Abstract
Rationale
Peripheral blood biomarkers are needed to identify and determine the extent of idiopathic pulmonary fibrosis (IPF). Current physiologic and radiographic prognostic indicators diagnose IPF too late in the course of disease. We hypothesize that peripheral blood biomarkers will identify disease in its early stages, and facilitate monitoring for disease progression.
Methods
Gene expression profiles of peripheral blood RNA from 130 IPF patients were collected on Agilent microarrays. Significance analysis of microarrays (SAM) with a false discovery rate (FDR) of 1% was utilized to identify genes that were differentially-expressed in samples categorized based on percent predicted DLCO and FVC.
Main Measurements and Results
At 1% FDR, 1428 genes were differentially-expressed in mild IPF (DLCO >65%) compared to controls and 2790 transcripts were differentially- expressed in severe IPF (DLCO >35%) compared to controls. When categorized by percent predicted DLCO, SAM demonstrated 13 differentially-expressed transcripts between mild and severe IPF (< 5% FDR). These include CAMP, CEACAM6, CTSG, DEFA3 and A4, OLFM4, HLTF, PACSIN1, GABBR1, IGHM, and 3 unknown genes. Principal component analysis (PCA) was performed to determine outliers based on severity of disease, and demonstrated 1 mild case to be clinically misclassified as a severe case of IPF. No differentially-expressed transcripts were identified between mild and severe IPF when categorized by percent predicted FVC.
Conclusions
These results demonstrate that the peripheral blood transcriptome has the potential to distinguish normal individuals from patients with IPF, as well as extent of disease when samples were classified by percent predicted DLCO, but not FVC.
Introduction
Idiopathic Pulmonary Fibrosis (IPF) is categorized as an interstitial lung disease (ILD) and is the most common subtype of idiopathic interstitial pneumonias (IIP) comprising nearly 71% of the total cases [1]. Of the IIPs, IPF has the poorest prognosis with a 50% mortality rate 3 years following diagnosis [2]. Prognostic indicators of IPF include progressive deterioration of clinical symptoms such as dyspnea, pulmonary function, and extent of disease on high-resolution chest CT [3]–[7]. While dyspnea scores have been used as a predictor of survival in IPF patients [8], it remains an ambiguous prognostic indicator since it is highly subjective. Pulmonary function tests such as diffusing capacity for carbon monoxide (DLCO) and forced vital capacity (FVC) have been utilized as predictive indicators [9], [10]. Studies demonstrate that a DLCO of <35% or a decline in DLCO >15% within a year period are associated with an increased mortality. Similarly, a decline of >10% in FVC over a six month period also indicated an earlier mortality [8], [11]. The overall extent of fibrosis on high-resolution chest CT (HRCT) characterized by a honeycomb pattern and reticulation predict survival [12]. Randomized prospective controlled clinical trials in IPF have demonstrated significant differences in the rate of decline in FVC and DLCO among the placebo arms of the trials indicating there is substantial disease heterogeneity within IPF [13]. Current indicators of disease progression fail to capture the dynamic biology associated with IPF particularly patients at risk for acute exacerbations of IPF [14]. Biomarkers that measure disease stage and activity would be of benefit in understanding the effects of novel treatments, disease progression, and the design of clinical trials with homogenous placebo and treatment groups.
Rosas and coworkers observed a differential expression of MMP7, MMP1, MMP8, IGFBP1, and TNFRSF1A proteins in the peripheral blood between familial interstitial pulmonary fibrosis and controls [15]. However, the use of these biomarkers to differentiate disease severity or extent of disease within the IPF cohort was not addressed. We hypothesize that peripheral blood transcriptional profiles from patients with IPF would enable us to distinguish patients with IPF from controls, and mild from more advanced disease stage, and allow for monitoring of the progression of disease in either sporadic or familial IPF.
Methods
Study Populations
One hundred thirty peripheral blood RNA specimens were collected from individuals enrolled in either the Interstitial Lung Disease (ILD) or the Familial Pulmonary Fibrosis (FPF) Programs conducted at National Jewish Health and Duke University. Only one individual case per family was utilized from the FPF repository. Individual samples had a consensus diagnosis of probable or definite IPF, and this was based on the ATS/ERS/JRS/ALAT criteria [16]. Subjects were excluded from selection if they were current smokers, or currently treated with agents that could alter mRNA levels such as glucocorticoids, azathioprine, or other immunomodulators. One-hundred twenty three RNA samples passed quality assurance parameters after RNA extraction, probe synthesis, and hybridization for further analysis. 53 of these samples were from sporadic cases of IPF, and 70 samples were from familial IPF. Peripheral blood gene expression profiles were analyzed on groups of individuals based on disease severity. Two pulmonary function measurements, DLCO and FVC, were used to stratify the cases into severe and mild disease categories. Mild disease is defined as either percent predicted DLCO ≥65% (N = 16) or FVC ≥75% (N = 27). Severe disease is defined as either DLCO ≤35% (N = 15), FVC ≤50% (N = 13). All of these were also compared to age and gender matched non-diseased, healthy controls (N = 27). Eight patients categorized as severe were used in both the DLCO and FVC analysis, whereas the mild disease classified by either DLCO or FVC are all distinct cases. Healthy control subjects are family members who participated in screening for the presence of pulmonary fibrosis, and after evaluation of their medical history, lung function, and chest CT, they were found to have no evidence of lung disease. Individual institutional review boards approved this research. All participants in this study provided written IRB-approved informed consent.
Pulmonary Function Testing
FEV1 and DLCO measurement were obtained according to American Thoracic Society standards and guidelines [17].
Expression Profiling
Peripheral blood RNA isolation and purification
Peripheral blood samples were collected in PAXgene RNA tubes (PreAnalytiX, 762165). RNA extraction and purification was performed manually utilizing the PAXgene Blood RNA kit (PreAnalytiX, 762164) according to the manufacturer’s protocol.
Total RNA quantification and quality characterization
Quantification of total RNA was measured via the Nanodrop ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE). Quality of the RNA was assessed with a RNA 6000 NanoChip (Agilent, Palo Alto, CA) on the 2100 Bioanalyzer (Agilent, Palo Alto, CA) by ratio comparison of the 18 S and 28 S rRNA bands.
Microarrays
Agilent Whole Human Genome Oligonucleotide Microarrays (Agilent, Palo Alto, CA), were used to determine gene expression levels in peripheral blood. Total RNA was used as a template for synthesis of cDNA utilizing the One Color Low Input Agilent Quick Amp Labeling Kit utilizing the Spike-In Kit to provide positive controls. The Agilent one-color microarray based gene expression analysis was followed per manufacturer’s instructions, and passed Agilent’s quality control (QC).
Microarray data analysis
Analysis was performed utilizing the Multi-Experiment Viewer (MeV) software package [18]. Significance analysis of microarrays (SAM) with a false discovery rate (FDR) of 1% and 5%, assessed by performing 100 permutations, was performed within the program to identify genes that were differentially-expressed between IPF samples categorized by percent predicted DLCO and FVC. All IPF samples were compared to normal controls to identify differentially-expressed genes. Principal component analysis (PCA) was carried out on genes identified by SAM analyses to identify outliers, and gene-based hierarchical clustering was performed to identify relationships among differentially-expressed genes related to mild or severe disease categorized by DLCO. All microarray data is minimal information about a microarray experiment (MIAME) compliant, and raw data has been deposited into the GEO database (GSE33566).
Gene ontology and functional network analysis
Data were analyzed through the use of Ingenuity Pathways Analysis (Ingenuity Systems, www.ingenuity.com). The Canonical Pathways Analysis identified the pathways, and the significance of these was determined by Fischer’s exact test. Biomarker Analysis was employed to identify the most relevant molecular biomarker candidates.
