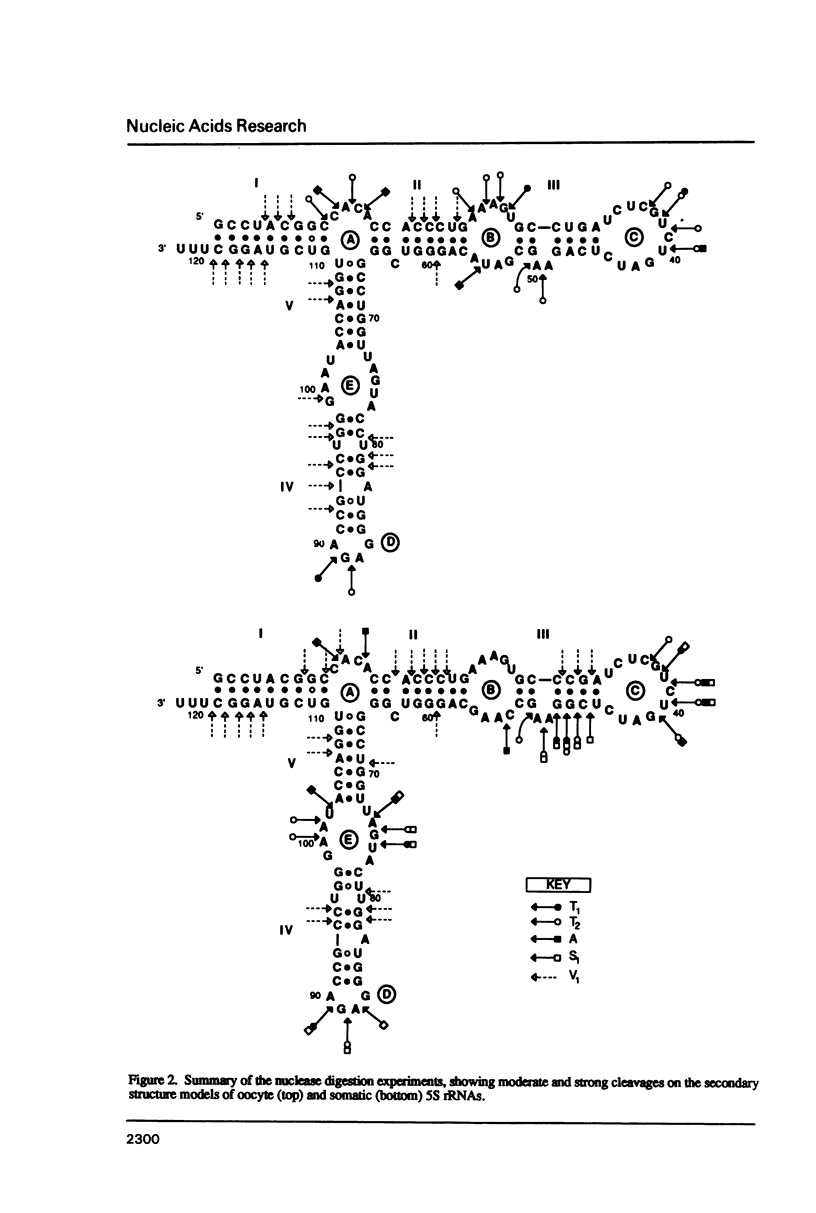
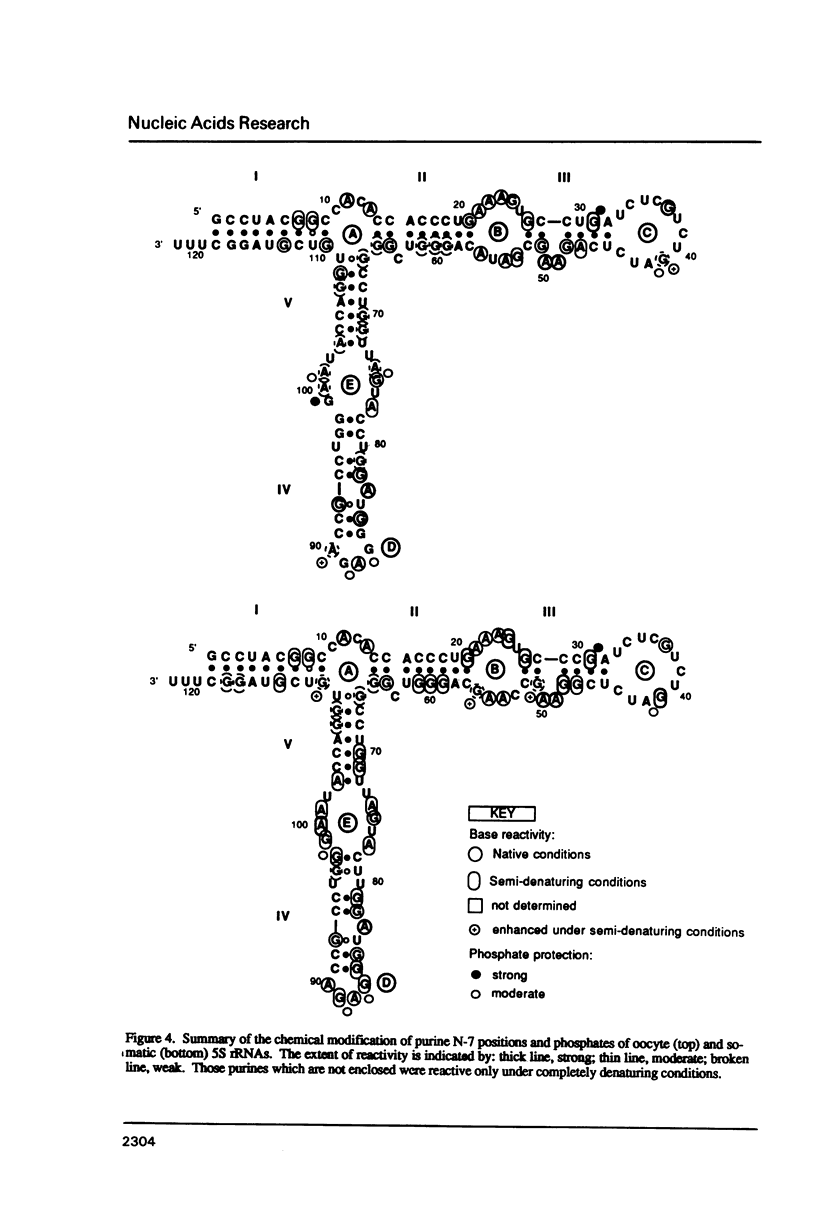
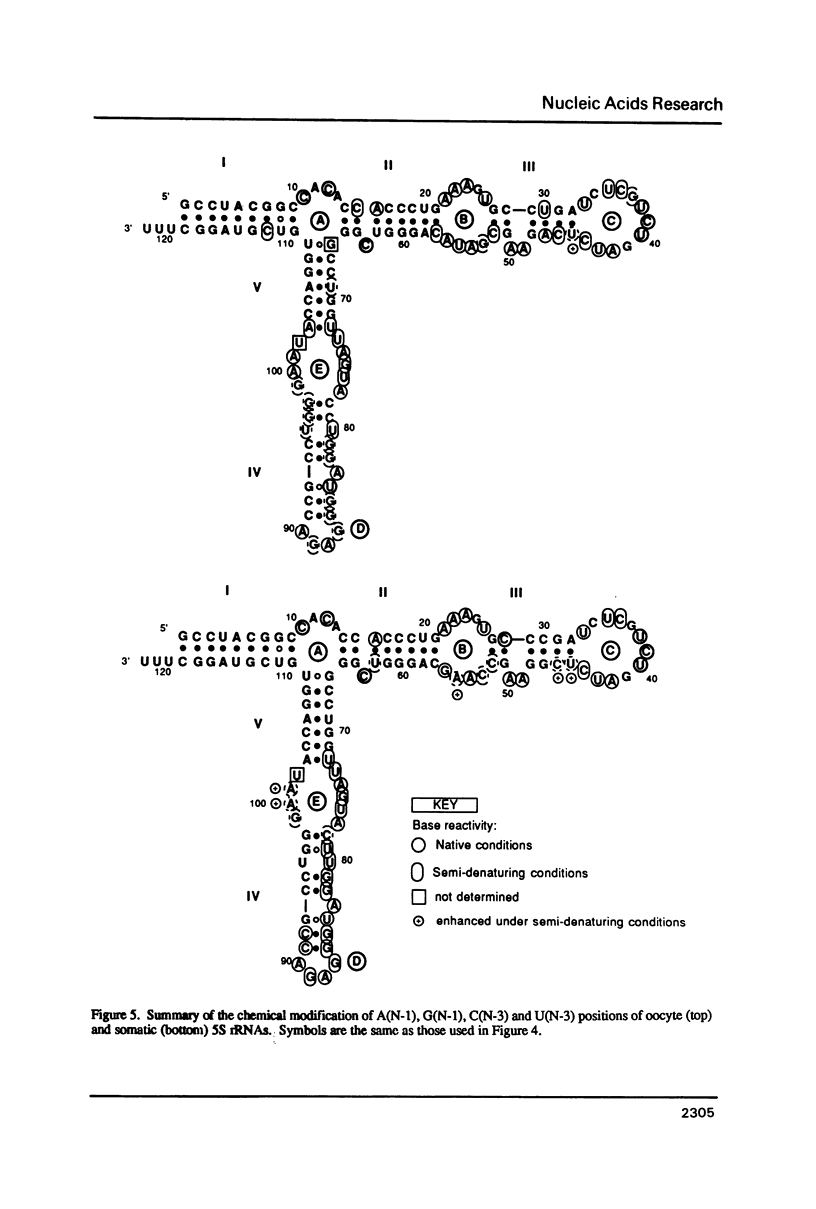
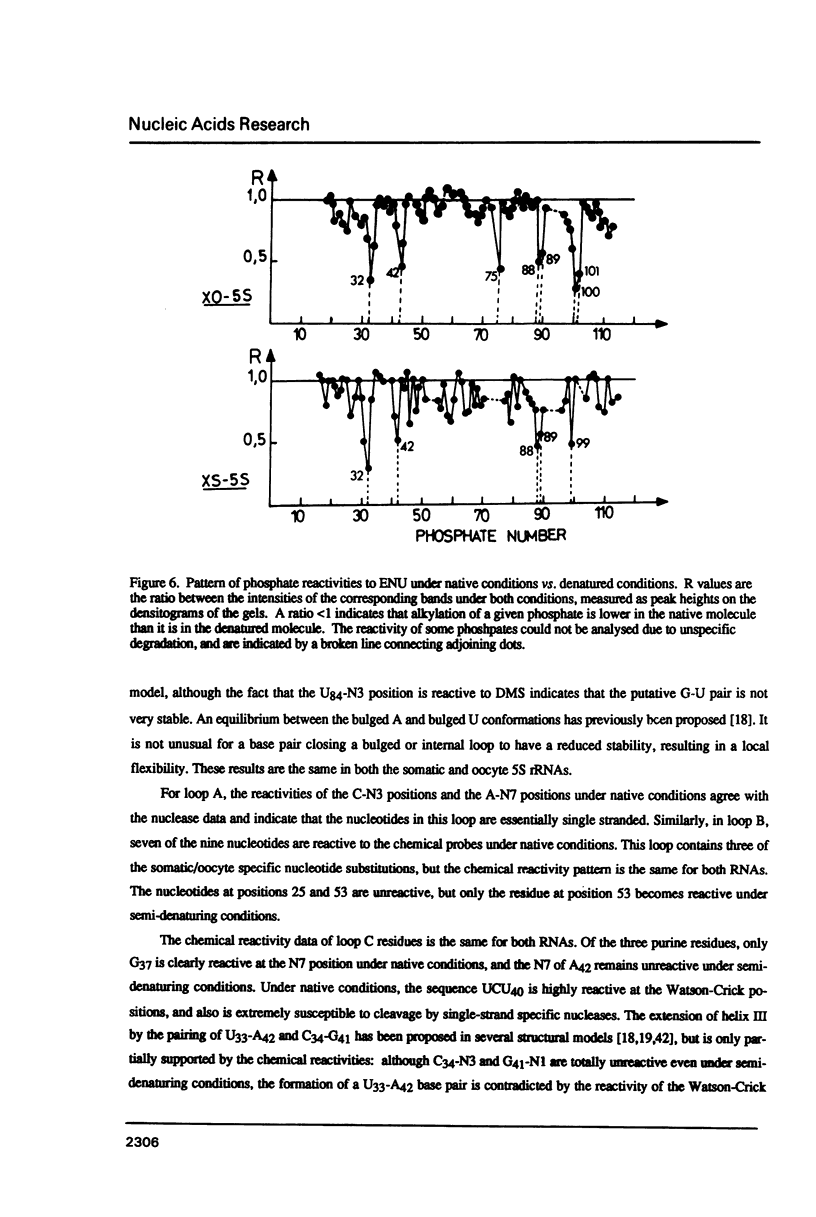
Abstract
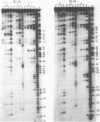
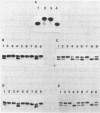
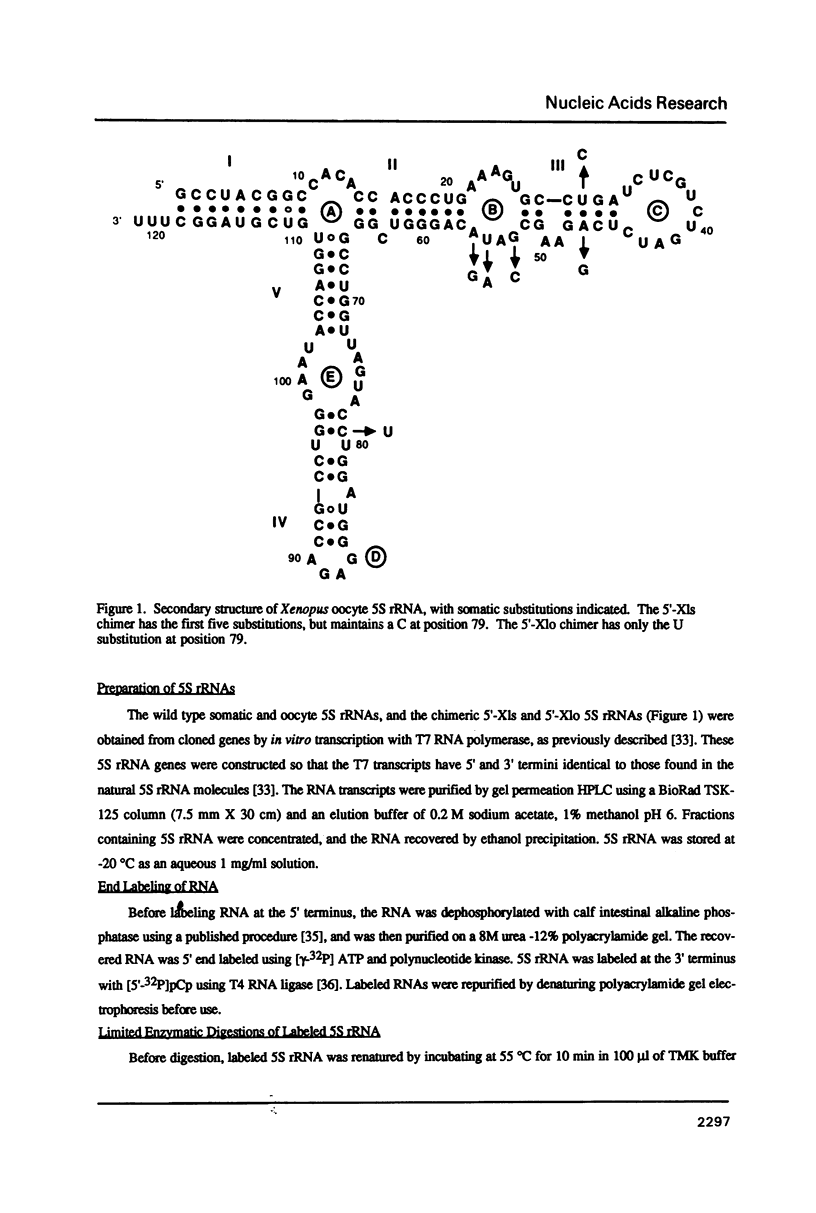
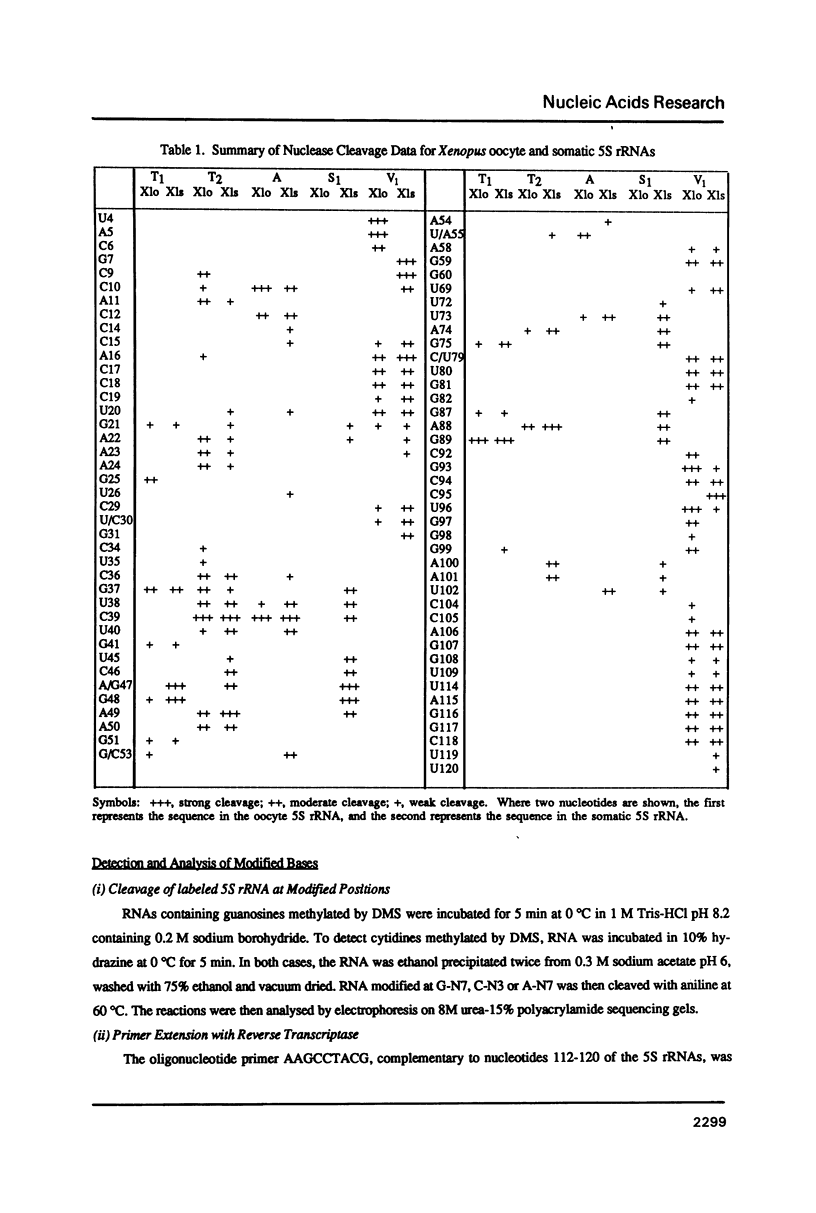
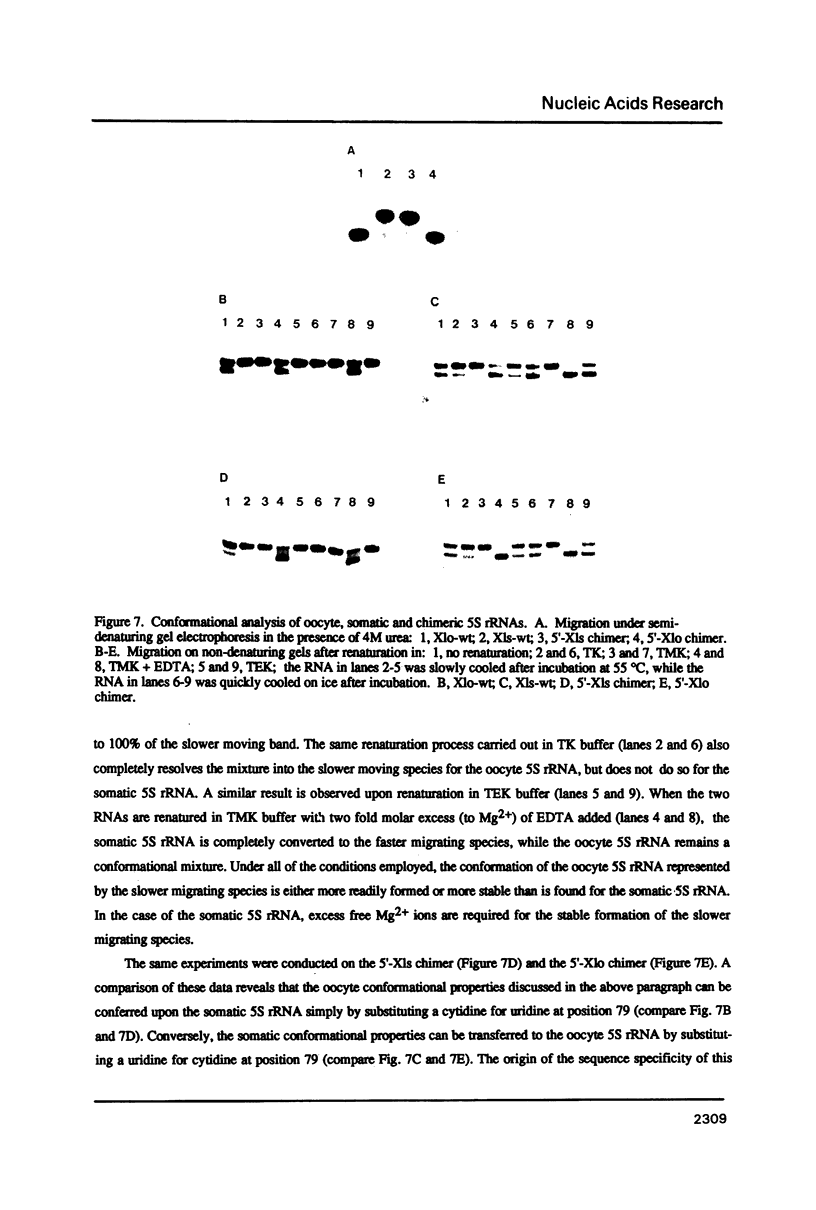
The secondary and tertiary structures of Xenopus oocyte and somatic 5S rRNAs were investigated using chemical and enzymatic probes. The accessibility of both RNAs towards single-strand specific nucleases (T1, T2, A and S1) and a helix-specific ribonuclease from cobra venom (RNase V1) was determined. The reactivity of nucleobase N7, N3 and N1 positions towards chemical probes was investigated under native (5 mM MgCl2, 100 mM KCl, 20 degrees C) and semi-denaturing (1 mM EDTA, 20 degrees C) conditions. Ethylnitrosourea was used to identify phosphates not reactive towards alkylation under native conditions. The results obtained confirm the presence of the five helical stems predicted by the consensus secondary structure model of 5S rRNA. The chemical reactivity data indicate that loops C and D are involved in a number of tertiary interactions, and loop E folds into an unusual secondary structure. A comparison of the data obtained for the two types of Xenopus 5S rRNA indicates that the conformations of the oocyte and somatic 5S rRNAs are very similar. However, the data obtained with nucleases