Abstract
In science, the guinea pig is known as one of the gold standards for modeling human disease. It is especially important as a molecular and cellular biology model for studying the human immune system, as its immunological genes are more similar to human genes than are those of mice. The utility of the guinea pig as a model organism can be further enhanced by further characterization of the genes encoding components of the immune system. Here, we report the genomic organization of the guinea pig immunoglobulin (Ig) heavy and light chain genes. The guinea pig IgH locus is located in genomic scaffolds 54 and 75, and spans approximately 6,480 kb. 507 VH segments (94 potentially functional genes and 413 pseudogenes), 41 DH segments, six JH segments, four constant region genes (μ, γ, ε, and α), and one reverse δ remnant fragment were identified within the two scaffolds. Many VH pseudogenes were found within the guinea pig, and likely constituted a potential donor pool for gene conversion during evolution. The Igκ locus mapped to a 4,029 kb region of scaffold 37 and 24 is composed of 349 Vκ (111 potentially functional genes and 238 pseudogenes), three Jκ and one Cκ genes. The Igλ locus spans 1,642 kb in scaffold 4 and consists of 142 Vλ (58 potentially functional genes and 84 pseudogenes) and 11 Jλ -Cλ clusters. Phylogenetic analysis suggested the guinea pig’s large germline VH gene segments appear to form limited gene families. Therefore, this species may generate antibody diversity via a gene conversion-like mechanism associated with its pseudogene reserves.
Introduction
The guinea pig (Cavia porcellus), also called the cavy, is a species of rodent belonging to the family Caviidae. This animal has been used in scientific experimentation since the 17th century. During the 19th and early 20th centuries, the guinea pig was a popular experimental animal for studying prevalent bacterial diseases such as tuberculosis and diphtheria [1], resulting in the epithet “guinea pig" being used to describe a test subject. Guinea pigs are currently still used in research, primarily as models for human diseases, including juvenile diabetes, tuberculosis, scurvy, and pregnancy complications [2], [3], [4], [5], [6], [7], [8].
Immunoglobulins (Igs) are only expressed by jawed vertebrates [9], [10], [11] and are usually composed of two identical heavy (H) chains and two identical light (L) chains. Exceptions include shark IgNAR and camelid IgGs, which are only comprised of heavy chains [9], [10], [11]. To date, mammalian Ig genes are organized into a ‘translocon’ configuration [12]. In the heavy chain locus, multiple variable (VH), diversity (DH), and joining (JH) gene segments are followed by μ, δ, γ, ε, and α genes [13]. In the kappa encoding locus, multiple joining (Jκ) region gene segments are present within a cluster, followed by a single constant (Cκ) gene, whereas in the lambda encoding locus, joining (Jλ) and constant (Cλ) genes occur as Jλ-Cλ blocks, which usually have multiple copies [14].
The word “guinea pig" is synonymous with scientific experimentation, but little is known about its Ig genes. We therefore used the recently available genome data of guinea pig provide as an opportunity to study the Ig genes of this species. Our study aimed to characterize the guinea pig IgH and IgL loci, in an effort to promote a better understanding of the immune system and evolutionary divergence of the Ig genes in placental mammals.
Materials and Methods
Guinea Pig Genome Sequence
Guinea pig (C. porcellus) genome data were obtained from the Ensembl database (http://www.ensembl.org), and the Broad Institute conducted genome sequencing and assembly (cavPor3, 6.79× coverage, Jul 2008). High-coverage ensured increased accuracy of the genome analysis results.
Identification of the Guinea Pig Ig Genes
Guinea pig Ig constant region genes were retrieved on the basis of comparing guinea pig and human Ig gene sequences (http://genome.ucsc.edu/). FUZZNUC, an online software (http://mobyle.pasteur.fr/cgi-bin/portal.py?#forms::fuzznuc), was used to find adjacent recombination signal sequences (RSSs) for the identification of variable, diversity, and joining gene segments. Five or more mismatched bases were allowed to cover all genes.
Total Ribonucleic Acid (RNA) Extract and 3′ RACE
Total RNA was extracted from spleen tissues of three male HARTLY guinea pigs using TRIzol Reagent (Invitrogen, USA) and pooled equally. Animals were treated in accordance with the China Agricultural University on the protection of animals used for experimental and other scientific purposes. The study was approved by animal welfare committee of China Agricultural University with approval number XK257.
Complementary deoxyribonucleic acid (cDNA) was synthesized from 1 µg of total RNA using 3′RACE-reverse transcription primers. The first polymerase chain reaction (PCR) amplification of each Ig heavy chain constant region gene (μ, γ, ε, and α) was performed using the corresponding sense primer and 3′RACE-antisense primer 1. While the second PCR amplification was conducted using the corresponding sense primer and 3′RACE-antisense primer 2. All primers are displayed in Table S1.
Southern Blotting Analysis of Genomic DNA Sample
Guinea pig liver genomic DNA was isolated according to the method as previously described [15]. The VH gene family-specific probes belonging to guinea pig VH1, VH2 and VH3 gene families were labeled using a PCR digoxygenin probe synthesis kit (Roche, Germany) with primers designed from VH1-95, VH2-17 and VH3-157 (The primer sequences are displayed in Table S1). Ten micrograms of genomic DNA digested separately with BamH I, EcoR I, Hind III and Xba I (New England Biolabs, USA) were loaded into each well of a 0.8% agarose gel and electrophoresed for 12 h, transferred to a positively charged nylon membrane (Roche, Germany). Hybridization and detection were conducted following the manufacturer’s instructions.
Sequence and Phylogenetic Analysis
Multiple sequence alignments were edited and handled with the Megalign software program [16], and the Clustal W and Clustal X algorithms [17], [18], before being analysed using BioEdit [19]. Comparative phylogenetic trees were constructed using the PHYLIP 3.67 [20] software and TreeView [21] based on the final nucleotide alignment. The neighbor-joining algorithm was used for phylogenetic analysis and bootstrap support was provided by 1000 replicates. Sequences from other species used in our phylogenetic analyses and sequence alignments are presented in Figures S1, S2, and S3 and Table S2.
Dot Matrix Analysis
Pairwise dot matrix comparisons were made using DNAMAN software (window size = 30-bp, mismatch limit = 9-bp) to identify potential alignment of nucleotide bases between the sequences.
Definition of the VH/VL Gene Families
In mammals, germline VH and VL genes are categorized into different families according to their amino acid or nucleotide sequences similarity [22]. Sequences with greater than 75% similarity are general considered to belong to the same family, while those with less than 70% similarity are placed in different gene families, and those possessing between 70% and 75% similarity are inspected on a case-by-case basis [23]. We placed potentially functional VH and VL gene segments sharing more than 70% similarity into the same family.
Results
Guinea Pig IgH Locus
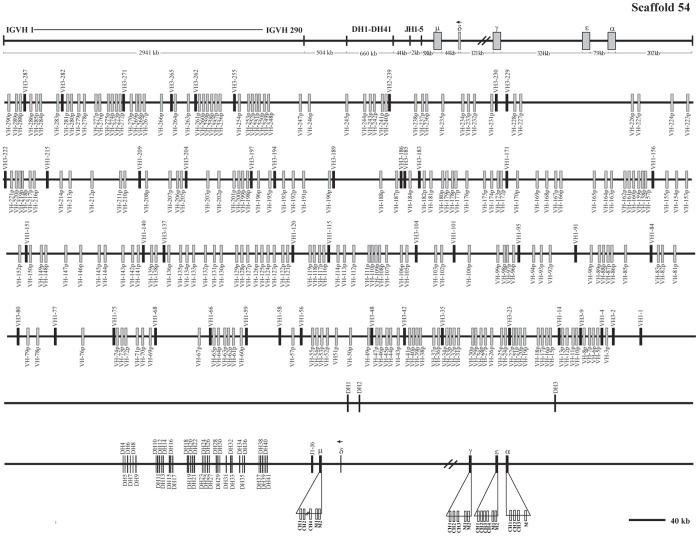
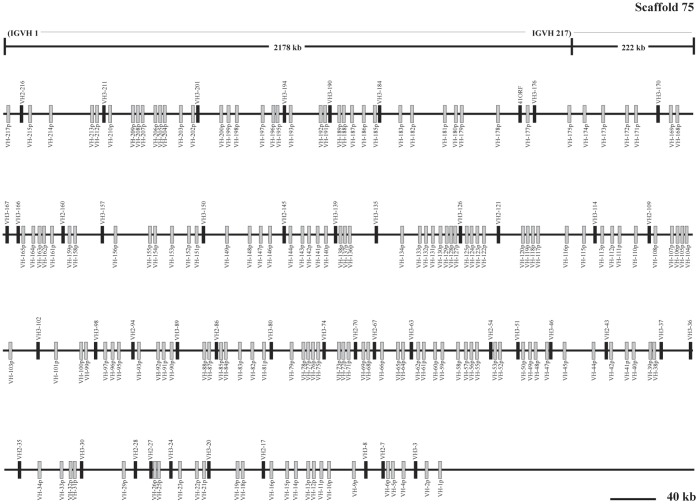
Analysis of the genomic sequence revealed that the guinea pig IgH locus is located within genomic scaffolds 54 and 75 (Figure 1, Figure 2). The entire IgH locus spans approximately 6,480 kb of the two scaffolds (4,302 kb in scaffold 54 and 2,178 kb in scaffold 75). The total length is an estimate due to the existence of sequence gaps (Figure 1, Figure 2). Six JH segments, 507 VH, 41 DH, four constant region genes (μ, γ, ε and α) and a reverse δ trace (marked with an arrow towards the left) were identified within the two scaffolds. Locations of the annotated IgH genes on the guinea pig genome are displayed in Table S3.
