terminus of the NMDA receptor (Kornau et al., 1995) and subsequent work led to the identification the subunit NR2B of the NMDAR complex (Moon et al., 1994), of Densin (Apperson et al., 1996), and of synGAP (Chen et al., 1998) showing the diversity of this fraction.
Only a few years later, Sheng and colleagues started a series of high-throughput yeast two-hybrid (Young, 1998) experiments to search for interaction partners for prominent PSD proteins such as PSD-95/SAP90 and the NMDA receptor, identifying the GKAP/SAPAP (Kim et al., 1997), and Shank (Naisbitt et al., 1999) protein families and Yotiao (Lin et al., 1998). Moreover, the yeast two-hybrid system proved to be an invaluable tool to look for additional information about a particular synaptic protein if this protein’s function is unclear as in the case of CASK/Lin-2 or Caldendrin. CASK/Lin-2 was shown to bind to the cell surface heparan sulfate proteoglycan Syndecan-2 in mammalian brain (Hsueh et al., 1998); whereas Caldendrin was found to be an interaction partner for the synapse-nucleus shuttle protein Jacob (Dieterich et al., 2008). From these experiments it became unambiguously clear that, in addition to acting as a scaffold for signaling enzymes at the postsynaptic membrane, the PSD tethers the receptive apparatus to signaling components in the extracellular space, and links it to signaling networks that extend from the synapse to the nucleus.
Simultaneously with antibody and yeast two-hybrid-based library screens, developments in the area of high-throughput mass spectrometry and associated genome and proteome projects allowed a faster identification and characterization of newly discovered synaptic components at the end of the 1990s. Therefore, large-scale studies for synaptic proteins started in 2000 (Walikonis et al., 2000), and recent subproteomic efforts, in particular neuroproteomics, are contributing to a large extend to complete our knowledge of the molecular composition of chemical synapses. These proteomic efforts have given an insight into the complexity of synaptic junctions and associated cellular components (Walikonis et al., 2000; Husi and Grant, 2001; Yamauchi, 2002; Jordan et al., 2004; Li et al., 2004; Peng et al., 2004; Yoshimura et al., 2004; Collins et al., 2005). More recent studies have begun to address the crucial question of the abundance of each protein in the detergent-resistant synaptic junction fraction and their stoichiometrical ratio (Peng et al., 2004) and in future the interactions of these proteins within the PSD and associated subcellular compartments (Li et al., 2010). All of these proteomic analyses have produced a vast collection of proteins that comprise several hundred to over a thousand individual proteins (Sheng and Hoogenraad, 2007; Shinohara, 2012).
However, there is no specialized and comprehensive collection of these data as of now. The portal for synaptic protein databases (SynProt) was initially developed to store and organize proteomic information on the postsynaptic, presynaptic and surrounding extracellular and astrocytic endfeet compartments that was retrieved from individual research articles. Now the SynProt database hosts the experimental results obtained from 12 independent proteomic studies of the detergent-resistant synaptic junction fraction. In addition, other well-known PSD proteins, or associated proteins, that have not been identified by proteomic analyses have been included. We distinguish the following proteins in SynProt: (i) proteins residing directly in the PSD (established PSD proteins such as PSD-95), (ii) proteins which are associated with the PSD by means of functional interaction with PSD proteins (such as Cortactin; Racz and Weinberg, 2004), (iii) proteins that are associated with either the presynaptic compartment, the synaptic extracellular matrix or astroglial endfeet contacting or tethered to the postsynaptic membrane, and (iv) co-fractionating proteins that behave like PSD proteins due to similar physico-chemical properties during the fractionation procedure. Note, that mainly established PSD proteins are annotated as such in SynProt with the corresponding immunogold citation (work in progress). The primary goal of the SynProt database is to extract, integrate, and classify the wealth of proteomic information into a user friendly environment. Although specific protein databases such as UniProtKB and KEGG often contain information regarding proteins including their subcellular localization and molecular function, the SynProt database was created to complement the existing databases and to include only proteins validated by scientists.
Materials and Methods
Data sources
The following proteomic profiling studies were used to generate SynProt: Jordan et al. (2004), Li et al. (2004, 2005), Peng et al. (2004), Phillips et al. (2004), Yoshimura et al. (2004), Collins et al. (2005, 2006), Trinidad et al. (2005, 2006), Cheng et al. (2006), Dosemeci et al. (2006). In addition, we manually added proteins taken from Langnaese et al. (1996), Husi et al. (2000) as well as other well-known constituents of the PSD.
Data processing and quality control
Data on detergent-resistant synaptic junction proteins identified via proteomic screening or alternative approaches (see Table 2) were manually combined into a mySQL-based database due to the different data formats in the publications. Initial data obtained from large-scale proteomic screens included name and/or synonym if stated in the proteomics-paper, UniProt or NCBI identifier, species information on the proteins, and the reference(s) to the proteomic paper identifying the candidate protein as itself. A special web-front end was created to put data from the tables within these papers into the database, which allows making changes simultaneously on multiple datasets. This frontend is not part of the public version of the web-interface of the SynProt database, because its primary goal was quick manual curation.
Table 2.
Contributions of the selected 12 proteomic publications to the SynProt database.
| Paper | Methods | Proteins contributed to database |
|---|---|---|
| Li et al. (2004) | MALDI TOF/TOF; ICAT | 152 |
| Cheng et al. (2006) | ICAT; LC-MS/MS | 287 |
| Phillips et al. (2004) | MudPIT | 25 |
| Dosemeci et al. (2006) | MudPIT | 117 |
| Collins et al. (2005) | IMAC; LC-MS/MS | 213 |
| Yoshimura et al. (2004) | MudPIT | 448 |
| Jordan et al. (2004) | LC-MS/MS | 937 |
| Peng et al. (2004) | LC-MS/MS | 353 |
| Li et al. (2005) | MALDI TOF/TOF; ICAT; LC-MS/MS | 124 |
| Trinidad et al. (2005) | SCX Chromatography; IMAC; LC-MS/MS | 41 |
| Collins et al. (2006) | LC-MS/MS | 1,118 |
| Trinidad et al. (2006) | SCX Chromatography; IMAC; LC-MS/MS | 1,233 |
| Homologous proteins | Retrieved from Swiss-Prot by homology | 2,772 |
| Manually added | Yeast two-hybrid screens, antibody-based cDNA library screens, … | 49 |
The second column shows the used proteomic methods, the last column depicts the number of proteins, which contributed to the database. About 1,168 proteins were found in more than one study. The entire database (entries from proteomics studies, manually added and retrieved from Swiss-Prot) contains 5,560 proteins: 2,067 records from mouse, 1,359 records from rat, and 2,134 records are assigned to human.
We removed any duplicates of proteins identified in more than one study using in-house developed Perl scripts. In addition to proteins identified in the 12 proteomic approaches, we included proteins that were previously identified and verified as established PSD proteins by antibody-based investigations such as immunogold electron microscopy. Established PSD proteins are indicated in the field “ultrastructural localization.” Moreover, protein data entries were complemented by automatically retrieving the following additional information on candidate proteins from UniProt (UniProt-Consortium, 2010; Magrane and Consortium, 2011)1: protein sequence, molecular weight, gene name, UniProt accession identifier, primary references, and comments (e.g., function, subcellular localization, tissue specificity, or similarity) if available using additional in-house built Perl scripts. Further, we manually assigned one or more functional categories to each protein and manually curated all data sets once more to eliminate mistakes. The functional categories include 32 different classifications; the complete list is given in Table 3. Finally, homologous proteins of the three species mouse, rat, and human are included in SynProt for all proteins retrieved via the approaches mentioned above insofar as their information is available from UniProt. To distinguish these homologous proteins from their identified SynProt counterparts, they appear in orange in the database.
Table 3.