under native conditions, and chemical probes under semi-denaturing conditions, reveal that helices III and IV in the somatic 5S rRNA are less stable than the same structures in oocyte 5S rRNA. Using chimeric 5S rRNAs, it was possible to demonstrate that the relative resistance of oocyte 5S rRNA to partial denaturation in 4 M urea is conferred by the five oocyte-specific nucleotide substitutions in loop B/helix III. In contrast, the superior stability of oocyte 5S rRNA in the presence of EDTA is related to a single C substitution at position 79.
Full text
PDF










Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Andersen J., Delihas N., Hanas J. S., Wu C. W. 5S RNA structure and interaction with transcription factor A. 1. Ribonuclease probe of the structure of 5S RNA from Xenopus laevis oocytes. Biochemistry. 1984 Nov 20;23(24):5752–5759. doi: 10.1021/bi00319a013. [DOI] [PubMed] [Google Scholar]
- Azad A. A. Intermolecular base-paired interaction between complementary sequences present near the 3' ends of 5S rRNA and 18S (16S) rRNA might be involved in the reversible association of ribosomal subunits. Nucleic Acids Res. 1979 Dec 11;7(7):1913–1929. doi: 10.1093/nar/7.7.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christiansen J., Brown R. S., Sproat B. S., Garrett R. A. Xenopus transcription factor IIIA binds primarily at junctions between double helical stems and internal loops in oocyte 5S RNA. EMBO J. 1987 Feb;6(2):453–460. doi: 10.1002/j.1460-2075.1987.tb04775.