Figure 1. The guinea pig IgH locus in scaffold 54.
The guinea pig IgH locus length is approximately 4,302 kb from the most 5′ VH segment (VH-290p) to the most 3′ end of α gene in genomic scaffold 54. Filled bars: potentially functional VH genes; open bars: VH pseudogenes and V-p; DH: diversity genes; JH: joining genes; μ: IgM coding gene; δ: IgD fragment; γ: IgG coding gene; ε: IgE coding gene and α: IgA coding gene. VH and DH genes are numbered based on the families and their order. The number before the dash (VHn) indicates the family, while the number after dash represents the genomic position. JH genes are numbered based on the order of their locations in the locus. Double slashes indicate gaps >10 kb. The unidirectional arrowheads above δ gene segments in scaffold 54 indicate that its direction is opposite to the other heavy chain constant genes transcriptional direction. The scale does not apply the upper frame diagram.
Figure 2. The guinea pig IgH locus in scaffold 75.
The guinea pig IgH locus length is approximately 2,177 kb from the most VH (VH-217p) to the most end (VH-1p) in scaffold 75. Filled bars: potentially functional VH genes; open bars: VH pseudogenes. The scale does not apply the upper frame diagram.
Analysis of the Guinea Pig Constant Region Genes
To acquire the cDNA sequence for four heavy chain isotypes (μ, γ, ε, and α), 3′RACE were performed on splenic RNA using specific primers. Using this strategy we successfully identified genes encoding the constant region exons for IgM, IgG, IgE, and IgA (Figure S4). As predicted from the analysis of the guinea pig Ig genomic genes, the domain structure for the four isotypes was typical of that for other mammalian species (Figure S5).
Within the guinea pig genome, CH3 and parts of the CH4 segments of the μ gene were determined to be missing because of the existence of gaps (Figure 1). By using the 3′ RACE method, the complete secreted IgM, including four exons, was successfully cloned (Figure S4–S5).
Most mammals also express the δ gene, with the exception of the rabbit [24] and opossum [25]. The area predicted to contain an IgD C region, and in particular the 3′ region of the IgM exons between IgM and IgG, as well as the whole cavPor3 assembly, was thoroughly searched in two different orientations for coding sequences that might correspond to a putative IgD. These searches detected δ trace fragments that are homologous with mammalian IgD (Figure 3) and showed an opposite transcription direction to the upstream μ gene. Based on sequence alignment, the three-fragment δ trace was found to belong to the CH2 and CH3 domains (Figure 3).
Figure 3. Alignment of the guinea pig IgD remnants with other mammalian IgD domains.
Amino acid residues that are identical to the top counterpart in every panel are shown as dots; Gaps and missing data are indicated by hyphens.
Although two IgG isotypes (IgG1 and IgG2) were previously identified in domestic guinea pig serum [26], we only identified one IgG gene in the guinea pig genome perhaps due to sequence gaps. Our sequence alignment further showed that the recognized IgG in guinea pig genome shared the highest similarity with IgG1 (six amino acid difference) (Figure S6). Interestingly, we were just able to clone the IgG2 mRNA transcript by 3′ RACE. To address this question, a further Southern blotting experiment using the CH1 exon (high similarity between IgG1 and IgG2) as a probe, which showed that there were more than one γ genes in the guinea pig genome (Figure S7). Taken together, these data suggested that the guinea pig had two γ genes in its genome but only one was preferentially expressed.
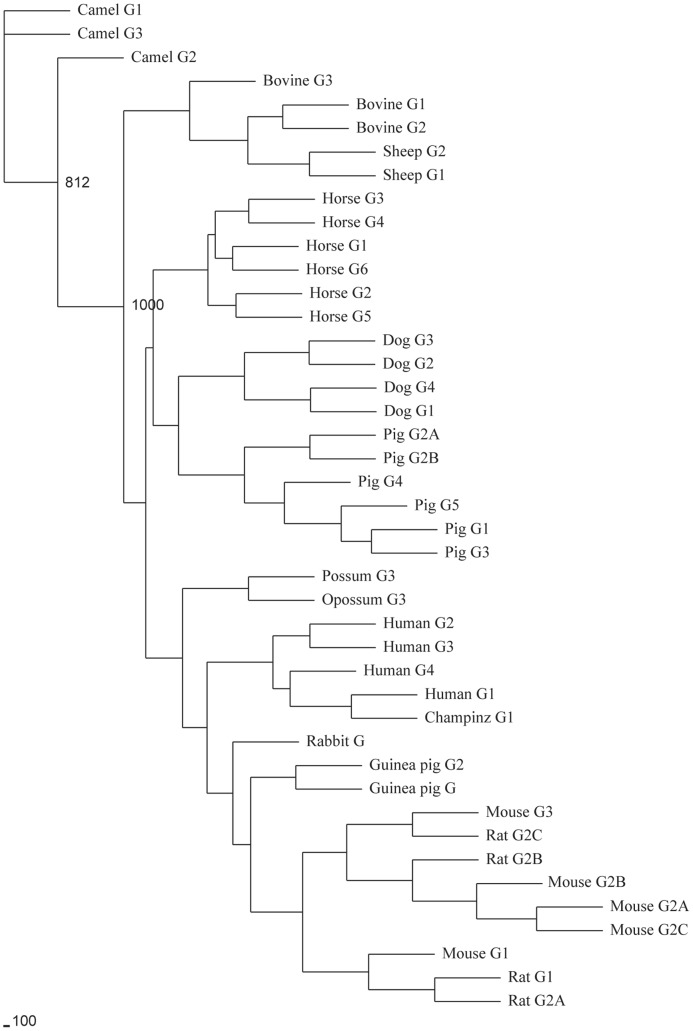
A phylogenetic tree constructed with CH2 and CH3 exons of IgG from different mammalian species revealed that the guinea pig IgG genes were clustered with rodents IgG genes (Figure 4). IgG2 and IgA also exhibit a hinge region, which is thought to have evolved by condensation of the CH2 exon in an ancestral isotype such as IgY in birds [25], [27]. The hinge region of IgA is encoded by the 5′ end of the CH2 exon, as observed in other eutherian mammals [28], [29], [30] (Figure S4–S5).
Figure 4. Phylogenetic analysis of mammalian immunoglobulin gamma genes.
The phylogenetic tree was constructed from the amino acid sequences of the CH2 and CH3 exons with various mammalian species immunoglobulin gamma gene. The guinea pig IgG genes were clustered with rodents IgG genes.
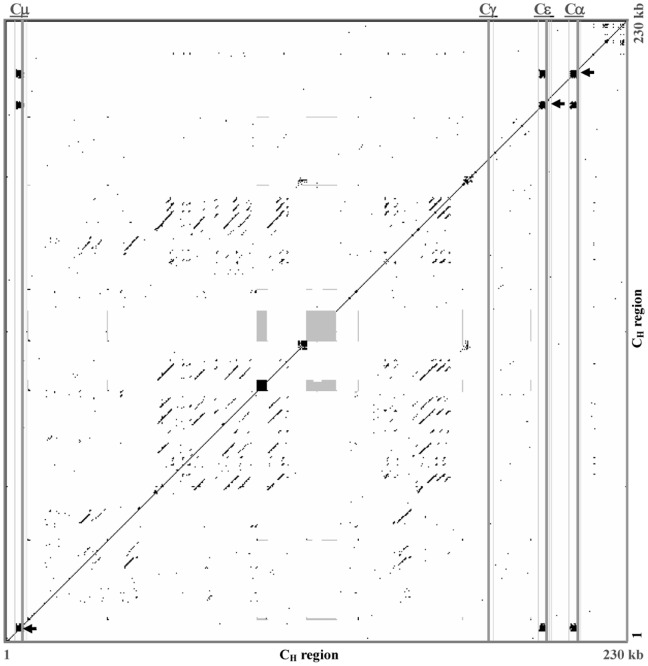
In mammals, especially in humans and mice, a pentameric tandem repeat sequence is found upstream of the heavy chain constant region, which acts as a switch or S region. S regions have previously been mapped and sequenced, and are relevant in Ig class switch recombination. Such characteristic tandem repeats were also found within the upstream Cμ, Cε and Cα gene sequences of the guinea pig. Three putative S regions span 2.2 kb to 3 kb, and exhibit similar repeats (GAGCT and GGGCT) to those observed in humans and mice. However, the characteristic sequence of the switch regions could not be identified within the γ gene, most likely due to sequence gaps. Dot matrix analysis of the guinea pig S region revealed substantial nucleotide similarity with those of humans and mice (Figure 5).