Validation
Quantitative real-time PCR was utilized to confirm differential expression of genes discovered by microarray analysis with an ABI 7900 HT Fast Real-Time PCR Detection System (Applied Biosystems, Foster City, CA) using forty cycles of amplification. All assays were performed in duplicate and data were analyzed by the ΔΔCt method utilizing glyceraldehyde 3 phosphate dehydrogenase (GAPDH) as an endogenous control [19].
Results
Demographics and Disease Severity
Age, gender, and smoking history of the study population stratified into mild and severe disease groups based on pulmonary function status (Tables 1 and 2) show a mean FVC of 85% predicted and DLCO 77.1% predicted in the mild disease group, and a mean FVC 42.5% predicted and 27.5% predicted DLCO in the severe disease group. The mean age is similar between groups when classified by either % predicted FVC (P = 0.18), or % predicted DLCO (P = 0.87), but the mean age of either disease group is slightly older compared to controls (P = 0.15). The majority of patients are male in both mild and severe disease categories, and similar between both disease categories when classified by either % predicted FVC (P = 0.48) or % predicted DLCO (P = 0.29). There are no significant differences between mild and severe disease categories with respect to prior tobacco use. The diagnosis of IPF was confirmed by surgical lung biopsy in 50.7% of the study subjects (seeTables S1, S2, S3, and S4). A highly confident diagnosis of definite IPF was obtained in 90% of patients (64 of 71, Table S1, S2, S3, and S4). Five patients with mild disease by DLCO (≥ 65% predicted) failed to meet definite HRCT criteria for IPF due to minimal honeycombing, and are categorized as probable IPF since the predominate HRCT findings are sub-pleural, bilateral, bi-basilar reticulation, and traction bronchiectasis, and clinical features supported the diagnosis of IPF. Of these 5 cases, the mean age is 70 (range 63–75), 4 of the 5 cases are male, and is consistent with IPF.
Table 1. Clinical and demographic IPF variables categorized by FVC.
| Variable | Characteristics | Mild IPF (N = 27) | Severe IPF (N = 13) | Controls (N = 27) |
| % Predicted FVC | 85.0±8.1 | 42.5±6.6 | NR | |
| Age | Mean±SD | 69.8±8.4 | 65.3±12.7 | 60.1±14.1 |
| Sex | Male/Female | 19/8 | 10/3 | 11/17 |
| Smoking Status | Current | 0 | 0 | 0 |
| Former | 7 | 7 | 14 | |
| Never | 18 | 6 | 13 | |
| Not Reported | 2 | 0 | 0 | |
Abbreviations: Idiopathic Pulmonary Fibrosis (IPF); Not Reported (NR); Diffusing Capacity for Carbon Monoxide (DLCO); Forced Vital Capacity (FVC).
Table 2. Clinical and demographic IPF variables categorized by DLCO.
| Variable | Characteristics | Mild IPF (N = 16) | Severe IPF (N = 15) | Controls (N = 27) |
| % Predicted DLCO | 77.1±11.9 | 27.4±5.3 | NR | |
| Age | Mean±SD | 67.4±6.0 | 66.8±13.7 | 60.1±14.1 |
| Sex | Male/Female | 11/5 | 11/4 | 11/17 |
| Smoking Status | Current | 0 | 0 | 0 |
| Former | 7 | 10 | 14 | |
| Never | 8 | 5 | 13 | |
| Not Reported | 1 | 0 | 0 | |
Abbreviations: Idiopathic Pulmonary Fibrosis (IPF); Not Reported (NR); Diffusing Capacity for Carbon Monoxide (DLCO); Forced Vital Capacity (FVC).
Identification of a Peripheral Blood Signature that Distinguishes Presence of Disease
Principal component analysis (PCA) was performed to determine outliers in the gene expression data based on disease categorization (mild or severe IPF vs. normal). The majority of mild cases (DLCO ≥65% predicted), both probable and definite, cluster along the first principal component (Figure 1). Two cases of probable IPF distribute with controls, and one control distributes with cases along the first principal component. Both of the probable cases that distributed with controls have very early disease, and they had mild disease on HRCT. The first case had a normal DLCO of 99% predicted, only a mildly reduced FVC of 71% predicted, and the second case case had a DLCO of 66% and FVC of 86% predicted. Severe cases of IPF (DLCO ≤35%), both probable and definite, distribute together along the first principal component (Figure 2). Three cases of advanced pulmonary fibrosis that by pathology review were deemed unclassifiable fibrotic lung disease due to advanced honeycomb lung but most consistent with IPFwere included in the analysis (Tables S1, S2, S3), and are similar to other cases of severe definite IPF. This demonstrates the potential of the peripheral blood molecular signature to diagnose patients when the interpretation of surgical lung biopsy is ambiguous. One control distributes with cases and is misclassified by PCA, and one control is not readily classified based on the first and second principal components. Thus, it is unlikely that the peripheral blood signature will achieve 100% accuracy to predict the presence of IPF.
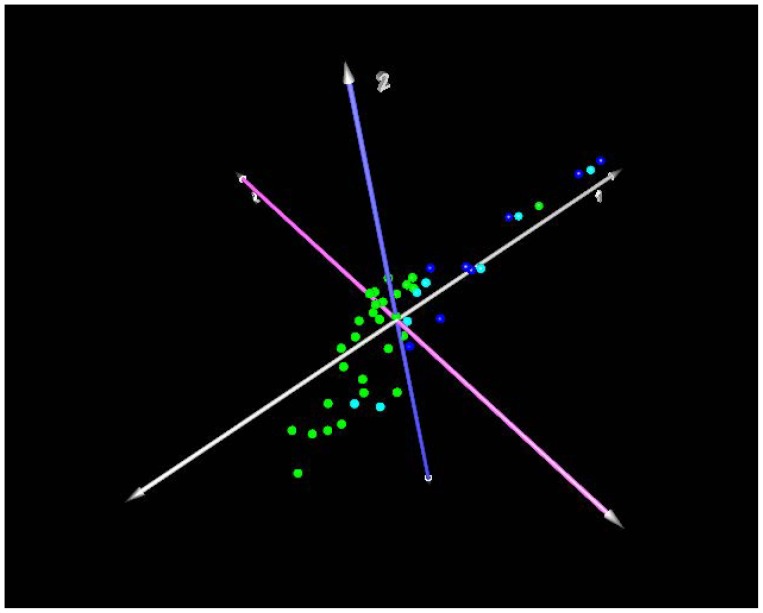
Figure 1. Principal component analysis of mild IPF cases (DLCO ≥65% predicted) compared to controls.
Dark blue spheres: definite IPF; Cyan/light blue spheres: probable IPF; Green spheres: healthy controls. Axis labels: white-first principal component; blue-second principal component; lavender- third principal component. The majority of cases, both probable and definite, cluster along the first principal component. Two cases of probable IPF distribute with controls, and one control distributes with cases along the first principal component.
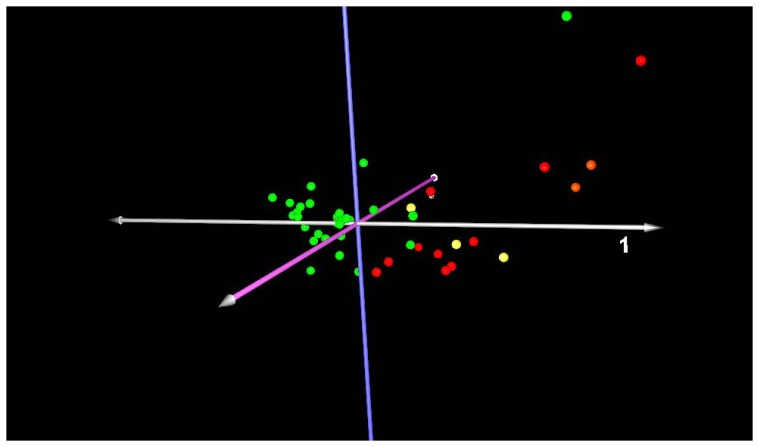
Figure 2. Principal component analysis of severe IPF (DLCO ≤35% predicted).