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davanloo P., Rosenberg A. H., Dunn J. J., Studier F. W. Cloning and expression of the gene for bacteriophage T7 RNA polymerase. Proc Natl Acad Sci U S A. 1984 Apr;81(7):2035–2039. doi: 10.1073/pnas.81.7.2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Wachter R., Chen M. W., Vandenberghe A. Conservation of secondary structure in 5 S ribosomal RNA: a uniform model for eukaryotic, eubacterial, archaebacterial and organelle sequences is energetically favourable. Biochimie. 1982 May;64(5):311–329. doi: 10.1016/s0300-9084(82)80436-7. [DOI] [PubMed] [Google Scholar]
- Delihas N., Andersen J. Generalized structures of the 5S ribosomal RNAs. Nucleic Acids Res. 1982 Nov 25;10(22):7323–7344. doi: 10.1093/nar/10.22.7323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Digweed M., Pieler T., Kluwe D., Schuster L., Walker R., Erdmann V. A. Improved procedure for the isolation of a double-strand-specific ribonuclease and its application to structural analysis of various 5S rRNAs and tRNAs. Eur J Biochem. 1986 Jan 2;154(1):31–39. doi: 10.1111/j.1432-1033.1986.tb09355.x. [DOI] [PubMed] [Google Scholar]
- Douthwaite S., Garrett R. A. Secondary structure of prokaryotic 5S ribosomal ribonucleic acids: a study with ribonucleases. Biochemistry. 1981 Dec 8;20(25):7301–7307. doi: 10.1021/bi00528a039. [DOI] [PubMed] [Google Scholar]