Statistical analysis of the 32 manual assigned functional classifications.
| Functional category | Number |
|---|---|
| Cell adhesion molecules | 43 |
| Lipid binding proteins | 56 |
| Transporter proteins | 187 |
| Membrane trafficking proteins | 249 |
| Glial proteins | 28 |
| Scaffolding and adaptor proteins | 144 |
| Ionotropic receptors and ion channels | 96 |
| Metabotropic receptors – GPCRs | 24 |
| Metabotropic receptors – RTKs and others | 18 |
| Cytoskeleton/actin-associated proteins | 235 |
| Cytoskeleton/intermediate filament-associated proteins | 32 |
| Cytoskeleton/microtuble-associated proteins | 141 |
| GTPases and regulators | 199 |
| Phosphatases and regulators | 242 |
| Protein modification and degradation | 103 |
| Chaperones and heat shock proteins | 63 |
| Mitochondrial proteins – energy metabolism | 217 |
| Protein synthesis | 174 |
| Endocytic proteins | 111 |
| Nucleic acid-binding proteins | 339 |
| No classification | 347 |
| Extracellularly matrix components | 32 |
| Presynaptic vesicle proteins | 67 |
| Presynaptic active zone proteins | 25 |
| Regulatory proteins | 622 |
| Ca2+ binding proteins | 48 |
| Proteasome | 18 |
| Catabolic pathway | 55 |
| Mitochondrial proteins – others | 71 |
| Cyclic nucleotide catabolic process | 3 |
| Transcription regulators | 58 |
| Energy metabolism, e.g., glycolysis | 21 |
1,686 of the 2,788 proteins (without entries, which were retrieved from Swiss-Prot by homology) were assigned to one classification, 896 proteins to two classifications, and 145 proteins to three classifications. For 320 proteins no classification could be found. About 2,772 entries were retrieved from UniProt to incorporate homologous proteins. These proteins are classified by assigning the manual classification from the original proteins.
Web portal application
The public face of the SynProt protein database is incorporated into the SynProt web portal for synaptic databases. The access to the database is restricted to academic research purposes only so far. For an easy public access to the database, a web-based user interface was designed (Corona Labs, Magdeburg, Germany)2. After registration it allows users to search the database for certain proteins or groups of proteins. At this time, there is no function to download the content of the database as a flat file.
Data querying
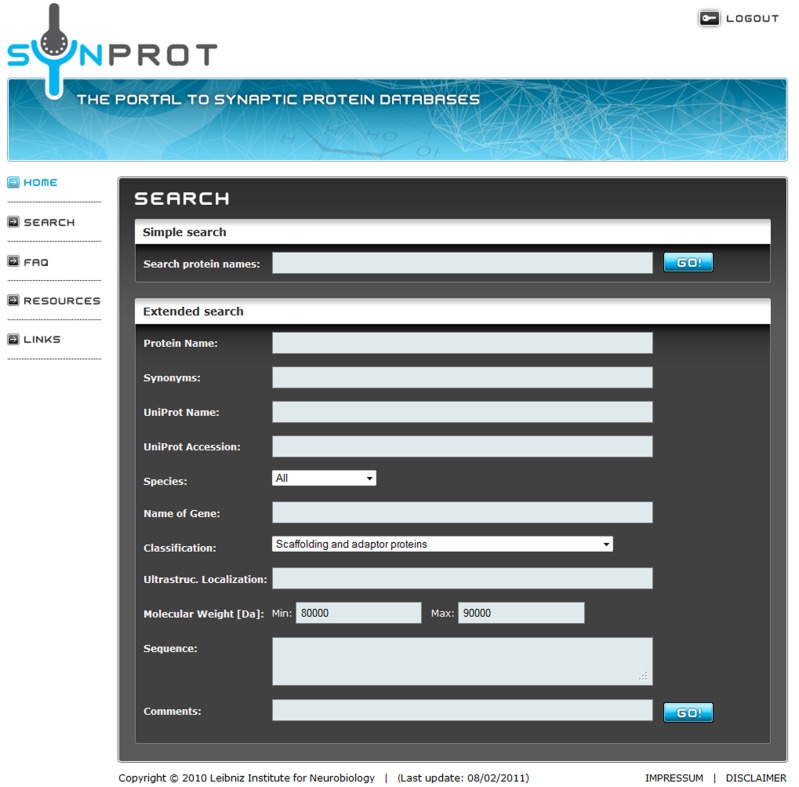
The user has the choice between a simple search and an extended search. In a simple search, the name of the protein is searched in all relevant fields like synonyms, whereas with an extended search the protein can be searched by properties, e.g., molecular weight or functions (Figure 1).
Figure 1.
Screenshot of a sample query. The user can choose between a simple search for a keyword in all fields or an extended search for one or more keywords in the specified fields. Here an extended search for a group of proteins is shown, which matches the following criteria: (1) classified as “Scaffolding and adaptor proteins” and (2) a molecular weight between 80,000 and 90,000 Da. The keywords are connected with a logical “AND” to accommodate detailed searches.
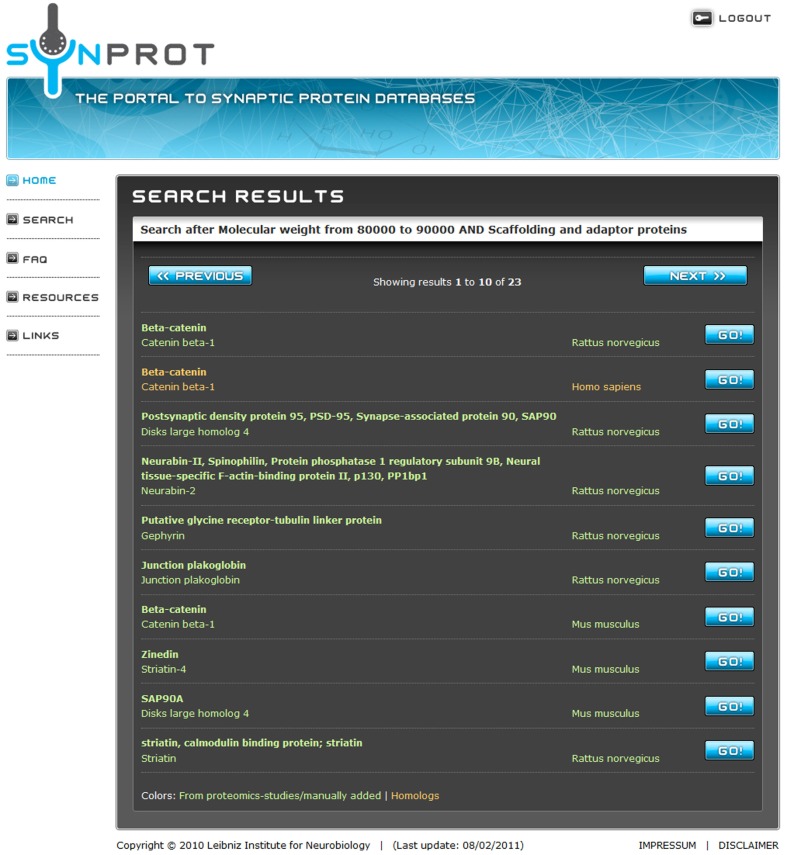
A group of proteins can be searched by a combination of properties in the extended search. The properties are connected with a logical “AND” to narrow the search results. After the search, a list of found data sets is presented (Figure 2). A maximum of 10 data sets can be displayed in one page. To get an overview of a certain data set, the protein name, synonyms, and the species are displayed. The user can now go back to refine the search or simply choose one of the data sets. The result page (Figure 3) shows the complete data set, the table shows fields with data, i.e., empty fields are not displayed.
Figure 2.
Screenshot after the extended search. The search retrieves 23 proteins matching the user-specified criteria. To prevent overloading of the browser, a maximum of 10 entries are shown on one page. Entries in green are derived from proteomic studies or manually added. Red entries are derived from UniProt due to homology. By clicking on the “Go!” Button, the user can choose the dataset of choice. The third and the ninth position show entries of the well-known postsynaptic protein PSD-95/SAP90 from different species.
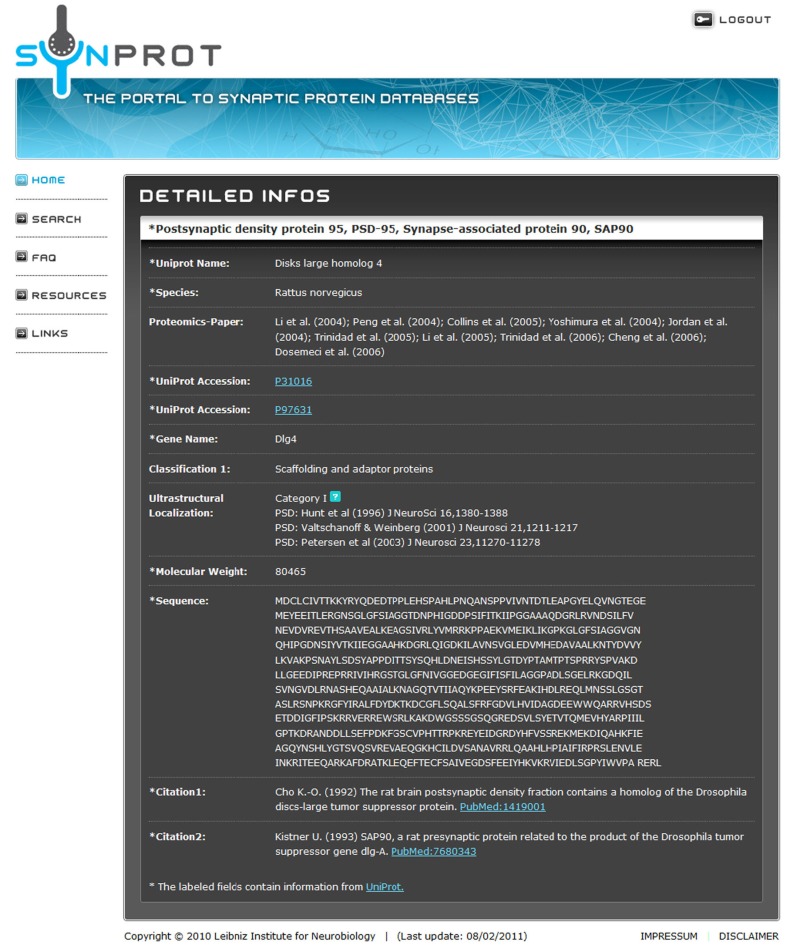
Figure 3.