Red spheres: definite IPF; Orange spheres: probable IPF; Yellow spheres: unclassifiable fibrosis; Green spheres: healthy controls. Axis labels: white-first principal component; blue-second principal component; lavender- third principal component. The majority of severe cases, both probable and definite, cluster along the first principal component. Three cases of unclassifiable fibrosis distribute with IPF cases.
Significant analysis of microarray (SAM) of the mild IPF cohort, when categorized by percent predicted DLCO (N = 16) compared to normal controls (N = 31) revealed 1,428 differentially-expressed transcripts (Table S6), and when categorized as severe IPF (N = 15) compared to normal controls (N = 31), 2,790 differentially-expressed transcripts (table S7) with <1% false discovery rate (FDR). Tables 3 and 4 list the differentially-expressed genes with at least a log2 2 fold change in expression (While a few genes are in common, there are different sets of genes have at least a log2 2-fold change in expression between mild cases compared to controls (table 3) compared to those that distinguish severe cases from control (table 4). Therefore, a molecular signature in the peripheral blood to detect IPF will be comprised of different genes depending on disease severity.
Table 3. Differentially-expressed genes that distinguish mild IPF from control.
| Gene Symbol | Description | Fold Change‡ |
| CEACAM4 | carcinoembryonic antigen-related cell adhesion molecule 4 | 4.02 |
| IL1R2 | interleukin 1 receptor, type II | 2.69 |
| FCN1 | ficolin (collagen/fibrinogen domain containing) 1 | 2.36 |
| GRN | granulin | 2.33 |
| PTGIR | prostaglandin I2 (prostacyclin) receptor (IP) | 2.30 |
| HLA-B | major histocompatibility complex, class I, B | 2.27 |
| DYSF | dysferlin, limb girdle muscular dystrophy 2 | 2.25 |
| LILRB3 | leukocyte immunoglobulin-like receptor, subfamily B (3) | 2.21 |
| TALDO1 | transaldolase 1 | 2.21 |
| CXCR2 | chemokine (C-X-C motif) receptor 2 | 2.19 |
| FKBP5 | FK506 binding protein 5 | 2.17 |
| SORL1 | sortilin-related receptor, L(DLR class) A repeats-containing | 2.16 |
| IMPDH1 | IMP (inosine monophosphate) dehydrogenase 1 | 2.15 |
| DAPK2 | death-associated protein kinase 2 | 2.14 |
| CA4 | carbonic anhydrase IV | 2.13 |
| MMP9 | matrix metallopeptidase 9 (gelatinase B) | 2.11 |
| PSAP | prosaposin | 2.09 |
| TUBA3D | tubulin, alpha 3d | −2.08 |
| RPL24 | ribosomal protein L24 | −2.17 |
| GPR78 | G protein-coupled receptor 78 | −2.68 |
Significance analysis of microarrays (SAM) of IPF samples when categorized by percent predicted DLCO ≥65% [N = 16]. Differentially- expressed transcripts with <1% false discovery rate and > 2-fold change in expression are represented. Fold changes are expressed as log2 ratio. See supplementary tables for a complete list of differentially-expressed genes and corresponding accession numbers.
Table 4. Differentially-expressed genes that distinguish severe IPF from control.
| Gene Symbol | Description | Fold Change |
| IL1R2 | interleukin 1 receptor, type II | 3.43 |
| DEFA3 | defensing, alpha 3, neutrophil specific | 3.39 |
| OLFM4 | olfactomedin 4 | 3.39 |
| MMP9 | matrix metallopeptidase 9 (gelatinase B) | 3.32 |
| GRB10 | growth factor receptor-bound protein 10 | 3.25 |
| DEFA4 | defensin, alpha 4, corticostatin | 3.00 |
| LTF | lactotransferrin | 2.97 |
| RAB8A | RAB8A, member RAS oncogene family | 2.76 |
| CTSG | cathepsin G | 2.64 |
| CAMP | cathelicidin antimicrobial peptide | 2.64 |
| CEACAMP8 | carcinoembryonic antigen-related cell adhesion (8) | 2.53 |
| VSIG4 | V-set and immunoglobulin domain containing 4 | 2.50 |
| PGLYRP1 | peptidoglycan recognition protein 1 | 2.45 |
| FKBP5 | FK506 binding protein 5 | 2.45 |
| LOC151438 | hypothetical protein LOC151438 | 2.43 |
| ECHDC3 | enoyl Coenzyme A hydratase domain containing 3 | 2.34 |
| LOC100130890 | similar to hCG2030844 | −2.34 |
| PRSS36 | protease, serine, 36 | −2.37 |
| MCAT | malonyl CoA:ACP acyltransferase (mitochondrial) | −2.42 |
| IGHM | immunoglobulin heavy constant mu | −3.0 |
Significance analysis of microarrays (SAM) of IPF samples when categorized by percent predicted DLCO ≤35% [N = 15]. Differentially- expressed transcripts with <1% false discovery rate and ≥ 2-fold change in expression are represented. Fold changes are expressed as log2 ratio. See supplementary tables for a complete list of differentially-expressed genes and corresponding accession numbers.
Identification of a Peripheral Blood Signature that Distinguishes Extent of Disease
Principal component analysis was performed to determine outliers in the data set based on severity of disease categorization. Results demonstrate that 1 severe IPF case appears to be clinically misclassified as a mild case of IPF, while other cases are correctly classified as mild or severe (Figure 3). Significance analysis of microarrays (SAM) of IPF samples, when categorized by percent predicted DLCO (DLCO ≥65% [N = 16] and DLCO ≤35% [N = 15]), demonstrated 13 differentially-expressed transcripts with less than a 5% false discovery rate. Table 5 lists all differentially-expressed genes found between mild and severe cases of IPF categorized by DLCO. When using a FDR of ≤1%, only defensin A3 (DEFA3) is differentially-expressed between both mild or severe cases compared to controls. SAM revealed no differentially-expressed transcripts with less than a 5% false discovery rate between peripheral blood samples when IPF patients were categorized by percent predicted FVC (N = 27 and N = 13, data not shown).
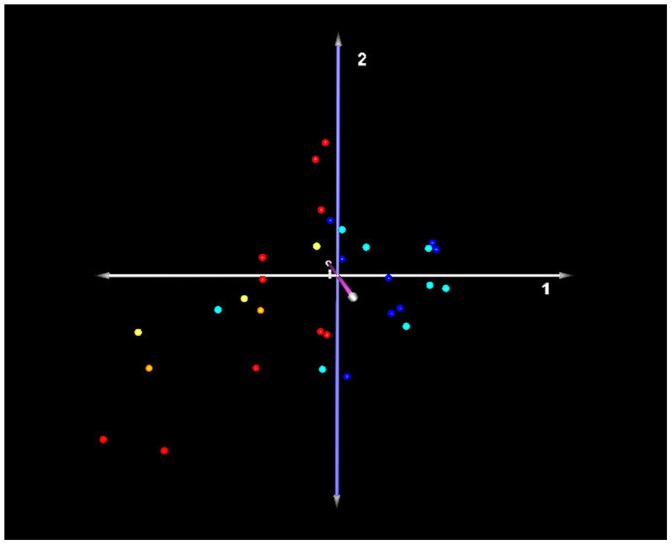
Figure 3. Principal component analysis of IPF samples grouped by extent of disease correlated with DLCO % predicted (mild ≥65%, severe ≤35%).