- England T. E., Uhlenbeck O. C. 3'-terminal labelling of RNA with T4 RNA ligase. Nature. 1978 Oct 12;275(5680):560–561. doi: 10.1038/275560a0. [DOI] [PubMed] [Google Scholar]
- Gaunt-Klöpfer M., Erdmann V. A. ATPase and GTPase activities associated with the 5-S RNA-protein complex of Escherichia coli ribosomes. Biochim Biophys Acta. 1975 May 1;390(2):226–230. doi: 10.1016/0005-2787(75)90343-3. [DOI] [PubMed] [Google Scholar]
- Göringer H. U., Szymkowiak C., Wagner R. Escherichia coli 5S RNA A and B conformers. Characterisation by enzymatic and chemical methods. Eur J Biochem. 1984 Oct 1;144(1):25–34. doi: 10.1111/j.1432-1033.1984.tb08426.x. [DOI] [PubMed] [Google Scholar]
- Hancock J., Wagner R. A structural model of 5S RNA from E. coli based on intramolecular crosslinking evidence. Nucleic Acids Res. 1982 Feb 25;10(4):1257–1269. doi: 10.1093/nar/10.4.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herr W., Noller H. F. A fragment of 23S RNA containing a nucleotide sequence complementary to a region of 5S RNA. FEBS Lett. 1975 May 1;53(2):248–252. doi: 10.1016/0014-5793(75)80030-5. [DOI] [PubMed] [Google Scholar]
- Huber P. W., Wool I. G. Use of the cytotoxic nuclease alpha-sarcin to identify the binding site on eukaryotic 5 S ribosomal ribonucleic acid for the ribosomal protein L5. J Biol Chem. 1986 Mar 5;261(7):3002–3005. [PubMed] [Google Scholar]