Screenshot of the result screen after a search for “PSD-95.” The field “Proteomics-Paper” shows the proteomic studies, in which this protein was identified. In this case, it was found in 10 out of 12 studies. The field “Classification 1” denotes the manually assigned classification of PSD-95/SAP90. The field “Ultrastructural Localization” shows studies, in which the localization of this protein was verified by immunogold-staining. In addition, some of the proteins were manually assigned to categories, which denote the localization (see Table 4). All other data were retrieved from UniProt.
Results – the SynProt Database
State-of-the-art proteomics obtains a large amount of information on protein ensembles in a relatively short-time. However, while mass spectrometry instruments routinely sequence single purified proteins with subfemtomolar sensitivity, the effective identification of low-abundance proteins is orders of magnitudes lower in complex mixtures due to limited dynamic range and sequencing speed (de Godoy et al., 2006). This fact may result in the exclusion of low-abundance established PSD and detergent-resistant synaptic junction proteins or proteins which are difficult to sequence from identification via MS. Such data need to be complemented and supplemented by other identification means including biochemical and molecular biological approaches such as high-sensitivity protein sequencing (Aebersold et al., 1987), antibody-based expression screening of cDNA libraries (Kistner et al., 1993; Müller et al., 1995; Seidenbecher et al., 1995; Langnaese et al., 1996), yeast two-hybrid interaction studies (Kim et al., 1997; Lin et al., 1998; Naisbitt et al., 1999), studies analyzing specific PSD-residing complexes such as the NMDA receptor complex (Husi and Grant, 2001), and candidate-based studies. Moreover, the use of different search algorithms may yield different results. Hence, there is only a partial overlap of identified proteins in these detergent-resistant synaptic junction fractions in the different published studies. Table 2 lists the contributions of the 12 main proteomic studies included in the SynProt database (Jordan et al., 2004; Li et al., 2004, 2005; Peng et al., 2004; Phillips et al., 2004; Yoshimura et al., 2004; Collins et al., 2005, 2006; Trinidad et al., 2005, 2006; Cheng et al., 2006; Dosemeci et al., 2006).
The manual retrieval of the main core of data from 12 selected primary proteomic profiling publications on the detergent-resistant synaptic junction fraction, as well as a supplementary study on the NMDA receptor complex (Husi et al., 2000), a study on expression screening of a rat cDNA library with the help of antisera raised against a rat brain synaptic protein preparation (Kistner et al., 1993; Müller et al., 1995; Seidenbecher et al., 1995; Langnaese et al., 1996), and also well-known manually added established PSD proteins from the literature resulted in a total of 9,298 records, including many proteins represented multiple times in these studies. Established PSD proteins are listed in Table 4. The most prominent proteins in the proteomic studies are PSD-93, which was identified in 11 studies, and PSD-95/SAP90 and ProSAP3/Shank1, which both were identified in 10 studies. In addition to these entries, 49 published constituents of the PSD have been manually added to the database, which have not been identified in the proteomic profiling experiments using mass spectrometry based methods but via other means, such as the synapto-nuclear messenger protein Jacob (Dieterich et al., 2008). After automatic removal of redundant protein entries and including any available information on homologous proteins from the three species human, mouse, and rat accessed from UniProt, the database currently holds a total of 5,560 records (mouse: 2,067 proteins, rat: 1,359 proteins, human: 2,134 proteins). In total 2,138 unique, homologous proteins can be searched by the user within the SynProt database. The majority of the proteins within the SynProt database are assigned in the proteomic identification procedures to the rodent brain (mouse: 1,741 records; rat: 922 records), fewer proteins were matched to human brain proteins (76 records).
Table 4.
Ultrastructural localization of proteins present in detergent-resistant synaptic junctions fractions.
| Protein name | Postsynaptic localization/distance to the synaptic cleft | Category | Reference |
|---|---|---|---|
| Pan-actin | Yes/109 ± 51 nm | (i), (ii) | Rostaing et al. (2006) |
| Alpha-actinin | Yes/20–120 nm | (i), (ii) | Racz and Weinberg (2004) |
| Bassoon | No, presynaptic | (iii) presynaptic | tom Dieck et al. (1998) |
| Beta-catenin | Yes/within 100 nm | (ii) | Petralia et al. (2005) |
| CaM-kinase II alpha | Yes/39 ± 31 nm | (i) | Rostaing et al. (2006) |
| Yes/within 100 nm | (i) | Petralia et al. (2005) | |
| CaM-kinase II alpha | Yes/approx. 30 nm | (i) | Petersen et al. (2003) |
| CASK | Yes, PSD-associated | (i), also presynaptic (iii) | Hsueh et al. (1998) |
| Cav1.2 | Yes, PSD-associated | (i) | Leitch et al. (2009) |
| Cortactin | Yes/100–150 nm | (ii) | Racz and Weinberg (2004) |
| CRIPT | Yes/40 nm | (i) | Niethammer et al. (1998) |
| Yes/27.9 ± 2.3 | (i) | Valtschanoff and Weinberg (2001) | |
| Dynein light chain 2 | Yes/40–120 nm; also spine apparatus | (i), also (iv) | Naisbitt et al. (2000) |
| Yes/31.5 ± 3.2; also spine apparatus | (i), also (iv) | Valtschanoff and Weinberg (2001) | |
| ERK1/2 | Yes, PSD-associated | (i) | Boggio et al. (2007) |
| GKAP1 | Yes/25.9 ± 2.1 | (i) | Valtschanoff and Weinberg (2001) |
| GKAP1 | Yes/within 100 nm | (i) | Petralia et al. (2005) |
| GluR1 | Yes/17 ± 13 nm | (i) | Rostaing et al. (2006) |
| Yes/PSD-associated | (i) | Racca et al. (2000) | |
| Yes/PSD-associated | (i) | Baude et al. (1995) | |
| GluR2 | Yes/PSD-associated | (i) | Racca et al. (2000) |
| GluR2/3 | Yes/PSD-associated | (i) | Racca et al. (2000) |
| Yes/PSD-associated | (i) | Baude et al. (1995) | |
| Yes/PSD-associated | (i) | Fux et al. (2003) | |
| GluR2/3 | Yes/approx. 20 nm | (i) | Kharazia and Weinberg (1999) |
| Yes/PSD-associated | (i) | Nusser et al. (1994) | |
| Yes/PSD-associated | (i) | Nusser et al. (1998) | |
| GluR2/3 | Yes/20 nm | (i) | Matsubara et al. (1996) |
| GluR4 | Yes/PSD-associated | (i) | Baude et al. (1995) |
| GluR4 | Yes/25 nm | (i) | Matsubara et al. (1996) |
| Homer 1b,c | Yes/within 100 nm | (i) | Petralia et al. (2005) |
| Yes/PSD-associated, perisynaptic | (i) | Tu et al. (1999) | |
| KA1 | Yes/PSD-associated | (i) | Darstein et al. (2003) |
| KA2 | Yes/PSD-associated | (i) | Darstein et al. (2003) |
| MCT2 | Yes/PSD-associated | (i) | Bergersen et al. (2002) |
| mGluR1 alpha | Yes/PSD-associated | (i) | Kuwajima et al. (2004) |
| Yes/PSD-associated, perisynaptic | (i) | Nusser et al. (1994) | |
| mGluR1 alpha | Yes/perisynaptic | (i) | Baude et al. (1993) |
| mGluR1 | Yes/PSD-associated, perisynaptic | (i) | Fux et al. (2003) |
| mGluR1 | Yes/perisynaptic | (i) | Lujan et al. (1996) |
| mGluR5 | Yes/PSD-associated, perisynaptic | (i) | Tu et al. (1999) |
| mGluR5 | Yes/PSD-associated | (i) | Kuwajima et al. (2004) |
| Yes/perisynaptic | (i) | Lujan et al. (1996) | |
| N-Cadherin | Yes/within 100 nm | (i) | Petralia et al. (2005) |
| NCAM180 | Yes/PSD-associated | (i) | Fux et al. (2003) |
| Neuroligin-1 | Yes/synaptic cleft and PSD-associated | (i) | Song et al. (1999) |
| nNOS | Yes/18.1 ± 2.1 | (i) | Valtschanoff and Weinberg (2001) |
| NR1 | Yes/PSD-associated | (i) | Racca et al. (2000) |
| NR1 | Yes/approx. 20 nm | (i) | Kharazia and Weinberg (1999) |
| NR2A/B | Yes/PSD-associated | (i) | Racca et al. (2000) |