Red spheres: definite IPF; Orange spheres: probable IPF; Yellow spheres: unclassifiable fibrosis;Blue spheres: mild IPF; Green spheres: severe cases of IPF. Axis labels: white-first principal component; blue-second principal component; lavender- third principal component.
Table 5. Differentially-expressed genes between mild and severe cases of IPF.
| Symbol | Entrez Gene Name | Probe ID | Accession Number | Fold Change | Location |
| CAMP | cathelicidin antimicrobial peptide | A_23_P253791 | NM_004345 | 2.591 | Cytoplasm |
| CEACAM6 (includes EG:4680) | carcinoembryonic antigen-related cell adhesion molecule 6 | A_23_P421483 | BC005008 | 2.353 | Plasma Membrane |
| CTSG | cathepsin G | A_23_P140384 | NM_001911 | 2.703 | Cytoplasm |
| DEFA3 (includes EG:1668) | defensin, alpha3, neutrophil-specific | A_23_P31816 | NM_005217 | 2.379 | Extracellular Space |
| DEFA4 (includes EG:1669) | defensin, alpha 4, corticostatin | A_23_P326080 | NM_001925 | 3.713 | Extracellular Space |
| OLFM4 | olfactomedin 4 | A_24_P181254 | NM_006418 | 3.807 | unknown |
| HLTF | helicase-like transcription factor | A32_P210798 | BF513730 | 1.413 | unknown |
| PACSIN1 | protein kinase C and casein kinase substrate in neurons 1 | A_23_P258088 | NM_020804 | −1.511 | Cytoplasm |
| FLJ11710 | hypothetical protein FLJ11710 | A_23_P3921 | AK021772 | −1.798 | unknown |
| GABBR1 | gamma-aminobutyric acid (GABA) B receptor, 1 | A_23_P93302 | NM_001470 | −1.471 | Plasma Membrane |
| IGHM | immunoglobulin heavy constant mu | A_24_P417352 | BX161420 | −2.451 | Plasma Membrane |
| unknown | Unknown | A_23_P91743 | unknown | −1.884 | unknown |
| unknown | Unknown | A_24_P481375 | AK021668 | −1.706 | unknown |
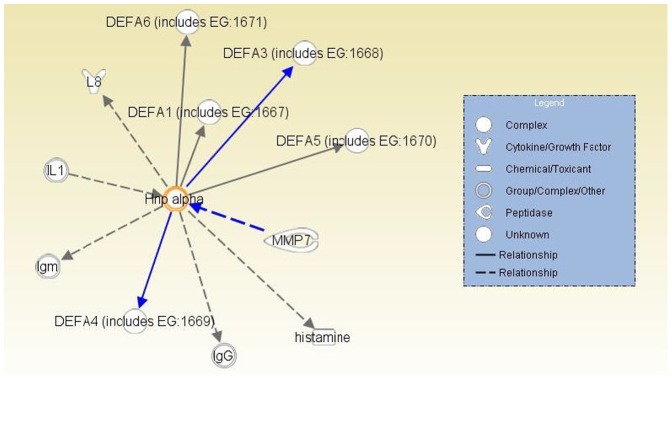
Alpha defensins activation pathway. Solid lines (direct relationship); Dashed lines (indirect relationship).
Peripheral Blood Signature Disease Progression Analysis
The general comparison analysis tool of the Ingenuity Pathway Analysis (IPA) software was utilized to identify the intersection or common differentially-expressed transcripts between normal and mild IPF, normal and severe IPF, and mild versus severe IPF when classified by DLCO for the purpose of discovery common potential biomarkers. This analysis showed that at FDR <5% only 2 differentially expressed transcripts, A3 (DEFA3) and hypothetical protein FLJ11710 , were differentially-expressed between control vs mild, mild vs severe, and control vs severe disease (DLCO ≤35%). There is up-regulation in DEFA3 expression compared to controls for severe IPF, while hypothetical protein FLJ11710 demonstrates a down regulation compared to controls for severe IPF. Between the mild and severe IPF cohort , and between normal and severe disease, there is up-regulation of several other host defense genes including defensin A4 (DEFA4), cathelicidin antimicrobial peptide (CAMP), cathespin G (CTSG), and down-regulation of immunoglobulin heavy chain constant mu (IGHM). The greatest increase found distinguishing controls from severe disease, and mild from severe disease is olfactinmedin 4 (OLFM4). We randomly selected 4 of the 13 differentially-expressed genes for qPCR validation, and qPCR confirmed the differential expression of these genes identified by microarray experiments (Table S8).
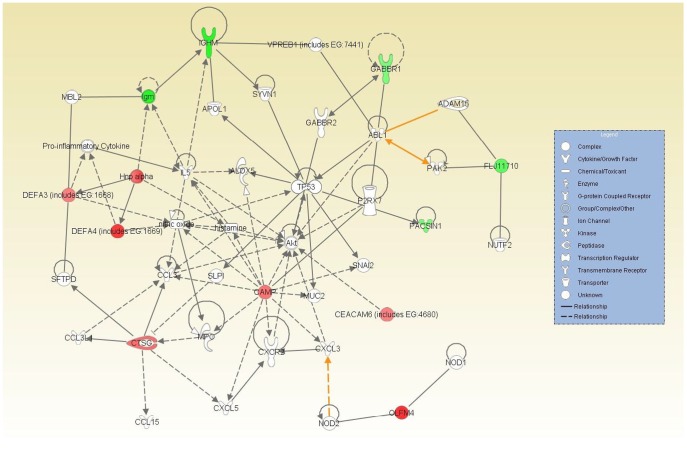
We also subjected the list of 13 differentially expressed genes based on DLCO (5%FDR, Table 5) to a functional analysis utilizing the Ingenuity Pathway Analysis (IPA) software. The functional analysis tool also calculates a significance value that is a measure for the likelihood that the association between a set of genes and a given process is due to random chance. Ten of the 13 differentially-expressed genes had annotations representing a gene, protein or chemical that was able to be mapped to an associated network. The associated network functions (Table S5) included: 1) inflammatory response (P<0.05), 2) cellular movement (P<0.05), 3) immune trafficking, 4) genetic disorder (P<0.05); and 4) cell-to-cell signaling (P<0.05). Figure 4 shows an overlay of all three associated networks illustrating both direct and indirect relationship pathways of the differentially-expressed genes. Finally, given that more than one α-defensin and other host defense-related proteins that distinguish normal, mild, and severe disease, we analyzed pathway interactions with α-defensins (Figure 5), and note an interaction with metalloproteinase 7 (MMP7) which activates α-defensins by proteolytic cleavage.
Figure 4. Overlaid networks and associated pathway analysis.
. Solid lines (direct relationship); Dashed lines (indirect relationship); Red filled (up-regulation); and Green filled (down-regulation).
Figure 5. Alpha defensins activation pathway.
Solid lines (direct relationship); Dashed lines (indirect relationship).