- Kao T. H., Crothers D. M. A proton-coupled conformational switch of Escherichia coli 5S ribosomal RNA. Proc Natl Acad Sci U S A. 1980 Jun;77(6):3360–3364. doi: 10.1073/pnas.77.6.3360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kime M. J., Moore P. B. NMR evidence for the existence of two native conformations of 5S RNA. Nucleic Acids Res. 1982 Aug 25;10(16):4973–4983. doi: 10.1093/nar/10.16.4973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leontis N. B., Moore P. B. NMR evidence for dynamic secondary structure in helices II and III of the RNA of Escherichia coli. Biochemistry. 1986 Jul 1;25(13):3916–3925. doi: 10.1021/bi00361a027. [DOI] [PubMed] [Google Scholar]
- Li S. J., Wu J. J., Marshall A. G. 500-MHz proton homonuclear Overhauser evidence for additional base pair in the common arm of eukaryotic ribosomal 5S RNA: wheat germ. Biochemistry. 1987 Mar 24;26(6):1578–1585. doi: 10.1021/bi00380a014. [DOI] [PubMed] [Google Scholar]
- Lowman H. B., Draper D. E. On the recognition of helical RNA by cobra venom V1 nuclease. J Biol Chem. 1986 Apr 25;261(12):5396–5403. [PubMed] [Google Scholar]
- McDougall J., Nazar R. N. Tertiary structure of the eukaryotic ribosomal 5 S RNA. Accessibility of phosphodiester bonds to ethylnitrosourea modification. J Biol Chem. 1983 Apr 25;258(8):5256–5259. [PubMed] [Google Scholar]
- Mougel M., Eyermann F., Westhof E., Romby P., Expert-Bezançon A., Ebel J. P., Ehresmann B., Ehresmann C. Binding of Escherichia coli ribosomal protein S8 to 16 S rRNA. A model for the interaction and the tertiary structure of the RNA binding site. J Mol Biol. 1987 Nov 5;198(1):91–107. doi: 10.1016/0022-2836(87)90460-8. [DOI] [PubMed] [Google Scholar]