| NR2A/B | Yes/11.2 ± 1.5 | (i) | Valtschanoff and Weinberg (2001) |
| NR2A | Yes/within 100 nm | (i) | Petralia et al. (2005) |
| Yes/PSD-associated | (i) | Fux et al. (2003) | |
| NR2B | Yes/within 100 nm | (i) | Petralia et al. (2005) |
| NrCAM | Yes/within 100 nm | (i) | Petralia et al. (2005) |
| Pan-cortactin | Yes/59 ± 36 nm | (i), (ii) | Rostaing et al. (2006) |
| Pan-GluR | Yes/PSD-associated | (i) | Racca et al. (2000) |
| Pan-GluR | Yes/PSD-associated | (i) | Nusser et al. (1998) |
| Pan-neuroligin | Yes/within 100 nm | (i) | Petralia et al. (2005) |
| Prickle-2 | Yes/approx. 30 nm | (i) | Hida et al. (2011) |
| ProSAP2/Shank3 | Yes/41 ± 25 nm | (i) | Rostaing et al. (2006) |
| Yes/within 100 nm | (i) | Petralia et al. (2005) | |
| Shank2/3 | Yes/PSD-associated | (i) | Tu et al. (1999) |
| PSD-95 | Yes/approx. 15 nm | (i) | Petersen et al. (2003) |
| Yes/11.9 ± 1.2 | Valtschanoff and Weinberg (2001) | ||
| Yes/PSD-associated | Hunt et al. (1996) | ||
| SAP102 | Yes/within 100 nm | (i) | Petralia et al. (2005) |
| Septin-4 | No/presynaptic, also astrocytic | (iii) presynaptic, astrocytic | Kinoshita et al. (2000) |
| Septin-5 | No/presynaptic | (iii) presynaptic, on vesicles | Kinoshita et al. (2000) |
| Septin-6 | No/presynaptic | (iii) presynaptic, on vesicles | Kinoshita et al. (2000) |
| Septin-7 | No/presynaptic also astrocytic | (iii) presynaptic, astrocytic | Kinoshita et al. (2000) |
| Septin-11 | Yes/PSD-associated | (i)/(ii) difficult to determine | Li et al. (2009) |
| Septin-3 | No/presynaptic | (iii) presynaptic, on vesicles | Xue et al. (2004) |
| Shank-1 | Yes/20–50 nm | (i) | Naisbitt et al. (1999) |
| Yes/24.1 ± 1.7 | (i) | Valtschanoff and Weinberg (2001) | |
| SPAR | Yes/within 100 nm | (i), (ii) | Petralia et al. (2005) |
| Synaptopodin | Yes/spine apparatus | (iv) | Niesmann et al. (2011) |
| Syndecan-2 | Yes/PSD-associated | (i), also presynaptic (iii) | Hsueh et al. (1998) |
| SynGAP | Yes/within 100 nm | (i), (ii) | Petralia et al. (2005) |
| TrkB | Yes/within 100 nm | (i), also presynaptic (iii) | Petralia et al. (2005) |
All data are collected from studies based on immunogold-stainings. Proteins are categorized as suggested in the introduction: (i) proteins residing directly in the PSD (within 100 nm from the postsynaptic membrane), (ii) proteins associated with the postsynaptic density by means of functional interaction with PSD proteins, (iii) proteins associated with either presynaptic compartments, synaptic extracellular matrix or astrocytic endfeet contacting or tethered to the postsynaptic membrane, (iv) co-fractionating proteins that behave like PSD proteins due to similar physico-chemical properties during the fractionation procedure. Some proteins belong to more than one category. If possible, quantitative data for the distance range or the postsynaptic distance from the synaptic cleft with highest number of gold particles are given in the column “Postsynaptic localization/distance to the synaptic cleft.”
The initially included datasets for 2,788 proteins were manually assigned to 32 different functional categories (three categories maximum per protein) totaling to 3,721 assignments. About 1,686 proteins were assigned to one category, 896 proteins to two categories, and 145 proteins to three categories.
Most notably, the majority of records were assigned to the functional classes of regulatory proteins (622 proteins), nucleic acid-binding proteins (339; Suzuki et al., 2007), and membrane trafficking proteins (249). The complete statistical analysis of the manual assignment is shown in Table 3.
Sample query
SynProt can be queried for single proteins as well as for groups of proteins matching certain, user-specified criteria. A simple search is used for a quick check if a distinct protein is included in the database by searching of the name in “synonyms,” “protein name,” and “UniProt name.” To showcase an example for an extended search; the SynProt database is queried for a group of proteins using two criteria: (i) “Scaffolding and adaptor proteins” and (ii) having a molecular weight of 80–90 kDa (Figure 1). The query returns a group of 23 proteins (Figure 2). To provide a better overview on the browser, a maximum of 10 proteins are shown at the same time. The list includes entries of the established PSD protein PSD-95/SAP90 from different species. From this list the user can select the hit of choice to obtain further information on this entry by clicking the appropriate “Go!” button. In case of PSD-95/SAP90 from rat a new window opens displaying the following detailed information (Figure 3): synonyms, full name, species, proteomics-papers in which this protein was identified, UniProt accession number(s), gene name, classification, molecular weight, sequence, and citation(s).
External user contributions
To complement the SynProt database external users are encouraged to submit any yet missing PSD protein by contacting the authors or the webmaster of SynProt along with a solid proof that the protein in question is indeed a true component either in the PSD itself or in the detergent-resistant synaptic junction fraction, i.e., by providing data on immunogold EM synaptic localization or by Western Blot analysis of these fractions, respectively.
Discussion
The identity and dynamics of the molecular constituents of synaptic junctions and the PSD in particular have been in the focus of extensive research for over 50 years now, aiming not only to decipher higher cognitive functions such as learning and memory, but also to ultimately disclose ways to fight brain diseases including Alzheimer’s disease, Morbus Parkinson, or Schizophrenia. Thus, it is not surprising that a simple Google search for “PSD protein” results in approx. 480,000 hits, and in combination with “disease” in approx. 288,000 hits underlining the huge amount of information in the internet about the PSD, which makes it difficult to retrieve particular information. On the other hand a query for “postsynaptic AND density” in the life-science relevant protein database UniProt retrieves a total of 577 results, and 193 hits combined with the restriction “subcellular location,” clearly not registering all resident proteins in the PSD. Therefore, we compiled the first comprehensive and manually curated database on detergent-resistant synaptic junctional proteins and established PSD proteins from rat, mouse, and human species using proteomic, biochemical, immunogold EM and additional bioinformatics data sources. The core of data sets of the SynProt database is derived from 12 proteomic profiling approaches on biochemically purified detergent-resistant synaptic junction fractions with subsequent MS analysis. This has been supplemented with additional PSD proteins as not all proteins are accessible by MS analysis. Next to the original data source, we provide additional information for all data entries, including the UniProt accession number, synonyms, gene name, sequence, molecular weight, primary publication citation(s), and to which biological classifications the protein can be assigned. The user can choose between two search modes: A “simple” search using either a particular protein name or synonymous name, or a more extensive search using a keyword, a string of search criteria for a group of proteins based on a particular biological classification, or even just a snippet of sequence information. Furthermore, the user can choose information on a particular protein either based on the originally identified data entry or its homologous counterparts if this information is available from UniProt.