Figure 5. Dot plot comparison of the guinea pig CH (μ, ε, γ and α) region.
A dot matrix representing repetitive sequences of guinea pig CH (μ, ε, γ and α) genes. Switch regions are indicated by black-squared boxes and marked with arrowheads, and gaps are indicated by grey-squared boxes. Positions of CH genes are indicated as grey vertical lines. The dots represent homologies with a search length of 30 bp and maximum of 9 bp mismatches.
Analysis of the Guinea Pig VH Gene Segments
A total of 507 VH segments were identified in scaffolds 54 and 75 (Figure 1, Figure 2). Ninety-four of these appeared to be potentially functional because they contained leader exons (L), uninterrupted open reading frames (ORF), downstream RSS, and a V gene domain (framework regions and complementarity determining regions). The remaining 413 segments that contain either in-frame stop codons or are partial sequences were designated as pseudogenes (Figure 1, Figure 2 and Table S3). Given that gaps existed within the assembly, it is possible that as yet unidentified VH genes are also present in the guinea pig genome.
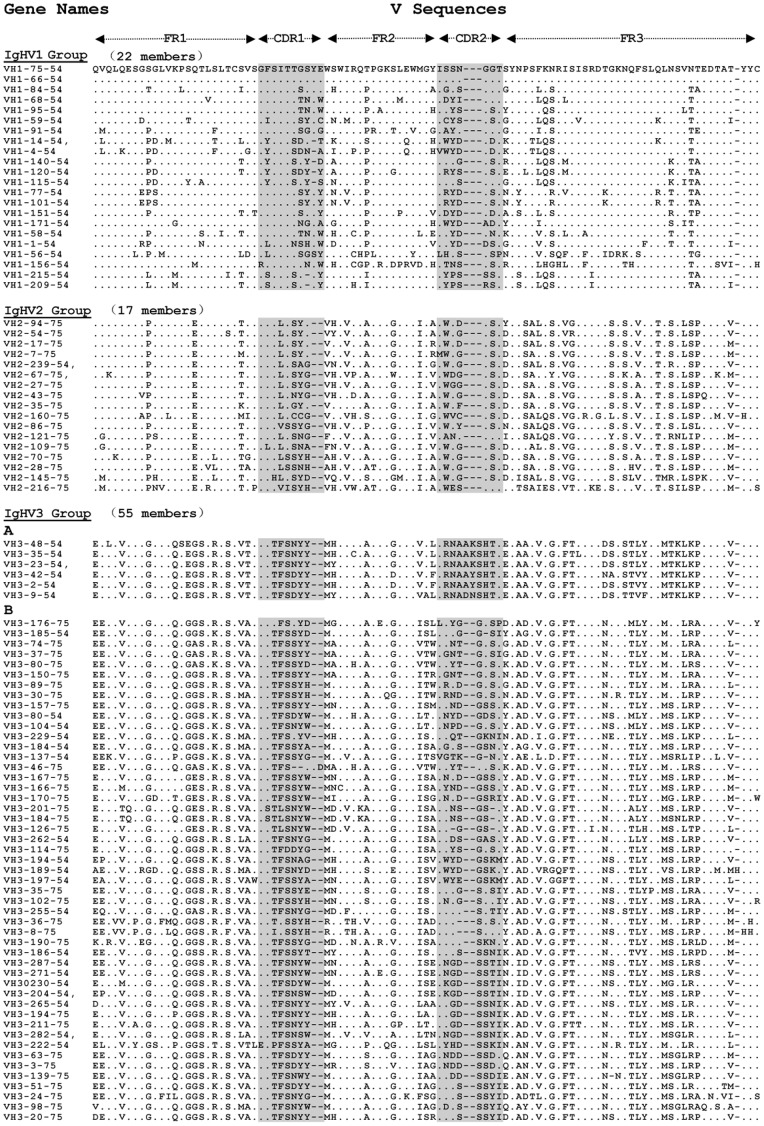
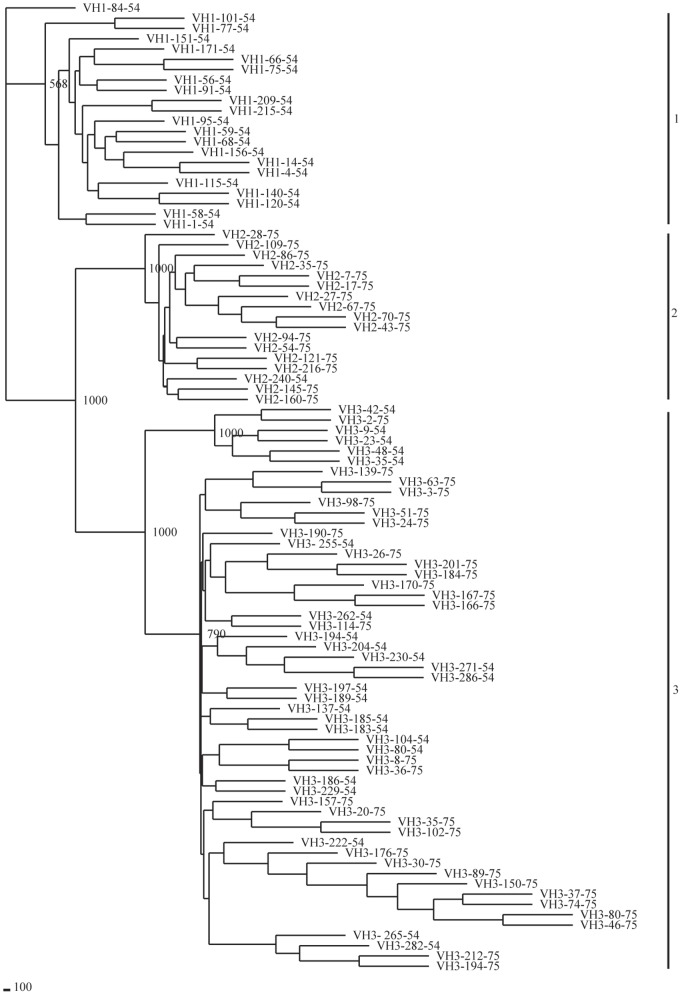
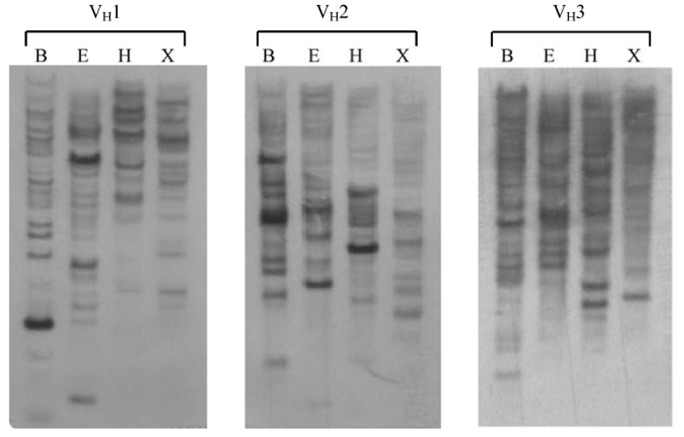
Phylogenetic analysis and multiple sequence alignments, including all functional guinea pig germline VH gene segments, revealed the VH gene families 1, 2 and 3, which were comprised of 22, 17 and 55 members, respectively (Figure 6, Figure 7). The largest family, VH3, could be further divided into two subfamilies (Figure 6, Figure 7). We also performed Southern blotting to verify the multiple numbers of VH genes of different families in the guinea pig genome (Figure 8).
Figure 6. The alignment of deduced amino acid sequences of guinea pig 94 functional VH genes.
The sequences of functional V genes were analyzed by Megalign software and BioEdit programe. The Designation of framework regions (FR) and complementarity determining regions (CDR) referred to IMGT numbering system, and the CDR region are indicated by grey-squared background. amino acid sequences that are the same as the top segment, VH1-75-54, are indicated with dots. Dashes mean gaps introduced to make the alignment.
Figure 7. Phylogenetic analysis of 94 guinea pig VH genes.