Discussion
This is the first study to investigate the IPF peripheral blood transcriptome based on disease severity defined by lung functional parameters. Our findings provide evidence that the peripheral blood transcriptome can be used to develop signatures that identify the presence and disease extent of IPF. We demonstrate a composite signature that distinguishes normal controls from IPF, 13 genes that distinguish mild from severe IPF utilizing DLCO values. Importantly, when a stringent false discovery rate of ≤1% is used, the genes that demonstrate the greatest differential-expression between control and mild disease, and control and severe disease are different. These data indicate that molecular signatures from the peripheral blood transcriptome that are intended to predict the presence of IPF will need to be take into account disease severity. There was differential-expressionof several inflammatory response and immune trafficking genes including α-defensins that are proteolytic substrates of MMP7. MMP7 has previously been shown to be over-expressed in IPF lung and serum, and its increased expression is associated with decreased survival [20]. These findings suggest that decreases in DLCO in IPF may be related to immune trafficking genes such as α-defensins.
We have identified that the peripheral blood transcriptome has the potential to discriminate controls from mild or severe IPF, and distinguish mild from severe disease using a cohort of both sporadic and familial IPF. Our previous gene expression studies indicate sporadic and familial IPF have similar gene expression patterns in lung [21]. Additionally, we have previously identified a promoter polymorphism in MUC5B that increase the risk of developing either sporadic or familial IPF [22]. Consequently, we believe both sporadic and familial IPF can be considered to be more similar than different, and justifies combining these in the current analysis.
Recent studies using plasma or serum protein markers have identified several proteins that might indicate disease presence or survival. Reduced survival has been associated with elevated serum levels of mucin 1 (MUC1/KL6) [23], chemokine (C-C motif) ligand 18 (CCL-18) [24], or surfactant protein A [25]. Rosas et al used a multi-analyte approach to screen a panel of 49 serum proteins and identified a combinatorial signature of 5 proteins that distinguished IPF patients from controls. MMP7 and MMP1 were the main components of this signature [15]. In their study, higher levels of MMP7 appeared to be associated with more severe disease. Most recently, Richards et al identified 140 patients with IPF and then validated in another 101 patients increased serum protein levels of MMP-7, ICAM-1, IL-8, VCAM-1, and S100A12 to be associated with decreased survival (XX). These studiesand ours demonstrate the emerging trend to identify relevant novel diagnostic and prognostic biomarkers for IPF in blood. There are no studies that evaluate on a genome-wide basis the peripheral blood transcriptome of IPF which relate to diagnostic or prognostic signatures.
Four of the 13 genes up-regulated in the current study have functions pertaining to host defense. This raises the question of an association of advanced disease and sub-clinical infection, or host-microbe interactions. All of these patients were stable when blood was drawn without overt signs of infection. Viral infection has been put forth as a possible trigger of acute exacerbations of IPF [12]. Two of the 13 genes are α-defensins which are small, cationic, cysteine-rich antimicrobial peptides that have important roles in host defense against bacteria, fungi and enveloped viruses [26]. In humans, α-defensins 1–4 are primarily found in neutrophils, and β-defensins are found in the epithelia of mucosal surfaces. Both are up-regulated by bacterial and viral infection [27], [28]. α-defensins are synthesized as inactive precursors consisting of 29–42 amino acid residues and are activated by proteolytic cleavage both by MMP7 [29], [30]. α-defensin levels in bronchoalveolar lavage and/or plasma are increased in fibrotic lung diseases, and significant amounts of α-defensins can be found outside neutrophils in fibroblastic foci in the lungs of patients with IPF [31]. Increased α-defensins levels are detected in the lung and blood of patients with acute exacerbations of IPF [32]. Elevated serum MMP-7 protein distinguish IPF from other types of diffuse parenchymal lung disease, and higher serum levels of MMP-7 in patients with IPF is associated with worse lung function [33]. MMP-7 expression is also up-regulated in lungs of patients with IPF [34]. In inflammatory lung disease complicated by fibroproliferation, it has been reported that α-defensins may contribute to the fibrotic response [12], [35], [36]. These data support the hypothesis that elevated levels of α-defensins may be an important substrate for MMP-7, and this interaction may be related to worsening physiologic function (DLCO).
Differential-expression analysis demonstrates up-regulation of the cathelicidin antimicrobial peptide (CAMP) between the severe IPF subgroup compared to controls, and between mild and severe disease groups. Cathelicidin is an antimicrobial protein of the innate immune system stored in peroxidase-negative granules of neutrophils [37]. CAMP is widely distributed, expressed in lung tissue, and detected in peripheral blood, plasma as well as bronchoalveolar lavage fluid (BAL) [38]. CAMP is another molecule of innate immunity that distinguishes mild from severe disease. While hypothetical protein FLJ11710 was down regulated in IPF groups compared to controls, little is known about its molecular functionality. CEACAM proteins have previously been shown to bind gram negative bacteria and are also over-expressed in lung cancers, associated with anti-apoptotic properties and tumor metastases. The CEACAM-1 gene encodes for glycosylated, glycosylphosphatidylinositol (GPI) anchored proteins that are expressed in alveolar epithelial cells [39]. Olfactomedin 4 is expressed primarily in bone marrow cells, but also in prostate, small intestine, colon, and stomach, and is upregulated in cancers of the stomach, colon, breast, and lung [40], [41]. Olfactomedin 4 appears to promote S-phase transition, and is a marker of intestinal stem cells [42]. An explanation for the increased expression of olfactomedin 4 in the peripheral blood of IPF patients, and its ability to distinguish mild from severe disease, is difficult.
There are limitations to this study. Given the minimal disease burden and lack of clinical symptoms, five cases of mild IPF were not appropriate candidates for surgical lung biopsy, and therefore did not have surgical tissue. Due to the minimal disease burden, they did not meet HRCT criteria of definite IPF according to the 2011 ATS/ERS/JRS/ALAT consensus statement. However, in the five cases the mean age was 70, and 4 cases were male supporting the diagnosis of IPF. Furthermore, these cases cluster with definite mild cases along the principal components (figure 1). Therefore, while these five cases of do not meet definite disease by current criteria, they very likely represent an early stage of IPF when all the definite criteria of disease cannot be met. Furthermore, the fact that these cases cluster with definite cases based on the peripheral blood signature demonstrates the feasibility of the peripheral blood gene expression signature as a diagnostic tool for early stage disease when HRCT findings are often ambiguous. There is no longitudinal data on the early cases to determine disease progression or other clinical variables. We have identified a signature that distinguishes disease extent (% predicted DLCO), but not when correlated with by FVC % predicted. The reason for this difference is unclear. FVC is traditionally used to stage the severity of pulmonary fibrosis. However, it has been recognized that a single measurement of the FVC is not useful for predicting disease progression or mortality of IPF [43]. In contrast, a 10 % decrease in FVC is associated with increased mortality in IPF [43]. Had we been able to obtain serial measurements of FVC, then perhaps we might have found an association with peripheral blood gene expression, disease severity, and FVC. In contrast, a single measurement of DLCO <50% predicts an increased mortality [43]. Therefore, it is possible that the peripheral blood signatures described correlate with disease progression as measured by a single DLCO% predicted but not a single FVC% predicted measurement due to the better performance of a single measurement of DLCO compared to FVC. A lack of correlation between FVC and severity of IPF can be due to concomitant emphysema, however review of the subject’s HRCT in this study does not demonstrate significant amounts of emphysema. It is possible the signature is a consequence of impaired oxygen transport, but this seems unlikely since two of the thirteen genes also distinguish controls from mild disease where oxygen transport is not a factor. Also, none of the 13 genes have functions related to oxygen transport, aerobic or anaerobic metabolism, or other pathways that might be implicated in the presence of hypoxemia or impaired oxygenation.