The statistical analysis of classifications shows the main function of the proteins of the core PSD and the detergent-resistant synaptic junction proteins is to form a signal transduction and modification machinery. The most prominent functional category of all proteins contained in SynProt concerns regulation of the signaling protein network, which transfers the signal on a short-time-scale. Interestingly, SynProt contains a variety of nucleic acid-binding proteins and components of the translation machinery identified in the detergent-resistant synaptic junction fraction. These are likely responsible for transduction or modification of the signal on a longer time-scale by local protein synthesis. However, one has to take into account that proteins with similar physico-chemical properties may co-fractionate with detergent-resistant synaptic junction proteins, one class being histone proteins. Notably a total of 28 proteins in SynProt are assigned to the class of glia-derived proteins, which points to the emerging concept of the “tripartite synapse.” Glial endfeet have recently been established as functional requirements for a proper synapse function and, therefore, for neuronal function and development (for a review see Araque et al., 1999).
The SynProt database is a convenient tool for a quick check for the occurrence of a specific protein in the PSD or in the detergent-resistant synaptic junction fraction. But more importantly, it allows the specific search for a group of proteins. The retrieval of proteins, which share a certain sequence (for example PDZ motifs) in combination with the curated information about the functional category, benefits our understanding of synaptic function in general and is a valuable source of information for students and researchers new to the field. The focus on postsynaptic proteins in the SynProt database enhances the quality of this information, which cannot be achieved by other existing protein databases or text-mining tools.
The quality and sustainability of a database depends on the long-term maintenance. Many labs investigate the constituents and the function of postsynaptic densities, and, therefore, a huge amount of data has been produced to date. Note, that the main core of protein entries in SynProt is derived from the detergent-resistant synaptic junctional protein fraction prepared from native animals, i.e., any proteins that transiently redistribute into the synaptic compartment due to a change in synaptic activity might not be included. The Leibniz Institute for Neurobiology will provide a permanent up-date of the database and all interfaces to the user. The SynProt database is the first one of specific databases for different classes of brain proteins, which will be unified under the SynProt web portal for synaptic proteins.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by Joint Initiative for Research and Innovation of the Leibniz Association; the Center for Behavioral Brain Sciences (CBBS/ERDF (2007–2013): NeuroNetworks ZVOH TP6/1); Center for Dynamic Systems at the Otto-von-Guericke University Magdeburg; and Deutsche Forschungsgemeinschaft (Emmy Noether Program to Daniela C. Dieterich). The authors want to express their gratitude toward the first reviewer of this paper for helpful comments and suggestions. The authors thank Ulrike Herrmann and Christine Freitag for their help with data extraction.
Footnotes
References
- Aebersold R. H., Leavitt J., Saavedra R. A., Hood L. E., Kent S. B. H. (1987). Internal amino acid sequence analysis of proteins separated by one- or two-dimensional gel electrophoresis after in situ proteasedigestion on nitrocellulose. Proc. Natl. Acad. Sci. U.S.A. 84, 6970–6974 10.1073/pnas.84.20.6970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akert K., Moor H., Pfenninger K., Sandri C. (1969). Contributions of new impregnation methods and freeze etching to the problems of synaptic fine structure. Prog. Brain Res. 31, 223–240 10.1016/S0079-6123(08)63241-0 [DOI] [PubMed] [Google Scholar]
- Apperson M. L., Moon I. S., Kennedy M. B. (1996). Characterization of densin-180, a new brain-specific synaptic protein of the O-sialoglycoprotein family. J. Neurosci. 16, 6839–6852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araque A., Parpura V., Sanzgiri R. P., Haydon P. G. (1999). Tripartite synapses: glia, the unacknowledged partner. Trends Neurosci. 22, 208–215 10.1016/S0166-2236(98)01349-6 [DOI] [PubMed] [Google Scholar]
- Baude A., Nusser Z., Molnar E., McIlhinney R. A., Somogyi P. (1995). High-resolution immunogold localization of AMPA type glutamate receptor subunits at synaptic and non-synaptic sites in rat hippocampus. Neuroscience 69, 1031–1055 10.1016/0306-4522(95)00350-R [DOI] [PubMed] [Google Scholar]
- Baude A., Nusser Z., Roberts J. D., Mulvihill E., McIlhinney R. A., Somogyi P. (1993). The metabotropic glutamate receptor (mGluR1 alpha) is concentrated at perisynaptic membrane of neuronal subpopulations as detected by immunogold reaction. Neuron 11, 771–787 10.1016/0896-6273(93)90086-7 [DOI] [PubMed] [Google Scholar]
- Bergersen L., Rafiki A., Ottersen O. P. (2002). Immunogold cytochemistry identifies specialized membrane domains for monocarboxylate transport in the central nervous system. Neurochem. Res. 27, 89–96 10.1023/A:1014806723147 [DOI] [PubMed] [Google Scholar]
- Boeckers T. M., Kreutz M. R., Winter C., Zuschratter W., Smalla K. H., Sanmarti-Vila L., Wex H., Langnaese K., Bockmann J., Garner C. C., Gundelfinger E. D. (1999). Proline-rich synapse-associated protein-1/cortactin binding protein 1 (ProSAP1/CortBP1) is a PDZ-domain protein highly enriched in the postsynaptic density. J. Neurosci. 19, 6506–6518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boggio E. M., Putignano E., Sassoe-Pognetto M., Pizzorusso T., Giustetto M. (2007). Visual stimulation activates ERK in synaptic and somatic compartments of rat cortical neurons with parallel kinetics. PLoS ONE 2, e604. 10.1371/journal.pone.0000604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlin R. K., Grab D. J., Cohen R. S., Siekevitz P. (1980). Isolation and characterization of postsynaptic densities from various brain regions: enrichment of different types of postsynaptic densities. J. Cell Biol. 86, 831–845 10.1083/jcb.86.3.831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cases-Langhoff C., Voss B., Garner A. M., Appeltauer U., Takei K., Kindler S., Veh R. W., De Camilli P., Gundelfinger E. D., Garner C. C. (1996). Piccolo, a novel 420 kDa protein associated with the presynaptic cytomatrix. Eur. J. Cell Biol. 69, 214–223 [PubMed] [Google Scholar]
- Chen H. J., Rojas-Soto M., Oguni A., Kennedy M. B. (1998). A synaptic Ras-GTPase activating protein (p135 SynGAP) inhibited by CaM kinase II. Neuron 20, 895–904 10.1016/S0896-6273(00)80471-7 [DOI] [PubMed] [Google Scholar]
- Cheng D., Hoogenraad C. C., Rush J., Ramm E., Schlager M. A., Duong D. M., Xu P., Wijayawardana S. R., Hanfelt J., Nakagawa T., Sheng M., Peng J. (2006). Relative and absolute quantification of postsynaptic density proteome isolated from rat forebrain and cerebellum. Mol. Cell. Proteomics 5, 1158–1170 10.1074/mcp.D500009-MCP200 [DOI] [PubMed] [Google Scholar]
- Cho K. O., Hunt C. A., Kennedy M. B. (1992). The rat brain postsynaptic density fraction contains a homolog of the Drosophila discs-large tumor suppressor protein. Neuron 9, 929–942 10.1016/0896-6273(92)90245-9 [DOI] [PubMed] [Google Scholar]
- Cohen R. S., Blomberg F., Berzins K., Siekevitz P. (1977). The structure of postsynaptic densities isolated from dog cerebral cortex. I. Overall morphology and protein composition. J. Cell Biol. 74, 181–203 10.1083/jcb.74.1.181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins M. O., Husi H., Yu L., Brandon J. M., Anderson C. N., Blackstock W. P., Choudhary J. S., Grant S. G. (2006). Molecular characterization and comparison of the components and multiprotein complexes in the postsynaptic proteome. J. Neurochem. 97(Suppl. 1), 16–23 10.1111/j.1471-4159.2005.03507.x [DOI] [PubMed] [Google Scholar]