- Nazar R. N., Willick G. E., Matheson A. T. The 5 S RNA.protein complex from an extreme halophile, Halobacterium cutirubrum. Studies on the RNA-protein interaction. J Biol Chem. 1979 Mar 10;254(5):1506–1512. [PubMed] [Google Scholar]
- Noller H. F., Garrett R. A. Structure of 5 S ribosomal RNA from Escherichia coli: identification of kethoxal-reactive sites in the A and B conformations. J Mol Biol. 1979 Aug 25;132(4):621–636. doi: 10.1016/0022-2836(79)90378-4. [DOI] [PubMed] [Google Scholar]
- Ofengand J., Henes C. The function of pseudouridylic acid in transfer ribonucleic acid. II. Inhibition of amino acyl transfer ribonucleic acid-ribosome complex formation by ribothymidylyl-pseudouridylyl-cytidylyl-guanosine 3'-phosphate. J Biol Chem. 1969 Nov 25;244(22):6241–6253. [PubMed] [Google Scholar]
- Picard B., Wegnez M. Isolation of a 7S particle from Xenopus laevis oocytes: a 5S RNA-protein complex. Proc Natl Acad Sci U S A. 1979 Jan;76(1):241–245. doi: 10.1073/pnas.76.1.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieler T., Erdmann V. A. Three-dimensional structural model of eubacterial 5S RNA that has functional implications. Proc Natl Acad Sci U S A. 1982 Aug;79(15):4599–4603. doi: 10.1073/pnas.79.15.4599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raacke I. D. A model for protein synthesis involving the intermediate formation of peptidyl-5S RNA. Proc Natl Acad Sci U S A. 1971 Oct;68(10):2357–2360. doi: 