In summary, our results demonstrate that the peripheral blood transcriptome can potentially distinguish extent of disease in individuals with IPF when samples are correlated with percent predicted DLCO, and distinguish IPF patients from normal. Our data is consistent with a role for MMP-7 interacting with α-defensins, and increased expression of host defense proteins in the peripheral blood being associated with deterioration of DLCO in IPF. The ability to use a peripheral blood biomarker to monitor disease progression for IPF could have a substantial impact on the diagnosis, treatment, and management of this disease, and perhaps be generally applicable to other subtypes of idiopathic interstitial pneumonias.
Supporting Information
†Definite IPF without surgical lung biopsy is defined by supporting clinical information and HRCT demonstrating sub-pleural and bibasilar predominate reticulation, honeycombing, and traction bronchiectasis without atypical features such as nodules, predominate ground glass opacities, pleural plaques, air-trapping, or lymphadenopathy. ‡Probable IPF without surgical lung biopsy is defined by supporting clinical information and HRCT demonstrating sub-pleural and bibasilar predominate reticulation, traction bronchiectasis without bilateral honeycombing, and without atypical features outlined above. Surgical lung biopsy (SLBx): definite IPF is defined as usual interstitial pneumonia requiring spatial and temporal heterogeneity; subpleurally accentuated microscopic honeycombing, fibroblastic foci without significant parenchymal, airway, or pleural mononuclear inflammation; definite IPF is also advanced honeycombing on lung biopsy with clinical and radiologic features supporting IPF. n/a is not available.
(DOCX)
†Definite IPF without surgical lung biopsy is defined by supporting clinical information and HRCT demonstrating sub-pleural and bibasilar predominate reticulation, honeycombing, and traction bronchiectasis without atypical features such as nodules, predominate ground glass opacities, pleural plaques, air-trapping, or lymphadenopathy. ‡Probable IPF without surgical lung biopsy is defined by supporting clinical information and HRCT demonstrating sub-pleural and bibasilar predominate reticulation, traction bronchiectasis without bilateral honeycombing, and without atypical features outlined above. Surgical lung biopsy (SLBx) :definite IPF is defined as usual interstitial pneumonia requiring spatial and temporal heterogeneity; subpleurally accentuated microscopic honeycombing, fibroblastic foci without significant parenchymal, airway, or pleural mononuclear inflammation; definite IPF is also advanced honeycombing on lung biopsy with clinical and radiologic features supporting IPF. n/a is not available.
(DOCX)
†Definite IPF without surgical lung biopsy is defined by supporting clinical information and HRCT demonstrating sub-pleural and bibasilar predominate reticulation, honeycombing, and traction bronchiectasis without atypical features such as nodules, predominate ground glass opacities, pleural plaques, air-trapping, or lymphadenopathy. ‡Probable IPF without surgical lung biopsy is defined by supporting clinical information and HRCT demonstrating sub-pleural and bibasilar predominate reticulation, traction bronchiectasis without bilateral honeycombing, and without atypical features outlined above. Surgical lung biopsy (SLBx) :definite IPF is defined as usual interstitial pneumonia requiring spatial and temporal heterogeneity; subpleurally accentuated microscopic honeycombing, fibroblastic foci without significant parenchymal, airway, or pleural mononuclear inflammation; definite IPF is also advanced honeycombing on lung biopsy with clinical and radiologic features supporting IPF. n/a is not available.
(DOCX)
†Definite IPF without surgical lung biopsy is defined by supporting clinical information and HRCT demonstrating sub-pleural and bibasilar predominate reticulation, honeycombing, and traction bronchiectasis without atypical features such as nodules, predominate ground glass opacities, pleural plaques, air-trapping, or lymphadenopathy. ‡Probable IPF without surgical lung biopsy is defined by supporting clinical information and HRCT demonstrating sub-pleural and bibasilar predominate reticulation, traction bronchiectasis without bilateral honeycombing, and without atypical features outlined above. Surgical lung biopsy (SLBx): definite IPF is defined as usual interstitial pneumonia requiring spatial and temporal heterogeneity; subpleurally accentuated microscopic honeycombing, fibroblastic foci without significant parenchymal, airway, or pleural mononuclear inflammation; definite IPF is also advanced honeycombing on lung biopsy with clinical and radiologic features supporting IPF. n/a is not available.
(DOCX)
Canonical pathway analysis using Ingenuity Pathway Analysis™ tool to identify associated networks. Network functions with their respective P values and the number of differentially-expressed genes identified are presented.
(DOCX)
Differentially-expressed genes between controls and mild IPF (DLCO >65%) using SAM at FDR of <1%. Agilent probe identifier, RefSeq number, and Entrez geneID number are presented. Fold change is log2. FDR is false discover rate (%).
(XLSX)
Differentially-expressed genes between controls and severe IPF (DLCO <35%) using SAM at FDR of <1%. Agilent probe identifier, RefSeq number, and Entrez geneID number are presented. Fold change is log2. FDR is false discover rate (%).
(XLSX)
Validation of selected differentially-expressed genes by Reatime™ PCR. Primers utilized for PCR and differential expression by Realtime PCR using ΔΔ Ct method.
(XLSX)
Footnotes
Competing Interests: I have read the journal’s policy and have the following conflicts. Dr. Kathy Boon is currently employed with Excerpta Medica. Excerpta Medica has no affiliations or interests in the research related to this manuscript. Excerpta Medica no funding, review, assistance, or any other relationship. At the time this research was conducted, Dr. Boon was not employed by Excerpta Medica. This does not alter the authors' adherence to all the PLoS ONE policies on sharing data and materials.
Funding: This work was funded by the National Heart Lung Blood Institute 5R01HL095393 and RC1HL09957. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.American Thoracic Society/European Respiratory Society International Multidisciplinary Consensus Classification of the Idiopathic Interstitial Pneumonias. This joint statement of the American Thoracic Society (ATS), and the European Respiratory Society (ERS) was adopted by the ATS board of directors, June 2001 and by the ERS Executive Committee, June 2001. Am J Respir Crit Care Med. 2002;165:277–304. doi: 10.1164/ajrccm.165.2.ats01. [DOI] [PubMed] [Google Scholar]
- 2.Bjorker JA, Ryu JH, Edwin MK, Myers JL, Tazelaar HD, et al. Prognostic Significance of Histopathologic Subsets in Idiopathic Pulmonary Fibrosis. Am J Respir Crit Care Med. 1998;157:199–203. doi: 10.1164/ajrccm.157.1.9704130. [DOI] [PubMed] [Google Scholar]
- 3.Collard HR, Pantilat SZ. Dyspnea in interstitial lung disease. Curr Opin Support Palliat Care. 2008;2:100–104. doi: 10.1097/SPC.0b013e3282ff6336. [DOI] [PubMed] [Google Scholar]
- 4.Tzilas V, Koti A, Papandrinopoulou D, Tsoukalas G. Prognostic Factors in Idiopathic Pulmonary Fibrosis. The American Journal of the Medical Sciences 338 481–485. 2009. [DOI] [PubMed]