- Collins M. O., Yu L., Coba M. P., Husi H., Campuzano I., Blackstock W. P., Choudhary J. S., Grant S. G. (2005). Proteomic analysis of in vivo phosphorylated synaptic proteins. J. Biol. Chem. 280, 5972–5982 10.1074/jbc.M411603200 [DOI] [PubMed] [Google Scholar]
- Cotman C. W., Banker G., Churchill L., Taylor D. (1974). Isolation of postsynaptic densities from rat brain. J. Cell Biol. 63, 441–455 10.1083/jcb.63.2.441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotman C. W., Taylor D. (1972). Isolation and structural studies on synaptic complexes from rat brain. J. Cell Biol. 55, 696–711 10.1083/jcb.55.3.696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotman C. W., Taylor D. (1974). Localization and characterization of concanavalin A receptors in the synaptic cleft. J. Cell Biol. 62, 236–242 10.1083/jcb.62.1.236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darstein M., Petralia R. S., Swanson G. T., Wenthold R. J., Heinemann S. F. (2003). Distribution of kainate receptor subunits at hippocampal mossy fiber synapses. J. Neurosci. 23, 8013–8019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Godoy L. M., Olsen J. V., De Souza G. A., Li G., Mortensen P., Mann M. (2006). Status of complete proteome analysis by mass spectrometry: SILAC labeled yeast as a model system. Genome Biol. 7, R50. 10.1186/gb-2006-7-6-r50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Robertis E. (1959). Submicroscopic morphology of the synapse. Int. Rev. Cytol. 8, 61–96 10.1016/S0074-7696(08)62728-X [DOI] [PubMed] [Google Scholar]
- de Robertis E., Pellegrino De Iraldi A., Rodriguez De Lores Arnaiz G., Salganicoff L. (1961). Electron microscope observations on nerve endings isolated from rat brain. Anat. Rec. 139, 220–221 [Google Scholar]
- de Robertis E. D. (1965). Some new electron microscopical contributions to the biology of neuroglia. Prog. Brain Res. 15, 1–11 10.1016/S0079-6123(08)60936-X [DOI] [PubMed] [Google Scholar]
- Dieterich D. C., Karpova A., Mikhaylova M., Zdobnova I., Konig I., Landwehr M., Kreutz M., Smalla K. H., Richter K., Landgraf P., Reissner C., Boeckers T. M., Zuschratter W., Spilker C., Seidenbecher C. I., Garner C. C., Gundelfinger E. D., Kreutz M. R. (2008). Caldendrin-Jacob: a protein liaison that couples NMDA receptor signalling to the nucleus. PLoS Biol. 6, e34. 10.1371/journal.pbio.0060034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dityatev A., Seidenbecher C. I., Schachner M. (2010). Compartmentalization from the outside: the extracellular matrix and functional microdomains in the brain. Trends Neurosci. 33, 503–512 10.1016/j.tins.2010.08.003 [DOI] [PubMed] [Google Scholar]
- Dosemeci A., Tao-Cheng J. H., Vinade L., Jaffe H. (2006). Preparation of postsynaptic density fraction from hippocampal slices and proteomic analysis. Biochem. Biophys. Res. Commun. 339, 687–694 10.1016/j.bbrc.2005.11.069 [DOI] [PubMed] [Google Scholar]
- Eccles J. C. (1957). The Physiology of Nerve Cells. Baltimore: John Hopkins Press [Google Scholar]
- Faissner A., Pyka M., Geissler M., Sobik T., Frischknecht R., Gundelfinger E. D., Seidenbecher C. (2010). Contributions of astrocytes to synapse formation and maturation – potential functions of the perisynaptic extracellular matrix. Brain Res. Rev. 63, 26–38 10.1016/j.brainresrev.2010.01.001 [DOI] [PubMed] [Google Scholar]
- Fux C. M., Krug M., Dityatev A., Schuster T., Schachner M. (2003). NCAM180 and glutamate receptor subtypes in potentiated spine synapses: an immunogold electron microscopic study. Mol. Cell. Neurosci. 24, 939–950 10.1016/j.mcn.2003.07.001 [DOI] [PubMed] [Google Scholar]
- Garner C. C., Kindler S., Gundelfinger E. D. (2000). Molecular determinants of presynaptic active zones. Curr. Opin. Neurobiol. 10, 321–327 10.1016/S0959-4388(00)00093-3 [DOI] [PubMed] [Google Scholar]
- Gray E. G. (1959). Axo-somatic and axo-dendritic synapses of the cerebral cortex: an electron microscope study. J. Anat. 93, 420–433 [PMC free article] [PubMed] [Google Scholar]
- Gray E. G. (1987). “Synapse morphology,” in Encyclopedia in Neuroscience, ed. Adelman G. (Boston: Birkhäuser; ), 1158–1162 [Google Scholar]
- Gray E. G., Whittaker V. P. (1962). The isolation of nerve endings from brain: an electron-microscopic study of cell fragments derived by homogenization and centrifugation. J. Anat. 96, 79–88 [PMC free article] [PubMed] [Google Scholar]
- Hida Y., Fukaya M., Hagiwara A., Deguchi-Tawarada M., Yoshioka T., Kitajima I., Inoue E., Watanabe M., Ohtsuka T. (2011). Prickle2 is localized in the postsynaptic density and interacts with PSD-95 and NMDA receptors in the brain. J. Biochem. 149, 693–700 10.1093/jb/mvr023 [DOI] [PubMed] [Google Scholar]
- Hollingsworth E. B., McNeal E. T., Burton J. L., Williams R. J., Daly J. W., Creveling C. R. (1985). Biochemical characterization of a filtered synaptoneurosome preparation from guinea pig cerebral cortex: cyclic adenosine 3′:5′-monophosphate-generating systems, receptors, and enzymes. J. Neurosci. 5, 2240–2253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsueh Y. P., Yang F. C., Kharazia V., Naisbitt S., Cohen A. R., Weinberg R. J., Sheng M. (1998). Direct interaction of CASK/LIN-2 and syndecan heparan sulfate proteoglycan and their overlapping distribution in neuronal synapses. J. Cell Biol. 142, 139–151 10.1083/jcb.142.1.139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt C. A., Schenker L. J., Kennedy M. B. (1996). PSD-95 is associated with the postsynaptic density and not with the presynaptic membrane at forebrain synapses. J. Neurosci. 16, 1380–1388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husi H., Grant S. G. (2001). Isolation of 2000-kDa complexes of N-methyl-D-aspartate receptor and postsynaptic density 95 from mouse brain. J. Neurochem. 77, 281–291 10.1046/j.1471-4159.2001.t01-1-00248.x [DOI] [PubMed] [Google Scholar]
- Husi H., Ward M. A., Choudhary J. S., Blackstock W. P., Grant S. G. (2000). Proteomic analysis of NMDA receptor-adhesion protein signaling complexes. Nat. Neurosci. 3, 661–669 10.1038/76615 [DOI] [PubMed] [Google Scholar]
- Jordan B. A., Fernholz B. D., Boussac M., Xu C., Grigorean G., Ziff E. B., Neubert T. A. (2004). Identification and verification of novel rodent postsynaptic density proteins. Mol. Cell. Proteomics 3, 857–871 10.1074/mcp.M400045-MCP200 [DOI] [PubMed] [Google Scholar]
- Kennedy M. B. (1993). The postsynaptic density. Curr. Opin. Neurobiol. 3, 732–737 10.1016/0959-4388(93)90145-O [DOI] [PubMed] [Google Scholar]
- Kennedy M. B. (1997). The postsynaptic density at glutamatergic synapses. Trends Neurosci. 20, 264–268 10.1016/S0166-2236(96)01033-8 [DOI] [PubMed] [Google Scholar]
- Kennedy M. B., Bennett M. K., Erondu N. E. (1983). Biochemical and immunochemical evidence that the “major postsynaptic density protein” is a subunit of a calmodulin-dependent protein kinase. Proc. Natl. Acad. Sci. U.S.A. 80, 7357–7361 10.1073/pnas.80.23.7357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharazia V. N., Weinberg R. J. (1999). Immunogold localization of AMPA and NMDA receptors in somatic sensory cortex of albino rat. J. Comp. Neurol. 412, 292–302 [DOI] [PubMed] [Google Scholar]
- Kim E., Naisbitt S., Hsueh Y. P., Rao A., Rothschild A., Craig A. M., Sheng M. (1997). GKAP, a novel synaptic protein that interacts with the guanylate kinase-like domain of the PSD-95/SAP90 family of channel clustering molecules. J. Cell Biol. 136, 669–678 10.1083/jcb.136.1.19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita A., Noda M., Kinoshita M. (2000). Differential localization of septins in the mouse brain. J. Comp. Neurol. 428, 223–239 [DOI] [PubMed] [Google Scholar]