10.1073/pnas.68.10.2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romaniuk P. J., de Stevenson I. L., Wong H. H. Defining the binding site of Xenopus transcription factor IIIA on 5S RNA using truncated and chimeric 5S RNA molecules. Nucleic Acids Res. 1987 Mar 25;15(6):2737–2755. doi: 10.1093/nar/15.6.2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segall J., Matsui T., Roeder R. G. Multiple factors are required for the accurate transcription of purified genes by RNA polymerase III. J Biol Chem. 1980 Dec 25;255(24):11986–11991. [PubMed] [Google Scholar]
- Serenkova T. I., Mazo A. M., Mashkova T. D., Toots I., Nigul A., Timofeeva MYa, Kisselev L. L. The secondary structure of oocyte and somatic 5S ribosomal RNAs of the fish Misgurnus fossilis L. from nuclease hydrolyses and chemical modification data. Nucleic Acids Res. 1984 Jul 11;12(13):5385–5404. doi: 10.1093/nar/12.13.5385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shastry B. S., Ng S. Y., Roeder R. G. Multiple factors involved in the transcription of class III genes in Xenopus laevis. J Biol Chem. 1982 Nov 10;257(21):12979–12986. [PubMed] [Google Scholar]
- Silberklang M., Gillum A. M., RajBhandary U. L. The use of nuclease P1 in sequence analysis of end group labeled RNA. Nucleic Acids Res. 1977 Dec;4(12):4091–4108. doi: 10.1093/nar/4.12.4091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl D. A., Pace B., Marsh T., Pace N. R. The ribonucleoprotein substrate for a ribosomal RNA-processing nuclease. J Biol Chem. 1984 Sep 25;259(18):11448–11453. [PubMed] [Google Scholar]