A phylogenetic tree of nucleotide sequences of 94 guinea pig potentially functional VH segments was constructed. The three identified VH gene families are labeled with Arabic numerals.
Figure 8. Southern blotting analysis of guinea pig genomic DNA.
Southern blotting analysis of the guinea pig heavy chain variable region genes. Genomic DNA was digested with BamH I (B), EcoR I (E), Hind III (H) and Xba I (X), and hybridized with probes for each of three guinea pig families.
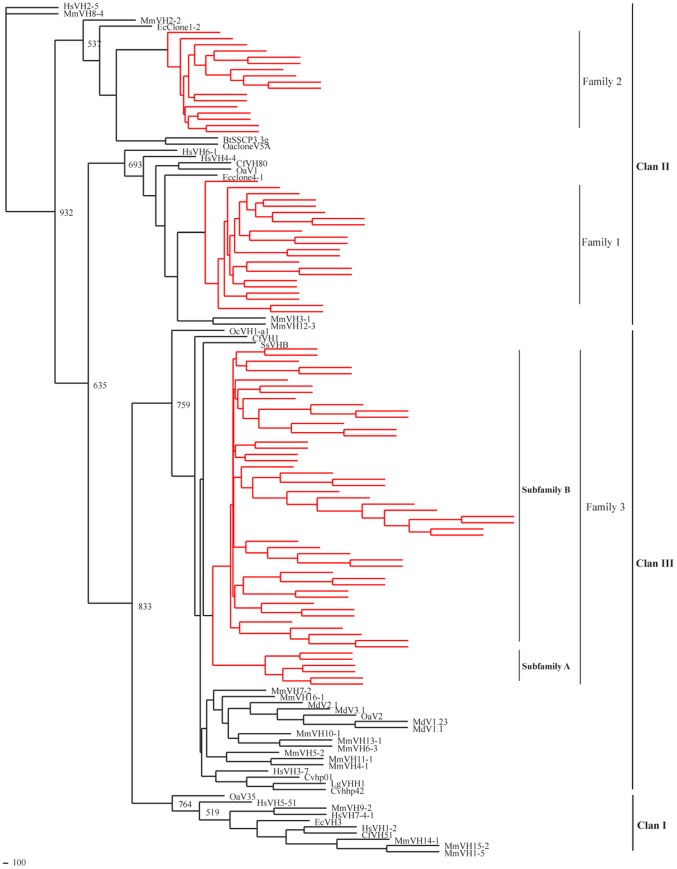
We chose all potentially functional guinea pig VH sequences of and VH sequences that represented previously reported gene families from other mammalian species to construct a neighbor-joining phylogenetic tree [31], [32] (Figure 9). The phylogenetic tree indicated that the mammalian VH gene families were classified into three clans [33]. The guinea pig VH families 1 and 2 belonged to clan II, and family 3 belonged to clan III.
Figure 9. Phylogenetic analysis of mammalian VH genes.
Guinea pig sequences are represented by red branches, other species sequences are represented by black branches. Three VH families of guinea pig were clustered respectively with other mammalian VH families, and the VH families of guinea pig belong to ClanII and ClanIII. The VH3 family could be divided into two subfamilies.
Analysis of the Guinea Pig DH and JH Gene Segments
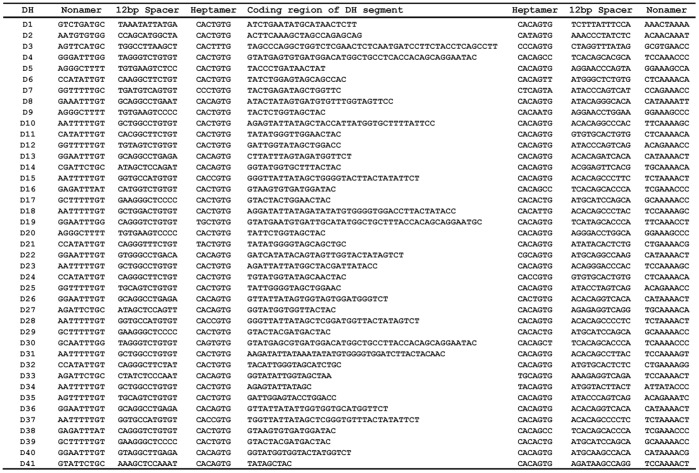
Approximately 504 kb downstream from the last VH segment (VH1-1), we identified 41 DH segments (i.e., DH1–DH41). They spanned a 660 kb region of DNA in scaffold 54 (Figure 1). Each DH segment was flanked by conserved RSS elements composed of heptamers and nonamers separated by 12 bp spacers and existed within at last one alternative reading frame (Figure 10 and Figure S8), suggesting that they are potentially functional.
Figure 10. The nucleotide sequences of DH genes.
DH coding region nucleotide sequences are represented together the RSS elements (nonamer and heptamer sequences).
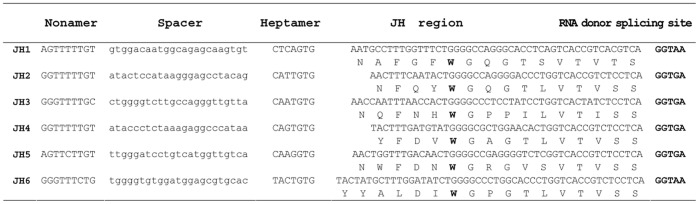
JH region contained six genes (designated JH1 to JH6) spanning approximately 2 kb. Each JH gene had an upstream RSS element with a 22–23 bp spacer, ORF, and a downstream RNA donor-splicing site at the 3′ end, suggesting that they are potentially functional (Figure 11).
Figure 11. The nucleotide sequences of JH genes.
JH nucleotide sequences are represented together the RSS elements (nonamer and heptamer sequences) and RNA donor splicing site.
Guinea Pig Igκ Locus
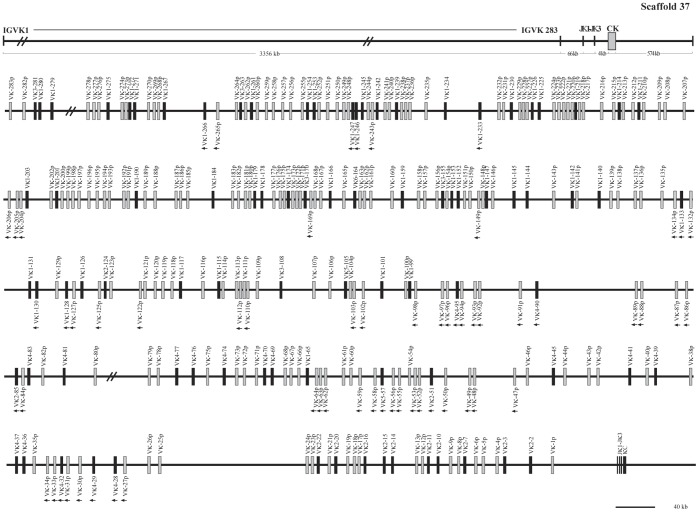
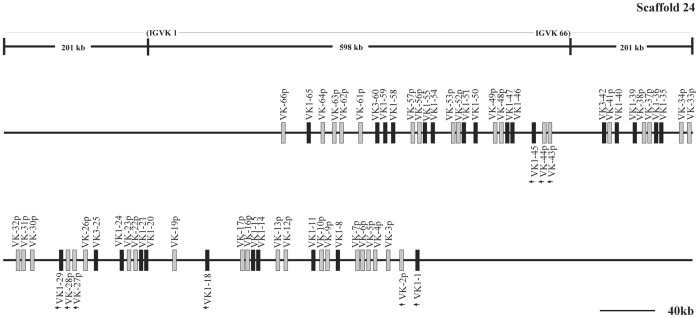
The guinea pig Igκ chain is located in scaffolds 37 and 24, and spans an approximately 4,029 kb genomic region (Figure 12, Figure 13 and Table S4). A total of 349 Vκ genes were identified. Further analysis revealed that 111 Vκ genes might be potentially functional genes, given that they contained an L sequence, ORF, RSS and V domain. The remaining 238 segments contain either in-frame stop codons or frameshifts, and are thus designated as pseudogenes. Based on sequence analysis, the 111 potentially functional Vκ genes were divided into six families (Vκ1–Vκ6), which contained 69, 15, 7, 17, 2, and 1 member/s, respectively (Figure 14). In addition, 222 Vκ genes were arranged in the same transcriptional orientation and exhibited downstream Jκ and Cκ, and 61 Vκ segments and a reverse transcriptional direction in scaffold 37. Downstream of the Vκ genes, three Jκ gene segments were identified that spanned 0.6 kb. Furthermore, approximately 4 kb downstream from the last Jκ, a single Cκ gene was identified.
Figure 12. The guinea pig Igκ locus in scaffold 37.