- 5.King TE, Jr, Tooze JA, Schwarz MI, Brown KR, Cherniack RM. Predicting survival in idiopathic pulmonary fibrosis: scoring system and survival model. Am J Respir Crit Care Med. 2001;164:1171–1181. doi: 10.1164/ajrccm.164.7.2003140. [DOI] [PubMed] [Google Scholar]
- 6.Flaherty KR, Mumford JA, Murray S, Kazerooni EA, Gross BH, et al. Prognostic implications of physiologic and radiographic changes in idiopathic interstitial pneumonia. Am J Respir Crit Care Med. 2003;168:543–548. doi: 10.1164/rccm.200209-1112OC. [DOI] [PubMed] [Google Scholar]
- 7.Lynch DA, Godwin JD, Safrin S, Starko KM, Hormel P, et al. High-resolution computed tomography in idiopathic pulmonary fibrosis: diagnosis and prognosis. American Journal of Respiratory and Critical Care Medicine. 2005;172:488–493. doi: 10.1164/rccm.200412-1756OC. [DOI] [PubMed] [Google Scholar]
- 8.Collard HR, King TE, Jr, Bartelson BB, Vourlekis JS, Schwarz MI, et al. Changes in Clinical and Physiologic Variables Predict Survival in Idiopathic Pulmonary Fibrosis. Am J Respir Crit Care Med. 2003;168:538–542. doi: 10.1164/rccm.200211-1311OC. [DOI] [PubMed] [Google Scholar]
- 9.Jegal Y, Kim DS, Shim TS, Lim C-M, Do Lee S, et al. Physiology Is a Stronger Predictor of Survival than Pathology in Fibrotic Interstitial Pneumonia. Am J Respir Crit Care Med. 2005;171:639–644. doi: 10.1164/rccm.200403-331OC. [DOI] [PubMed] [Google Scholar]
- 10.Latsi PI, du Bois RM, Nicholson AG, Colby TV, Bisirtzoglou D, et al. Fibrotic Idiopathic Interstitial Pneumonia: The Prognostic Value of Longitudinal Functional Trends. Am J Respir Crit Care Med. 2003;168:531–537. doi: 10.1164/rccm.200210-1245OC. [DOI] [PubMed] [Google Scholar]
- 11.Flaherty KR, Andrei A-C, Murray S, Fraley C, Colby TV, et al. Idiopathic Pulmonary Fibrosis: Prognostic Value of Changes in Physiology and Six-Minute-Walk Test. Am J Respir Crit Care Med. 2006;174:803–809. doi: 10.1164/rccm.200604-488OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Collard HR, Moore BB, Flaherty KR, Brown KK, Kaner RJ, et al. Acute exacerbations of idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. 2007;176:636–643. doi: 10.1164/rccm.200703-463PP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Noble PW, Albera C, Bradford WZ, Costabel U, Glassberg MK, et al. Pirfenidone in patients with idiopathic pulmonary fibrosis (CAPACITY): two randomised trials. Lancet. 2011;377:1760–1769. doi: 10.1016/S0140-6736(11)60405-4. [DOI] [PubMed] [Google Scholar]
- 14.Collard HR, Moore BB, Flaherty KR, Brown KK, Kaner RJ, et al. Acute exacerbations of idiopathic pulmonary fibrosis. American Journal of Respiratory and Critical Care Medicine. 2007;176:636–643. doi: 10.1164/rccm.200703-463PP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rosas IO, Richards TJ, Konishi K, Zhang Y, Gibson K, et al. MMP1 and MMP7 as Potential Peripheral Blood Biomarkers in Idiopathic Pulmonary Fibrosis. PLoS Medicine. 2008;5:623–633. doi: 10.1371/journal.pmed.0050093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Raghu G, Collard HR, Egan JJ, Martinez FJ, Behr J, et al. An Official ATS/ERS/JRS/ALAT Statement: Idiopathic Pulmonary Fibrosis: Evidence-based Guidelines for Diagnosis and Management. Am J Respir Crit Care Med. 2011;183:788–824. doi: 10.1164/rccm.2009-040GL. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Standardization of spirometry–1987 update. Statement of the American Thoracic Society. American Review of Respiratory Disease. 1987;136:1285–1298. doi: 10.1164/ajrccm/136.5.1285. [DOI] [PubMed] [Google Scholar]
- 18.Saeed A, Sharov V, White J, Li J, Liang W, et al. TM4: a free, open-source system for microarray data management and analysis. Biotechniques. 2003;34:374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- 19.Heid CA, Stevens J, Livak KJ, Williams PM. Real time quantitative PCR. Genome Res. 1996;6:986–994. doi: 10.1101/gr.6.10.986. [DOI] [PubMed] [Google Scholar]
- 20.Richards TJ, Kaminski N, Baribaud F, Flavin S, Brodmerkel C, et al. Peripheral blood proteins predict mortality in idiopathic pulmonary fibrosis. American Journal of Respiratory and Critical Care Medicine. 2012;185:67–76. doi: 10.1164/rccm.201101-0058OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang IV, Burch LH, Steele MP, Savov JD, Hollingsworth JW, et al. Gene expression profiling of familial and sporadic interstitial pneumonia. American Journal of Respiratory and Critical Care Medicine. 2007;175:45–54. doi: 10.1164/rccm.200601-062OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seibold MA, Wise AL, Speer MC, Steele MP, Brown KK, et al. A common MUC5B promoter polymorphism and pulmonary fibrosis. N Engl J Med. 2011;364:1503–1512. doi: 10.1056/NEJMoa1013660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yokoyama A, Kondo K, Nakajima M, Matsushima T, Takahashi T, et al. Prognostic value of circulating KL-6 in idiopathic pulmonary fibrosis. Respirology. 2006;11:164–168. doi: 10.1111/j.1440-1843.2006.00834.x. [DOI] [PubMed] [Google Scholar]
- 24.Prasse A, Probst C, Bargagli E, Zissel G, Toews GB, et al. Serum CC-chemokine ligand 18 concentration predicts outcome in idiopathic pulmonary fibrosis. American Journal of Respiratory and Critical Care Medicine. 2009;179:717–723. doi: 10.1164/rccm.200808-1201OC. [DOI] [PubMed] [Google Scholar]
- 25.Greene KE, King TE, Jr, Kuroki Y, Bucher-Bartelson B, Hunninghake GW, et al. Serum surfactant proteins-A and -D as biomarkers in idiopathic pulmonary fibrosis. Eur Respir J. 2002;19:439–446. doi: 10.1183/09031936.02.00081102. [DOI] [PubMed] [Google Scholar]
- 26.Bevins CL. Paneth cell defensins: key effector molecules of innate immunity. Antimicrobial Peptides. 2006;34:253–266. doi: 10.1042/BST20060263. [DOI] [PubMed] [Google Scholar]
- 27.Amenomori M, Mukae H, Ishimatsu Y, Sakamoto N, Kakugawa T, et al. Differential effects of human neutrophil peptide-1 on growth factor and interleukin-8 production by human lung fibroblasts and epithelial cells. Experimental Lung Research. 2010;36:411–419. doi: 10.3109/01902141003714049. [DOI] [PubMed] [Google Scholar]
- 28.Zitvogel L, Kepp O, Kroemer G. Decoding cell death signals in inflammation and immunity. Cell. 2010;140:798–804. doi: 10.1016/j.cell.2010.02.015. [DOI] [PubMed] [Google Scholar]
- 29.Wilson CL, Schmidt AP, Pirilä E, Valore EV, Ferri N, et al. Differential Processing of α- and β-Defensin Precursors by Matrix Metalloproteinase-7 (MMP-7). Biol Chem. 2009;284:8301–8311. doi: 10.1074/jbc.M809744200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wilson CL, Schmidt AP, Pirila E, Valore EV, Ferri N, et al. Differential Processing of {alpha}- and {beta}-Defensin Precursors by Matrix Metalloproteinase-7 (MMP-7). Journal of Biological Chemistry. 2009;284:8301–8311. doi: 10.1074/jbc.M809744200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mukae H, Iiboshi H, Nakazato M, Hiratsuka T, Tokojima M, et al. Raised plasma concentrations of alpha-defensins in patients with idiopathic pulmonary fibrosis. Thorax. 2002;57:623–628. doi: 10.1136/thorax.57.7.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Konishi K, Gibson KF, Lindell KO, Richards TJ, Zhang Y, et al. Gene expression profiles of acute exacerbations of idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. 2009;180:167–175. doi: 10.1164/rccm.200810-1596OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rosas IO, Ren P, Avila NA, Chow CK, Franks TJ, et al. Early interstitial lung disease in familial pulmonary fibrosis. Am J Respir Crit Care Med. 2007;176:698–705. doi: 10.1164/rccm.200702-254OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zuo F, Kaminski N, Eugui E, Allard J, Yakhini Z, et al. Gene expression analysis reveals matrilysin as a key regulator of pulmonary fibrosis in mice and humans. Proc Natl Acad Sci U S A. 2002;99:6292–6297. doi: 10.1073/pnas.092134099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Han W, Wang W, Mohammed K, Yunchao S. α-Defensins increase lung fibroblast proliferation and collagen synthesis via the β-catenin signaling pathway. FEBS J. 2009;276:6603–6614. doi: 10.1111/j.1742-4658.2009.07370.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Van Wetering S, Tjabringa GS, Hiemstra PS. Interactions between neutrophil-derived antimicrobial peptides and airway epithelial cells. J Leukoc Biol. 2005;77:444–450. doi: 10.1189/jlb.0604367. [DOI] [PubMed] [Google Scholar]