- Toots I., Misselwitz R., Böhm S., Welfle H., Villems R., Saarma M. Two distinct conformations of rat liver ribosomal 5S RNA. Nucleic Acids Res. 1982 Jun 11;10(11):3381–3389. doi: 10.1093/nar/10.11.3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlassov V. V., Giegé R., Ebel J. P. Tertiary structure of tRNAs in solution monitored by phosphodiester modification with ethylnitrosourea. Eur J Biochem. 1981 Sep;119(1):51–59. doi: 10.1111/j.1432-1033.1981.tb05575.x. [DOI] [PubMed] [Google Scholar]
- Wakefield L., Gurdon J. B. Cytoplasmic regulation of 5S RNA genes in nuclear-transplant embryos. EMBO J. 1983;2(9):1613–1619. doi: 10.1002/j.1460-2075.1983.tb01632.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westhof E., Dumas P., Moras D. Crystallographic refinement of yeast aspartic acid transfer RNA. J Mol Biol. 1985 Jul 5;184(1):119–145. doi: 10.1016/0022-2836(85)90048-8. [DOI] [PubMed] [Google Scholar]
- Woese C. R., Fox G. E. Phylogenetic structure of the prokaryotic domain: the primary kingdoms. Proc Natl Acad Sci U S A. 1977 Nov;74(11):5088–5090. doi: 10.1073/pnas.74.11.5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wormington W. M., Bogenhagen D. F., Jordan E., Brown D. D. A quantitative assay for Xenopus 5S RNA gene transcription in vitro. Cell. 1981 Jun;24(3):809–817. doi: 10.1016/0092-8674(81)90106-9. [DOI] [PubMed] [Google Scholar]