The guinea pig Igκ locus is in scaffold 37, the potentially functional Vκ gene segments are shown in filled bars, and pseudogenes are represented by open bars. The unidirectional arrowheads below Vκ gene segments on scaffold 37 indicate that their transcriptional direction is opposite to downstream Jκ segments. Double slashes indicate gaps >10 kb. The scale does not apply the upper frame diagram.
Figure 13. The guinea pig Igκ locus in scaffold 24.
The guinea pig Igκ locus is in scaffold 24, the potentially functional Vκ gene segments are shown in filled bars, and pseudogenes are represented by open bars. The unidirectional arrowheads below Vκ gene segments merely indicate a transcriptional direction different from that of the remaining Vκ gene segments in scaffold 24. The scale does not apply the upper frame diagram.
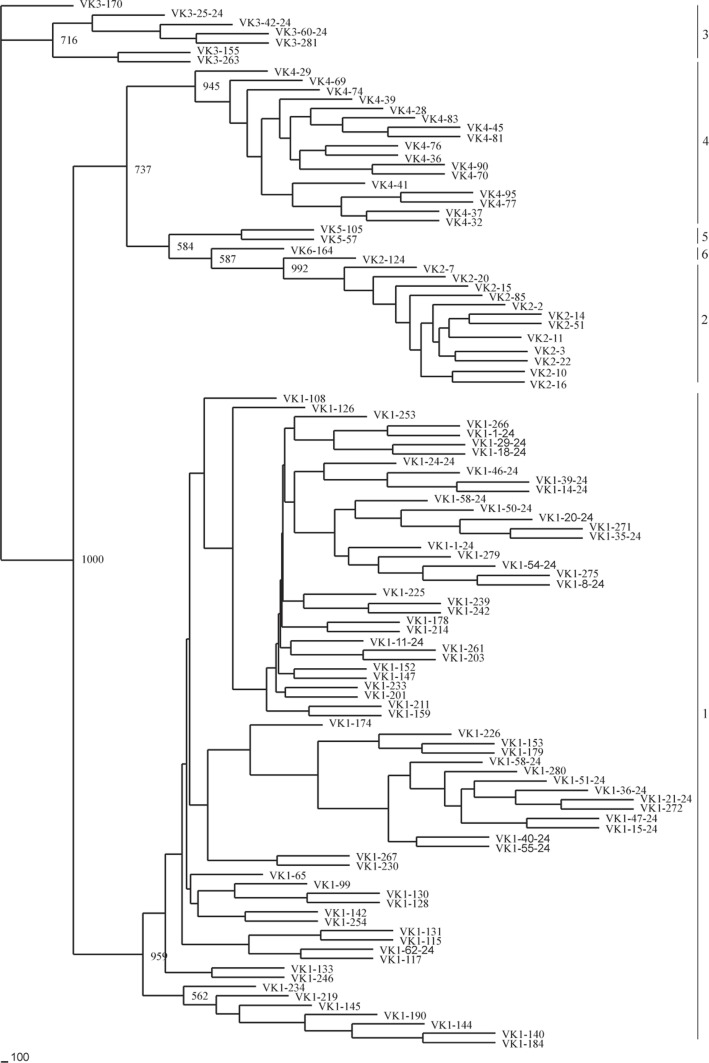
Figure 14. Phylogenetic analysis of the 111 guinea pig Vκ genes.
A phylogenetic tree of the nucleotide sequences of 111 guinea pig Vκ segments were constructed. The six Vκ gene families are labeled with Arabic numerals.
Guinea Pig Igλ Locus
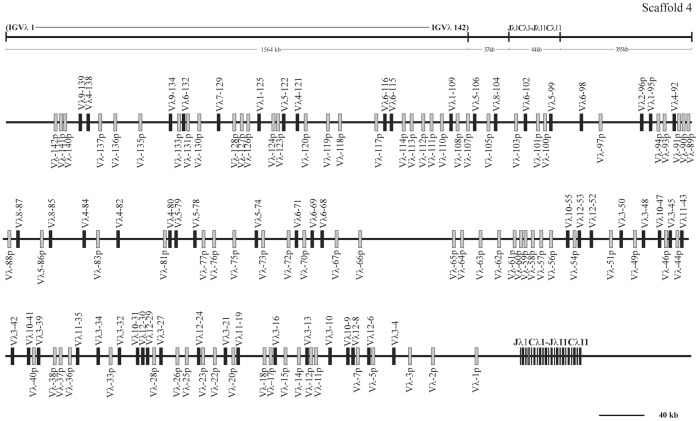
Guinea pig Igλ chain genes were identified in scaffold 4, and spanned an approximately 1,642 kb length (Figure 15, Table S5). Of 142 germline Vλ genes identified, 58 segments were categorized as potentially functional genes, and the remaining 84 were differentiated as pseudogenes. Based on sequence similarity analysis, the potentially functional guinea pig Vλ genes were assigned to twelve families (Figure 16), comprised of 3, 1, 13, 6, 6, 8, 1, 3, 2, 5, 3 and 7 member/s, respectively. In contrast to the Vκ genes, all the Vλ genes have the same transcriptional orientation as the downstream Jκ and Cκ regions. At the 3′ end of this scaffold, 11 J segments and C segments were organized in tandem and spanned 44 kb, while the Cλ2 exhibited less structural integrity owing to the presence of gaps. The sequence alignments of Jλ and Cλ gene segments are shown in Figure 17.
Figure 15. The guinea pig Igλ locus.
The guinea pig Igλ locus is in scaffold 4, the potentially functional Vλ gene segments are shown in filled bars, and pseudogenes are represented by open bars.
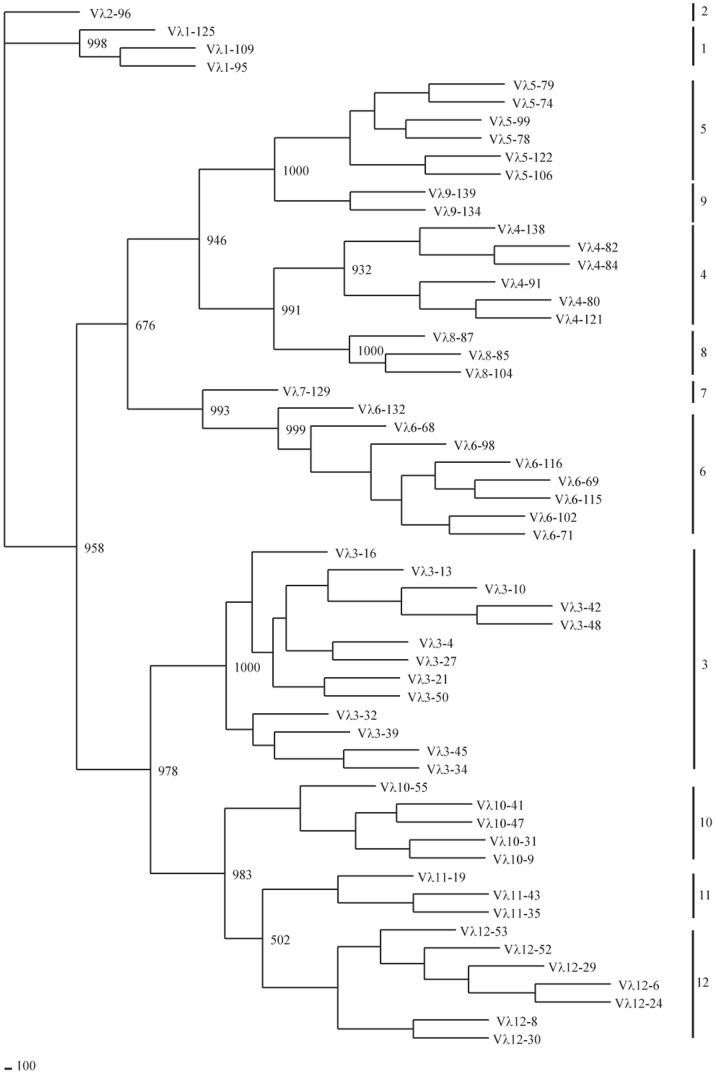
Figure 16. Phylogenetic tree analysis of the 58 guinea pig Vλ genes.
A phylogenetic tree of the nucleotide sequences of 58 guinea pig Vλ segments were constructed. The eleven Vλ gene families are labeled with Arabic numerals.
Figure 17. The alignment of amino acid sequences of J and C genes from guinea pig IgL chains.
A, Alignment of the deduced amino acid sequences of the three guinea pig Jκ gene segments. B, Alignment of the amino acid sequences of the Cκ proteins from human, mouse and rabbit. C, Alignment of the deduced amino acid sequences of the eleven guinea pig Jλ gene segments. D, Alignment of the amino acid sequences of the Cλ proteins from human, mouse and rabbit. Amino acid residues that are identical to the top counterpart in every panel are shown as dots; Gaps and missing data are indicated by hyphens.