- 37.Zanetti M. The role of cathelicidins in the innate host defenses of mammals. Curr Issues Mol Biol. 2005;7:179–196. [PubMed] [Google Scholar]
- 38.Ganz T. Antimicrobial polypeptides in host defense of the respiratory tract. J Clin Invest. 2002;109:693–697. doi: 10.1172/JCI15218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Venkatadri K, Gonzales LW, Bailey NA, Wang P, Angampalli S, et al. Ballard Carcinoembryonic cell adhesion molecule 6 in human lung: regulated expression of a multifunctional type II cell protein Physiol Lung Cell Mol Physiol 296 L1019-L1030. 2009. [DOI] [PMC free article] [PubMed]
- 40.Koshida S, Kobayashi D, Moriai R, Tsuji N, Watanabe N. Specific overexpression of OLFM4(GW112/HGC-1) mRNA in colon, breast and lung cancer tissues detected using quantitative analysis. Cancer Sci. 2007;98:315–320. doi: 10.1111/j.1349-7006.2006.00383.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu W, Lee HW, Liu Y, Wang R, Rodgers GP. Olfactomedin 4 is a novel target gene of retinoic acids and 5-aza-2′-deoxycytidine involved in human myeloid leukemia cell growth, differentiation, and apoptosis. Blood. 2010;116:4938–4947. doi: 10.1182/blood-2009-10-246439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van der Flier LG, van Gijn ME, Hatzis P, Kujala P, Haegebarth A, et al. Transcription factor achaete scute-like 2 controls intestinal stem cell fate. Cell. 2009;136:903–912. doi: 10.1016/j.cell.2009.01.031. [DOI] [PubMed] [Google Scholar]
- 43.King TE, Jr, Safrin S, Starko KM, Brown KK, Noble PW, et al. Analyses of efficacy end points in a controlled trial of interferon-gamma1b for idiopathic pulmonary fibrosis. Chest. 2005;127:171–177. doi: 10.1378/chest.127.1.171. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
†Definite IPF without surgical lung biopsy is defined by supporting clinical information and HRCT demonstrating sub-pleural and bibasilar predominate reticulation, honeycombing, and traction bronchiectasis without atypical features such as nodules, predominate ground glass opacities, pleural plaques, air-trapping, or lymphadenopathy. ‡Probable IPF without surgical lung biopsy is defined by supporting clinical information and HRCT demonstrating sub-pleural and bibasilar predominate reticulation, traction bronchiectasis without bilateral honeycombing, and without atypical features outlined above. Surgical lung biopsy (SLBx): definite IPF is defined as usual interstitial pneumonia requiring spatial and temporal heterogeneity; subpleurally accentuated microscopic honeycombing, fibroblastic foci without significant parenchymal, airway, or pleural mononuclear inflammation; definite IPF is also advanced honeycombing on lung biopsy with clinical and radiologic features supporting IPF. n/a is not available.
(DOCX)
†Definite IPF without surgical lung biopsy is defined by supporting clinical information and HRCT demonstrating sub-pleural and bibasilar predominate reticulation, honeycombing, and traction bronchiectasis without atypical features such as nodules, predominate ground glass opacities, pleural plaques, air-trapping, or lymphadenopathy. ‡Probable IPF without surgical lung biopsy is defined by supporting clinical information and HRCT demonstrating sub-pleural and bibasilar predominate reticulation, traction bronchiectasis without bilateral honeycombing, and without atypical features outlined above. Surgical lung biopsy (SLBx) :definite IPF is defined as usual interstitial pneumonia requiring spatial and temporal heterogeneity; subpleurally accentuated microscopic honeycombing, fibroblastic foci without significant parenchymal, airway, or pleural mononuclear inflammation; definite IPF is also advanced honeycombing on lung biopsy with clinical and radiologic features supporting IPF. n/a is not available.
(DOCX)
†Definite IPF without surgical lung biopsy is defined by supporting clinical information and HRCT demonstrating sub-pleural and bibasilar predominate reticulation, honeycombing, and traction bronchiectasis without atypical features such as nodules, predominate ground glass opacities, pleural plaques, air-trapping, or lymphadenopathy. ‡Probable IPF without surgical lung biopsy is defined by supporting clinical information and HRCT demonstrating sub-pleural and bibasilar predominate reticulation, traction bronchiectasis without bilateral honeycombing, and without atypical features outlined above. Surgical lung biopsy (SLBx) :definite IPF is defined as usual interstitial pneumonia requiring spatial and temporal heterogeneity; subpleurally accentuated microscopic honeycombing, fibroblastic foci without significant parenchymal, airway, or pleural mononuclear inflammation; definite IPF is also advanced honeycombing on lung biopsy with clinical and radiologic features supporting IPF. n/a is not available.
(DOCX)
†Definite IPF without surgical lung biopsy is defined by supporting clinical information and HRCT demonstrating sub-pleural and bibasilar predominate reticulation, honeycombing, and traction bronchiectasis without atypical features such as nodules, predominate ground glass opacities, pleural plaques, air-trapping, or lymphadenopathy. ‡Probable IPF without surgical lung biopsy is defined by supporting clinical information and HRCT demonstrating sub-pleural and bibasilar predominate reticulation, traction bronchiectasis without bilateral honeycombing, and without atypical features outlined above. Surgical lung biopsy (SLBx): definite IPF is defined as usual interstitial pneumonia requiring spatial and temporal heterogeneity; subpleurally accentuated microscopic honeycombing, fibroblastic foci without significant parenchymal, airway, or pleural mononuclear inflammation; definite IPF is also advanced honeycombing on lung biopsy with clinical and radiologic features supporting IPF. n/a is not available.
(DOCX)
Canonical pathway analysis using Ingenuity Pathway Analysis™ tool to identify associated networks. Network functions with their respective P values and the number of differentially-expressed genes identified are presented.
(DOCX)
Differentially-expressed genes between controls and mild IPF (DLCO >65%) using SAM at FDR of <1%. Agilent probe identifier, RefSeq number, and Entrez geneID number are presented. Fold change is log2. FDR is false discover rate (%).
(XLSX)
Differentially-expressed genes between controls and severe IPF (DLCO <35%) using SAM at FDR of <1%. Agilent probe identifier, RefSeq number, and Entrez geneID number are presented. Fold change is log2. FDR is false discover rate (%).
(XLSX)
Validation of selected differentially-expressed genes by Reatime™ PCR. Primers utilized for PCR and differential expression by Realtime PCR using ΔΔ Ct method.
(XLSX)