- Kistner U., Wenzel B. M., Veh R. W., Cases-Langhoff C., Garner A. M., Appeltauer U., Voss B., Gundelfinger E. D., Garner C. C. (1993). SAP90, a rat presynaptic protein related to the product of the Drosophila tumor suppressor gene dlg-A. J. Biol. Chem. 268, 4580–4583 [PubMed] [Google Scholar]
- Kornau H. C., Schenker L. T., Kennedy M. B., Seeburg P. H. (1995). Domain interaction between NMDA receptor subunits and the postsynaptic density protein PSD-95. Science 269, 1737–1740 10.1126/science.7569905 [DOI] [PubMed] [Google Scholar]
- Kuwajima M., Hall R. A., Aiba A., Smith Y. (2004). Subcellular and subsynaptic localization of group I metabotropic glutamate receptors in the monkey subthalamic nucleus. J. Comp. Neurol. 474, 589–602 10.1002/cne.20158 [DOI] [PubMed] [Google Scholar]
- Langnaese K., Seidenbecher C., Wex H., Seidel B., Hartung K., Appeltauer U., Garner A., Voss B., Müller B., Garner C. C., Gundelfinger E. D. (1996). Protein components of a rat brain synaptic junctional protein preparation. Brain Res. Mol. Brain Res. 42, 118–122 10.1016/S0169-328X(96)00147-7 [DOI] [PubMed] [Google Scholar]
- Leitch B., Szostek A., Lin R., Shevtsova O. (2009). Subcellular distribution of L-type calcium channel subtypes in rat hippocampal neurons. Neuroscience 164, 641–657 10.1016/j.neuroscience.2009.08.006 [DOI] [PubMed] [Google Scholar]
- Li K., Hornshaw M. P., Van Minnen J., Smalla K. H., Gundelfinger E. D., Smit A. B. (2005). Organelle proteomics of rat synaptic proteins: correlation-profiling by isotope-coded affinity tagging in conjunction with liquid chromatography-tandem mass spectrometry to reveal post-synaptic density specific proteins. J. Proteome Res. 4, 725–733 10.1021/pr049802+ [DOI] [PubMed] [Google Scholar]
- Li K. W., Hornshaw M. P., Van Der Schors R. C., Watson R., Tate S., Casetta B., Jimenez C. R., Gouwenberg Y., Gundelfinger E. D., Smalla K. H., Smit A. B. (2004). Proteomics analysis of rat brain postsynaptic density. Implications of the diverse protein functional groups for the integration of synaptic physiology. J. Biol. Chem. 279, 987–1002 10.1074/jbc.M312004200 [DOI] [PubMed] [Google Scholar]
- Li K. W., Klemmer P., Smit A. B. (2010). Interaction proteomics of synapse protein complexes. Anal. Bioanal. Chem. 397, 3195–3202 10.1007/s00216-010-3777-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Serwanski D. R., Miralles C. P., Nagata K., De Blas A. L. (2009). Septin 11 is present in GABAergic synapses and plays a functional role in the cytoarchitecture of neurons and GABAergic synaptic connectivity. J. Biol. Chem. 284, 17253–17265 10.1074/jbc.M109.056499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J. W., Wyszynski M., Madhavan R., Sealock R., Kim J. U., Sheng M. (1998). Yotiao, a novel protein of neuromuscular junction and brain that interacts with specific splice variants of NMDA receptor subunit NR1. J. Neurosci. 18, 2017–2027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lujan R., Nusser Z., Roberts J. D., Shigemoto R., Somogyi P. (1996). Perisynaptic location of metabotropic glutamate receptors mGluR1 and mGluR5 on dendrites and dendritic spines in the rat hippocampus. Eur. J. Neurosci. 8, 1488–1500 10.1111/j.1460-9568.1996.tb01611.x [DOI] [PubMed] [Google Scholar]
- Magrane M., Consortium U. (2011). UniProt Knowledgebase: a hub of integrated protein data. Database (Oxford) 2011, bar009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubara A., Laake J. H., Davanger S., Usami S., Ottersen O. P. (1996). Organization of AMPA receptor subunits at a glutamate synapse: a quantitative immunogold analysis of hair cell synapses in the rat organ of Corti. J. Neurosci. 16, 4457–4467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matus A., Ackermann M., Pehling G. (1981). Regularity and differentiation within the structure of brain postsynaptic densities. J. Neurocytol. 10, 889–896 10.1007/BF01258520 [DOI] [PubMed] [Google Scholar]
- Matus A. I., Walters B. B. (1975). Ultrastructure of the synaptic junctional lattice isolated from mammalian brain. J. Neurocytol. 4, 369–375 10.1007/BF01099094 [DOI] [PubMed] [Google Scholar]
- Moon I. S., Apperson M. L., Kennedy M. B. (1994). The major tyrosine-phosphorylated protein in the postsynaptic density fraction is N-methyl-D-aspartate receptor subunit 2B. Proc. Natl. Acad. Sci. U.S.A. 91, 3954–3958 10.1073/pnas.91.9.3954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller B. M., Kistner U., Veh R. W., Cases-Langhoff C., Becker B., Gundelfinger E. D., Garner C. C. (1995). Molecular characterization and spatial distribution of SAP97, a novel presynaptic protein homologous to SAP90 and the Drosophila discs-large tumor suppressor protein. J. Neurosci. 15, 2354–2366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naisbitt S., Kim E., Tu J. C., Xiao B., Sala C., Valtschanoff J., Weinberg R. J., Worley P. F., Sheng M. (1999). Shank, a novel family of postsynaptic density proteins that binds to the NMDA receptor/PSD-95/GKAP complex and cortactin. Neuron 23, 569–582 10.1016/S0896-6273(00)80809-0 [DOI] [PubMed] [Google Scholar]
- Naisbitt S., Valtschanoff J., Allison D. W., Sala C., Kim E., Craig A. M., Weinberg R. J., Sheng M. (2000). Interaction of the postsynaptic density-95/guanylate kinase domain-associated protein complex with a light chain of myosin-V and dynein. J. Neurosci. 20, 4524–4534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niesmann K., Breuer D., Brockhaus J., Born G., Wolff I., Reissner C., Kilimann M. W., Rohlmann A., Missler M. (2011). Dendritic spine formation and synaptic function require neurobeachin. Nat. Commun. 2, 557. 10.1038/ncomms1565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niethammer M., Valtschanoff J. G., Kapoor T. M., Allison D. W., Weinberg R. J., Craig A. M., Sheng M. (1998). CRIPT, a novel postsynaptic protein that binds to the third PDZ domain of PSD-95/SAP90. Neuron 20, 693–707 10.1016/S0896-6273(00)81009-0 [DOI] [PubMed] [Google Scholar]
- Nusser Z., Lujan R., Laube G., Roberts J. D., Molnar E., Somogyi P. (1998). Cell type and pathway dependence of synaptic AMPA receptor number and variability in the hippocampus. Neuron 21, 545–559 10.1016/S0896-6273(00)80565-6 [DOI] [PubMed] [Google Scholar]
- Nusser Z., Mulvihill E., Streit P., Somogyi P. (1994). Subsynaptic segregation of metabotropic and ionotropic glutamate receptors as revealed by immunogold localization. Neuroscience 61, 421–427 10.1016/0306-4522(94)90421-9 [DOI] [PubMed] [Google Scholar]
- Palay S. L., Palade G. E. (1955). The fine structure of neurons. J. Biophys. Biochem. Cytol. 1, 69–88 10.1083/jcb.1.1.69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J., Kim M. J., Cheng D., Duong D. M., Gygi S. P., Sheng M. (2004). Semiquantitative proteomic analysis of rat forebrain postsynaptic density fractions by mass spectrometry. J. Biol. Chem. 279, 21003–21011 10.1074/jbc.M402294200 [DOI] [PubMed] [Google Scholar]
- Peters A., Palay S., Webster H. (1991). “Synapses,” in The Fine Structure of the Nervous System, Chap. 5 (New York: Oxford University Press; ), 138–211 [Google Scholar]
- Petersen J. D., Chen X., Vinade L., Dosemeci A., Lisman J. E., Reese T. S. (2003). Distribution of postsynaptic density (PSD)-95 and Ca2+/calmodulin-dependent protein kinase II at the PSD. J. Neurosci. 23, 11270–11278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petralia R. S., Sans N., Wang Y. X., Wenthold R. J. (2005). Ontogeny of postsynaptic density proteins at glutamatergic synapses. Mol. Cell. Neurosci. 29, 436–452 10.1016/j.mcn.2005.03.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips G. R., Anderson T. R., Florens L., Gudas C., Magda G., Yates J. R., III, Colman D. R. (2004). Actin-binding proteins in a postsynaptic preparation: Lasp-1 is a component of central nervous system synapses and dendritic spines. J. Neurosci. Res. 78, 38–48 10.1002/jnr.20224 [DOI] [PubMed] [Google Scholar]