Discussion
Rodents are a ubiquitous group of species worldwide, representing nearly half of all mammalian species, which evolved from a common ancestor shared with the lagomorphs approximately 62–100 million years ago [34], [35]. The classification of the guinea pig within the family Caviidae and genus Cavia is somewhat controversial because the origin of this rodent is poorly known, with current classification data mostly relying on fossils or genetic relationships [36], [37], [38], [39], [40], [41], [42], [43], [44]. We therefore analyzed the Ig genes sequences of the guinea pig, not only to better understand the immune system of this species, but also to provide data for comparative studies of mammalian Ig genes.
The IgH locus of mammals is arranged in a “translocon" configuration [32], [45], [46], [47], [48], [49], [50], [51], [52]. In the present study, we characterized the guinea pig Ig genes based on recently released genomic data and our experimental results. The guinea pig IgH locus in a configuration of VH (507)-DH (41)-JH (6)-Cμ-ψδ-Cγ2-Cε-Cα spanning at least 6,480 kb in two scaffolds may be largest in all mammalian species studied so far.
On the basis of sequence analysis, we identified a single μ, γ, ε and α gene within the guinea pig genome. We also found three fragments of δ gene in an opposite direction downstream from the μ gene. Due to sequence gaps, it is not certain if an additional functional δ gene also exists in this species. We have also tried to confirm the sequence and orientation of the δ gene fragments by genomic PCR to eliminate assembly error. The sequences of the δ gene fragments were verified. Because two genomic fragments (approximately two kb and six kb) between μ and δ gene can not be successfully amplified, so the orientation of the δ gene fragments remains a question.
Many placental mammals, such as human, cow, sheep, horse and dog, have a single functional δ gene. Except for a functional δ gene which consists of ten CH domains, a reverse δ pseudogene was previously observed in the platypus IgH locus [53], while in the elephant genome, only one Cδ3 remnant fragment was identified [51]. In camelids, Cδ3 exon appears to be highly mutated [54]. In the rabbit [24] and opossum (marsupial) [25], the δ gene has clearly been shown to be missing in their genomes, indicating that the δ gene might be not as essential as the μ gene in the humoral immunity.
IgG is an important antibody molecule, which is believed to have initially evolved 600 million years ago [55]. The structure of γ gene usually contains three CH domains and one hinge domain. Different IgG subclasses have been reported in the majority of mammalian species, ranging from one in the rabbit [56], two in sheep [57], three in cattle [58], four in human and rat [49], [59], five in mouse [60], six in pig [61], seven in horse [62], up to nine in elephant [51]. In the guinea pig, two IgG subclasses are identified, and they share about 73% amino acid similarity in CH domains. It has been postulated that different γ subclasses are derived from gene duplications of ancestral γ gene in mammals [63]. Three ancestral γ genes of mouse and rat can evolve multiple γ genes by gene duplications [64], [65], and the divergence of the γ genes of human depends on one ancestral γ gene and duplicated Cγ-Cγ-Cε-Cα fragments [66].
In different mammalian species, the number of VH genes and the ratio between VH functional genes and VH pseudogenes vary significantly, even between closely related species, like mice and rats. For example, there are 60 pseudogenes and 44 functional VH genes in humans, whereas in cows, only 6 pseudogenes and 11 functional genes have been identified [49], [67], [68]. The guinea pig germline VH repertoire contains at least 507 VH gene segments, which is the largest number in mammals studied to date. A large number of these germline VH genes may greatly contribute to guinea pig antibody diversity. Another notable feature is the number of guinea pig VH pseudogenes, which amount to 81% of the total VH genes (413 pseudogenes vs. 507 total genes). The high frequency of gene duplication in variable region generated multiple VH copies, many of which became pseudogenes due to genomic drift [68]. The VH pseudogenes are not truly nonfunctional in some species, for example, in rabbits, the pseudogenes usually are used to generate immunoglobulin diversity by gene conversion [67].
Guinea pig VH gene segments were also divided into three gene families (families 1, 2 and 3), which were orthologous to human VH families 4, 2 and 3, respectively. The guinea pig exhibited a multiple gene family group, as observed in dogs (three families) [69], humans (seven families) [70], [71], horses (seven families) [32], [72] and mice (sixteen families) [73], yet different from the single family group observed in sheep [57], rabbits [74], camels [75], swine [76] and cattle [72]. The guinea pig VH genes were also classified into different clans as in other mammalian species [50], [52], [67], [72], [74], [77], [78], [79]. Some reports have revealed that representative VH genes belong to all three VH clans in the human, mouse, cat and dog [67], [77]. This characteristic is different in cattle, horses and sheep [50], [52], [72], [78], [79], which have lost much of their ancestral repertoire, and the VH genes only belong to clan II. Pigs and rabbits only have clan III genes [74], [77], [79]. VH genes of guinea pig are distributed in clans II and III, and the majority of VH genes (family 3) are most closely related to human VH3 (clan III), which has been proposed as the ancestral VH gene family [78], [80]. These features are also found in swine and rabbits [74], [76].
The precise evolutionary relationship among mammalian lineages has not yet been resolved [81]. Results of the relevant study show that marsupials and monotremes were estimated to have separated from the common ancestor of present-day placental mammals more than 130 million years ago, while the major radiation of the placental mammals occurred approximately 70–120 million years ago [82], [83], [84], [85], [86]. Certain V gene families which descended from common ancestor genes are orthologues between nonplacental mammals and placental mammals. For example, platypus (Monotreme), dog and human share the same ancestral gene families (VH3 and VH4), While the American short-tailed opossum (Monodelphis domestica), swine, rabbit and human share ancestral VH3 family, and artiodactyl species share ancestral VH4 family. With an older evolutionary origin in present day mammals [86], platypus have two ancestral VH gene families, while other mammals share one or two ancestral VH gene families. These could be explained by an inactivation or loss of VH gene members in these species during evolution [25]. For new VH gene families in human or mouse, the divergence of VH genes probably occurred after speciation.
The ratio of functional Vκ and Vλ is variable within mammalian species, with the germline Vκ genes being more abundant than Vλ genes in humans (40 Vκ genes vs. 30 Vλ genes) and mice (Vκ genes vs. Vλ genes over 95%) [87]. This is also the case for the guinea pig, in which Vκ germline genes are more dominant than Vλ (84 functional Vκ genes vs. 58 functional Vλ genes). It has been proposed that the priority of use of the light-chain gene isotypes at the protein level may be connected with the overall number of V gene segments [87]. It is possible that the κ chain preponderates over the λ chain at the protein level in guinea pigs. Also, multiple pseudogenes exist in the guinea pig VH (413), Vκ (238), and Vλ (84) loci, which may contribute to the Ig diversity in guinea pigs, similar to other species [47], [88], [89].
In conclusion, we have reported the characterization and annotation of the guinea pig Ig loci genomic maps for the first time. This information, together with the characterization of the guinea pig Ig germline gene repertoire currently being undertaken, should lay the foundations for further studies into the differentiation and structure of mammalian Ig genes, including those found in guinea pigs.
Supporting Information
Multiple sequence alignments of guinea pig VH genes.
(DOC)
Multiple sequence alignments of guinea pig Vκ genes.
(DOC)
Multiple sequence alignments of guinea pig Vλ genes.
(DOC)
Guinea pig immunoglobulin heavy chain constant region encoding gene amino acid sequences. Four guinea pig encoding gene amino acid sequences of constant region (IgM, IgG, IgE and IgA) are cloned by 3′RACE.
(TIF)
Analysis results of the guinea pig Ig μ, γ, ε and α genes in scaffold 54. Four guinea pig constant region encoding genes of tansmembrane type for IgM, IgG, IgE and IgA were identified by bioinformatics analysis.
(TIF)
Alignment of the IgG amino acid sequences of the guinea pig. Alignment of IgG1, IgG2 and IgG squence of guinea pig. Dots indicate similar residues as in G1 and G, whereas dashes indicate gaps introduced for optimal alignment. The diference is represented by grey background between G and G1.
(TIF)
Southern blotting analysis of guinea pig genomic DNA. Southern blotting analysis of guinea pig heavy chain constant region IgG genes. The genomic DNA was digested with BamH I (B), EcoR I (E), Hind III (H) and Xba I (X), and hybridized with probes for IgG-CH1 sequence.
(TIF)
Guinea pig germline DH segments. The nonamers (9-mer) and heptamers (7-mer) are displayed. Heptamer components that are different from the consensus (5′: CACTGTG and 3′: CACAGTG) are shadowed. The deduced amino acids of all three reading frames of the coding region of D segments are shown, all the DH segments were attached by 12 bp spacer.
(RAR)
Primers were designed for amplify guinea pig four classes immunoglobulin M, E, A, and G genes.
(DOC)
GenBank accession numbers or references of the gene sequences from other species.
(RAR)
The guinea pig immunoglobulin heavy chain DNA segments location in scaffolds 54 and 75.
(RAR)
The guinea pig immunoglobulin κ light chain DNA segments location in scaffolds 37 and 24.
(RAR)
The guinea pig immunoglobulin λ light chain DNA segments location in scaffold 4.
(RAR)
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: The authors are supported by the National Science Fund for Distinguished Young Scholars (30725029), the Tai shan Scholar Foundation of Shan dong Province, the National Basic Research Program of China (973 Program-2010CB945300), Program for New Century Excellent Talents in University of China and the Swedish Research Council. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Padilla-Carlin DJ, McMurray DN, Hickey AJ. The guinea pig as a model of infectious diseases. Comp Med. 2008;58:324–340. [PMC free article] [PubMed] [Google Scholar]
- 2.Mizutani N, Nabe T, Shimazu M, Yoshino S, Kohno S. Effect of Ganoderma lucidum on pollen-induced biphasic nasal blockage in a guinea pig model of allergic rhinitis. Phytother Res. 2011;26:325–332. doi: 10.1002/ptr.3557. [DOI] [PubMed] [Google Scholar]
- 3.Ikeda R, Nakaya K, Oshima H, Oshima T, Kawase T, et al. Effect of aspiration of perilymph during stapes surgery on the endocochlear potential of guinea pig. Otolaryngol Head Neck Surg. 2011;145:801–805. doi: 10.1177/0194599811409516. [DOI] [PubMed] [Google Scholar]
- 4.Sturm EM, Schuligoi R, Konya V, Sturm GJ, Heinemann A. Inhibitory effect of prostaglandin I2 on bone marrow kinetics of eosinophils in the guinea pig. J Leukoc Biol. 2011;90:285–291. doi: 10.1189/jlb.0211087. [DOI] [PubMed] [Google Scholar]
- 5.Helling K, Wodarzcyk K, Brieger J, Schmidtmann I, Li H, et al. Doxycycline reduces nitric oxide production in guinea pig inner ears. Auris Nasus Larynx. 2011;38:671–677. doi: 10.1016/j.anl.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 6.Andersson KE, Soler R, Fullhase C. Rodent models for urodynamic investigation. Neurourol Urodyn. 2011;30:636–646. doi: 10.1002/nau.21108. [DOI] [PubMed] [Google Scholar]
- 7.Kajiwara D, Aoyagi H, Shigeno K, Togawa M, Tanaka K, et al. Role of hematopoietic prostaglandin D synthase in biphasic nasal obstruction in guinea pig model of experimental allergic rhinitis. Eur J Pharmacol. 2011;667:389–395. doi: 10.1016/j.ejphar.2011.05.041. [DOI] [PubMed] [Google Scholar]
- 8.Andrianova EP, Krementsugskaia SR, Lugovskaia NN, Mayorova TK, Borisov VV, et al. Foot and mouth disease virus polyepitope protein produced in bacteria and plants induces protective immunity in Guinea pigs. Biochemistry (Mosc) 2011;76:339–346. doi: 10.1134/s0006297911030072. [DOI] [PubMed] [Google Scholar]
- 9.Greenberg AS, Avila D, Hughes M, Hughes A, McKinney EC, et al. A new antigen receptor gene family that undergoes rearrangement and extensive somatic diversification in sharks. Nature. 1995;374:168–173. doi: 10.1038/374168a0. [DOI] [PubMed] [Google Scholar]
- 10.Hamers-Casterman C, Atarhouch T, Muyldermans S, Robinson G, Hamers C, et al. Naturally occurring antibodies devoid of light chains. Nature. 1993;363:446–448. doi: 10.1038/363446a0. [DOI] [PubMed] [Google Scholar]
- 11.Freedman MH. Limit, logic, and computation. Proc Natl Acad Sci U S A. 1998;95:95–97. doi: 10.1073/pnas.95.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang YA, Nie P, Luo HY, Wang YP, Sun YH, et al. Characterization of cDNA encoding immunoglobulin light chain of the mandarin fish (Siniperca chuatsi). Vet Immunol Immunopathol. 2003;95:81–90. doi: 10.1016/s0165-2427(03)00105-3. [DOI] [PubMed] [Google Scholar]
- 13.Thomas RK, Re D, Wolf J, Diehl V. Part I: Hodgkin’s lymphoma–molecular biology of Hodgkin and Reed-Sternberg cells. Lancet Oncol. 2004;5:11–18. doi: 10.1016/s1470-2045(03)01319-6. [DOI] [PubMed] [Google Scholar]
- 14.Qin T, Ren L, Hu X, Guo Y, Fei J, et al. Genomic organization of the immunoglobulin light chain gene loci in Xenopus tropicalis: evolutionary implications. Dev Comp Immunol. 2008;32:156–165. doi: 10.1016/j.dci.2007.05.007. [DOI] [PubMed] [Google Scholar]
- 15.Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. 1989.
- 16.Clewley JP, Arnold C. MEGALIGN. The multiple alignment module of LASERGENE. Methods Mol Biol. 1997;70:119–129. [PubMed] [Google Scholar]
- 17.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Higgins DG, Thompson JD, Gibson TJ. Using CLUSTAL for multiple sequence alignments. Methods Enzymol. 1996;266:383–402. doi: 10.1016/s0076-6879(96)66024-8. [DOI] [PubMed] [Google Scholar]
- 19.Hall T. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series. 1990;41:95–98. [Google Scholar]
- 20.Felsenstein J. PHYLIP (Phylogeny Inference Package) Ver. 3.67 [Computer software and manual]. Seattle, Washington: Department of Genome Sciences and Department of Biology, University of Washington. 2007.
- 21.Page RD. TreeView: an application to display phylogenetic trees on personal computers. Comput Appl Biosci. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- 22.Schroeder HW, Jr, Hillson JL, Perlmutter RM. Structure and evolution of mammalian VH families. Int Immunol. 1990;2:41–50. doi: 10.1093/intimm/2.1.41. [DOI] [PubMed] [Google Scholar]
- 23.Brodeur PH, Riblet R. The immunoglobulin heavy chain variable region (Igh-V) locus in the mouse. I. One hundred Igh-V genes comprise seven families of homologous genes. Eur J Immunol. 1984;14:922–930. doi: 10.1002/eji.1830141012. [DOI] [PubMed] [Google Scholar]
- 24.Lanning DK, Zhai SK, Knight KL. Analysis of the 3′ Cmu region of the rabbit Ig heavy chain locus. Gene. 2003;309:135–144. doi: 10.1016/s0378-1119(03)00500-6. [DOI] [PubMed] [Google Scholar]
- 25.Wang X, Olp JJ, Miller RD. On the genomics of immunoglobulins in the gray, short-tailed opossum Monodelphis domestica. Immunogenetics. 2009;61:581–596. doi: 10.1007/s00251-009-0385-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Phizackerley RP, Wishner BC, Bryant SH, Amzel LM, Lopez de Castro JA, et al. Three dimensional structure of the pFc’ fragment of guinea pig IgG1. Mol Immunol. 1979;16:841–850. doi: 10.1016/0161-5890(79)90083-x. [DOI] [PubMed] [Google Scholar]
- 27.Warr GW, Magor KE, Higgins DA. IgY: clues to the origins of modern antibodies. Immunol Today. 1995;16:392–398. doi: 10.1016/0167-5699(95)80008-5. [DOI] [PubMed] [Google Scholar]
- 28.Tucker PW, Slightom JL, Blattner FR. Mouse IgA heavy chain gene sequence: implications for evolution of immunoglobulin hinge axons. Proc Natl Acad Sci U S A. 1981;78:7684–7688. doi: 10.1073/pnas.78.12.7684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kawamura S, Saitou N, Ueda S. Concerted evolution of the primate immunoglobulin alpha-gene through gene conversion. J Biol Chem. 1992;267:7359–7367. [PubMed] [Google Scholar]
- 30.Osborne BA, Golde TE, Schwartz RL, Rudikoff S. Evolution of the IgA heavy chain gene in the genus Mus. Genetics. 1988;119:925–931. doi: 10.1093/genetics/119.4.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sitnikova T, Su C. Coevolution of immunoglobulin heavy- and light-chain variable-region gene families. Mol Biol Evol. 1998;15:617–625. doi: 10.1093/oxfordjournals.molbev.a025965. [DOI] [PubMed] [Google Scholar]