- Phillips G. R., Huang J. K., Wang Y., Tanaka H., Shapiro L., Zhang W., Shan W. S., Arndt K., Frank M., Gordon R. E., Gawinowicz M. A., Zhao Y., Colman D. R. (2001). The presynaptic particle web: ultrastructure, composition, dissolution, and reconstitution. Neuron 32, 63–77 10.1016/S0896-6273(01)00450-0 [DOI] [PubMed] [Google Scholar]
- Racca C., Stephenson F. A., Streit P., Roberts J. D., Somogyi P. (2000). NMDA receptor content of synapses in stratum radiatum of the hippocampal CA1 area. J. Neurosci. 20, 2512–2522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Racz B., Weinberg R. J. (2004). The subcellular organization of cortactin in hippocampus. J. Neurosci. 24, 10310–10317 10.1523/JNEUROSCI.2080-04.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rostaing P., Real E., Siksou L., Lechaire J. P., Boudier T., Boeckers T. M., Gertler F., Gundelfinger E. D., Triller A., Marty S. (2006). Analysis of synaptic ultrastructure without fixative using high-pressure freezing and tomography. Eur. J. Neurosci. 24, 3463–3474 10.1111/j.1460-9568.2006.05234.x [DOI] [PubMed] [Google Scholar]
- Schoch S., Gundelfinger E. D. (2006). Molecular organization of the presynaptic active zone. Cell Tissue Res. 326, 379–391 10.1007/s00441-006-0244-y [DOI] [PubMed] [Google Scholar]
- Seidenbecher C. I., Langnaese K., Sanmarti-Vila L., Boeckers T. M., Smalla K. H., Sabel B. A., Garner C. C., Gundelfinger E. D., Kreutz M. R. (1998). Caldendrin, a novel neuronal calcium-binding protein confined to the somato-dendritic compartment. J. Biol. Chem. 273, 21324–21331 10.1074/jbc.273.33.21324 [DOI] [PubMed] [Google Scholar]
- Seidenbecher C. I., Richter K., Rauch U., Fassler R., Garner C. C., Gundelfinger E. D. (1995). Brevican, a chondroitin sulfate proteoglycan of rat brain, occurs as secreted and cell surface glycosylphosphatidylinositol-anchored isoforms. J. Biol. Chem. 270, 27206–27212 10.1074/jbc.270.45.27206 [DOI] [PubMed] [Google Scholar]
- Sheng M., Hoogenraad C. C. (2007). The postsynaptic architecture of excitatory synapses: a more quantitative view. Annu. Rev. Biochem. 76, 823–847 10.1146/annurev.biochem.76.060805.160029 [DOI] [PubMed] [Google Scholar]
- Shinohara Y. (2012). Quantification of postsynaptic density proteins: glutamate receptor subunits and scaffolding proteins. Hippocampus 22, 942–953 10.1002/hipo.20950 [DOI] [PubMed] [Google Scholar]
- Siekevitz P. (1985). The postsynaptic density: a possible role in long-lasting effects in the central nervous system. Proc. Natl. Acad. Sci. U.S.A. 82, 3494–3498 10.1073/pnas.82.10.3494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J. Y., Ichtchenko K., Sudhof T. C., Brose N. (1999). Neuroligin 1 is a postsynaptic cell-adhesion molecule of excitatory synapses. Proc. Natl. Acad. Sci. U.S.A. 96, 1100–1105 10.1073/pnas.96.21.11776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steward O., Davis L., Dotti C., Phillips L. L., Rao A., Banker G. (1988). Protein synthesis and processing in cytoplasmic microdomains beneath postsynaptic sites on CNS neurons. A mechanism for establishing and maintaining a mosaic postsynaptic receptive surface. Mol. Neurobiol. 2, 227–261 10.1007/BF02935634 [DOI] [PubMed] [Google Scholar]
- Suzuki T. (2011). “Isolation of synapse subdomains by subcellular fractionation using sucrose density gradient centrifugation,” in Neuroproteomics, ed. Li K. W. (New York: Springer; ), 47–61 [Google Scholar]
- Suzuki T., Tanaka R. (1984). Isolation and major components of core structure of postsynaptic density. Neurochem. Int. 6, 665–676 10.1016/0197-0186(84)90048-2 [DOI] [PubMed] [Google Scholar]
- Suzuki T., Tian Q. B., Kuromitsu J., Kawai T., Endo S. (2007). Characterization of mRNA species that are associated with postsynaptic density fraction by gene chip microarray analysis. Neurosci. Res. 57, 61–85 10.1016/j.neures.2006.09.009 [DOI] [PubMed] [Google Scholar]
- tom Dieck S., Sanmarti-Vila L., Langnaese K., Richter K., Kindler S., Soyke A., Wex H., Smalla K. H., Kampf U., Franzer J. T., Stumm M., Garner C. C., Gundelfinger E. D. (1998). Bassoon, a novel zinc-finger CAG/glutamine-repeat protein selectively localized at the active zone of presynaptic nerve terminals. J. Cell Biol. 142, 499–509 10.1083/jcb.142.2.499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinidad J. C., Specht C. G., Thalhammer A., Schoepfer R., Burlingame A. L. (2006). Comprehensive identification of phosphorylation sites in postsynaptic density preparations. Mol. Cell. Proteomics 5, 914–922 10.1074/mcp.T500041-MCP200 [DOI] [PubMed] [Google Scholar]
- Trinidad J. C., Thalhammer A., Specht C. G., Schoepfer R., Burlingame A. L. (2005). Phosphorylation state of postsynaptic density proteins. J. Neurochem. 92, 1306–1316 10.1111/j.1471-4159.2004.02943.x [DOI] [PubMed] [Google Scholar]
- Tu J. C., Xiao B., Naisbitt S., Yuan J. P., Petralia R. S., Brakeman P., Doan A., Aakalu V. K., Lanahan A. A., Sheng M., Worley P. F. (1999). Coupling of mGluR/Homer and PSD-95 complexes by the Shank family of postsynaptic density proteins. Neuron 23, 583–592 10.1016/S0896-6273(00)80810-7 [DOI] [PubMed] [Google Scholar]
- UniProt-Consortium. (2010). The Universal Protein Resource (UniProt) in 2010. Nucleic Acids Res. 38, D142–D148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valtschanoff J. G., Weinberg R. J. (2001). Laminar organization of the NMDA receptor complex within the postsynaptic density. J. Neurosci. 21, 1211–1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walikonis R. S., Jensen O. N., Mann M., Provance D. W., Jr., Mercer J. A., Kennedy M. B. (2000). Identification of proteins in the postsynaptic density fraction by mass spectrometry. J. Neurosci. 20, 4069–4080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh M. J., Kuruc N. (1992). The postsynaptic density: constituent and associated proteins characterized by electrophoresis, immunoblotting, and peptide sequencing. J. Neurochem. 59, 667–678 10.1111/j.1471-4159.1992.tb09421.x [DOI] [PubMed] [Google Scholar]
- Xue J., Tsang C. W., Gai W. P., Malladi C. S., Trimble W. S., Rostas J. A., Robinson P. J. (2004). Septin 3 (G-septin) is a developmentally regulated phosphoprotein enriched in presynaptic nerve terminals. J. Neurochem. 91, 579–590 10.1111/j.1471-4159.2004.02755.x [DOI] [PubMed] [Google Scholar]
- Yamauchi T. (2002). Molecular constituents and phosphorylation-dependent regulation of the post-synaptic density. Mass Spectrom. Rev. 21, 266–286 10.1002/mas.10033 [DOI] [PubMed] [Google Scholar]
- Yoshimura Y., Yamauchi Y., Shinkawa T., Taoka M., Donai H., Takahashi N., Isobe T., Yamauchi T. (2004). Molecular constituents of the postsynaptic density fraction revealed by proteomic analysis using multidimensional liquid chromatography-tandem mass spectrometry. J. Neurochem. 88, 759–768 10.1046/j.1471-4159.2003.02136.x [DOI] [PubMed] [Google Scholar]
- Young K. H. (1998). Yeast two-hybrid: so many interactions, (in) so little time. Biol. Reprod. 58, 302–311 10.1095/biolreprod58.2.302 [DOI] [PubMed] [Google Scholar]
- Ziff E. B. (1997). Enlightening the postsynaptic density. Neuron 19, 1163–1174 10.1016/S0896-6273(00)80409-2 [DOI] [PubMed] [Google Scholar]
- Zuber B., Nikonenko I., Klauser P., Muller D., Dubochet J. (2005). The mammalian central nervous synaptic cleft contains a high density of periodically organized complexes. Proc. Natl. Acad. Sci. U.S.A. 102, 19192–19197 10.1073/pnas.0509527102 [DOI] [PMC free article] [PubMed] [Google Scholar]