- 32.Almagro JC, Martinez L, Smith SL, Alagon A, Estevez J, et al. Analysis of the horse V(H) repertoire and comparison with the human IGHV germline genes, and sheep, cattle and pig V(H) sequences. Mol Immunol. 2006;43:1836–1845. doi: 10.1016/j.molimm.2005.10.017. [DOI] [PubMed] [Google Scholar]
- 33.Tomlinson IM, Walter G, Marks JD, Llewelyn MB, Winter G. The repertoire of human germline VH sequences reveals about fifty groups of VH segments with different hypervariable loops. J Mol Biol. 1992;227:776–798. doi: 10.1016/0022-2836(92)90223-7. [DOI] [PubMed] [Google Scholar]
- 34.Benton MJ, Donoghue PC. Paleontological evidence to date the tree of life. Mol Biol Evol. 2007;24:26–53. doi: 10.1093/molbev/msl150. [DOI] [PubMed] [Google Scholar]
- 35.Churakov G, Sadasivuni MK, Rosenbloom KR, Huchon D, Brosius J, et al. Rodent evolution: back to the root. Mol Biol Evol. 2010;27:1315–1326. doi: 10.1093/molbev/msq019. [DOI] [PubMed] [Google Scholar]
- 36.Horner DS, Lefkimmiatis K, Reyes A, Gissi C, Saccone C, et al. Phylogenetic analyses of complete mitochondrial genome sequences suggest a basal divergence of the enigmatic rodent Anomalurus. BMC Evol Biol. 2007;7:16. doi: 10.1186/1471-2148-7-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Graur D, Hide WA, Li WH. Is the guinea-pig a rodent? Nature. 1991;351:649–652. doi: 10.1038/351649a0. [DOI] [PubMed] [Google Scholar]
- 38.Li WH, Hide WA, Zharkikh A, Ma DP, Graur D. The molecular taxonomy and evolution of the guinea pig. J Hered. 1992;83:174–181. doi: 10.1093/oxfordjournals.jhered.a111188. [DOI] [PubMed] [Google Scholar]
- 39.D’Erchia AM, Gissi C, Pesole G, Saccone C, Arnason U. The guinea-pig is not a rodent. Nature. 1996;381:597–600. doi: 10.1038/381597a0. [DOI] [PubMed] [Google Scholar]
- 40.Reyes A, Pesole G, Saccone C. Long-branch attraction phenomenon and the impact of among-site rate variation on rodent phylogeny. Gene. 2000;259:177–187. doi: 10.1016/s0378-1119(00)00438-8. [DOI] [PubMed] [Google Scholar]
- 41.Cao Y, Adachi J, Yano T, Hasegawa M. Phylogenetic place of guinea pigs: no support of the rodent-polyphyly hypothesis from maximum-likelihood analyses of multiple protein sequences. Mol Biol Evol. 1994;11:593–604. doi: 10.1093/oxfordjournals.molbev.a040139. [DOI] [PubMed] [Google Scholar]
- 42.Kuma K, Miyata T. Mammalian phylogeny inferred from multiple protein data. Jpn J Genet. 1994;69:555–566. doi: 10.1266/jjg.69.555. [DOI] [PubMed] [Google Scholar]
- 43.Robinson-Rechavi M, Ponger L, Mouchiroud D. Nuclear gene LCAT supports rodent monophyly. Mol Biol Evol. 2000;17:1410–1412. doi: 10.1093/oxfordjournals.molbev.a026424. [DOI] [PubMed] [Google Scholar]
- 44.Lin YH, McLenachan PA, Gore AR, Phillips MJ, Ota R, et al. Four new mitochondrial genomes and the increased stability of evolutionary trees of mammals from improved taxon sampling. Mol Biol Evol. 2002;19:2060–2070. doi: 10.1093/oxfordjournals.molbev.a004031. [DOI] [PubMed] [Google Scholar]
- 45.Warr GW. The adaptive immune system of fish. Dev Biol Stand. 1997;90:15–21. [PubMed] [Google Scholar]
- 46.Bengten E, Wilson M, Miller N, Clem LW, Pilstrom L, et al. Immunoglobulin isotypes: structure, function, and genetics. Curr Top Microbiol Immunol. 2000;248:189–219. doi: 10.1007/978-3-642-59674-2_9. [DOI] [PubMed] [Google Scholar]
- 47.Vargas-Madrazo E, Almagro JC, Lara-Ochoa F. Structural repertoire in VH pseudogenes of immunoglobulins: comparison with human germline genes and human amino acid sequences. J Mol Biol. 1995;246:74–81. doi: 10.1006/jmbi.1994.0067. [DOI] [PubMed] [Google Scholar]
- 48.Chevillard C, Ozaki J, Herring CD, Riblet R. A three-megabase yeast artificial chromosome contig spanning the C57BL mouse Igh locus. J Immunol. 2002;168:5659–5666. doi: 10.4049/jimmunol.168.11.5659. [DOI] [PubMed] [Google Scholar]
- 49.Hendricks J, Terpstra P, Dammers PM, Somasundaram R, Visser A, et al. Organization of the variable region of the immunoglobulin heavy-chain gene locus of the rat. Immunogenetics. 2010;62:479–486. doi: 10.1007/s00251-010-0448-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Berens SJ, Wylie DE, Lopez OJ. Use of a single VH family and long CDR3s in the variable region of cattle Ig heavy chains. Int Immunol. 1997;9:189–199. doi: 10.1093/intimm/9.1.189. [DOI] [PubMed] [Google Scholar]
- 51.Guo Y, Bao Y, Wang H, Hu X, Zhao Z, et al. A preliminary analysis of the immunoglobulin genes in the African elephant (Loxodonta africana). PLoS One. 2011;6:e16889. doi: 10.1371/journal.pone.0016889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sun Y, Wang C, Wang Y, Zhang T, Ren L, et al. A comprehensive analysis of germline and expressed immunoglobulin repertoire in the horse. Dev Comp Immunol. 2010;34:1009–1020. doi: 10.1016/j.dci.2010.05.003. [DOI] [PubMed] [Google Scholar]
- 53.Gambon-Deza F, Sanchez-Espinel C, Magadan-Mompo S. The immunoglobulin heavy chain locus in the platypus (Ornithorhynchus anatinus). Mol Immunol. 2009;46:2515–2523. doi: 10.1016/j.molimm.2009.05.025. [DOI] [PubMed] [Google Scholar]
- 54.Achour I, Cavelier P, Tichit M, Bouchier C, Lafaye P, et al. Tetrameric and homodimeric camelid IgGs originate from the same IgH locus. J Immunol. 2008;181:2001–2009. doi: 10.4049/jimmunol.181.3.2001. [DOI] [PubMed] [Google Scholar]
- 55.Ellison JW, Berson BJ, Hood LE. The nucleotide sequence of a human immunoglobulin C gamma1 gene. Nucleic Acids Res. 1982;10:4071–4079. doi: 10.1093/nar/10.13.4071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Knight KL, Burnett RC, McNicholas JM. Organization and polymorphism of rabbit immunoglobulin heavy chain genes. J Immunol. 1985;134:1245–1250. [PubMed] [Google Scholar]
- 57.Dufour V, Malinge S, Nau F. The sheep Ig variable region repertoire consists of a single VH family. J Immunol. 1996;156:2163–2170. [PubMed] [Google Scholar]
- 58.Knight KL, Suter M, Becker RS. Genetic engineering of bovine Ig. Construction and characterization of hapten-binding bovine/murine chimeric IgE, IgA, IgG1, IgG2, and IgG3 molecules. J Immunol. 1988;140:3654–3659. [PubMed] [Google Scholar]
- 59.Flanagan JG, Lefranc MP, Rabbitts TH. Mechanisms of divergence and convergence of the human immunoglobulin alpha 1 and alpha 2 constant region gene sequences. Cell. 1984;36:681–688. doi: 10.1016/0092-8674(84)90348-9. [DOI] [PubMed] [Google Scholar]
- 60.Shimizu A, Takahashi N, Yaoita Y, Honjo T. Organization of the constant-region gene family of the mouse immunoglobulin heavy chain. Cell. 1982;28:499–506. doi: 10.1016/0092-8674(82)90204-5. [DOI] [PubMed] [Google Scholar]
- 61.Butler JE, Sun J, Wertz N, Sinkora M. Antibody repertoire development in swine. Dev Comp Immunol. 2006;30:199–221. doi: 10.1016/j.dci.2005.06.025. [DOI] [PubMed] [Google Scholar]
- 62.Wagner B, Miller DC, Lear TL, Antczak DF. The complete map of the Ig heavy chain constant gene region reveals evidence for seven IgG isotypes and for IgD in the horse. J Immunol. 2004;173:3230–3242. doi: 10.4049/jimmunol.173.5.3230. [DOI] [PubMed] [Google Scholar]
- 63.Honjo T, Matsuda F. Immunoglobulin heavy chain loci of mouse and man. Immunoglobulin Genes. Academic Press New York: Pp. 1995. pp. 145–171.
- 64.Morgado MG, Cam P, Gris-Liebe C, Cazenave PA, Jouvin-Marche E. Further evidence that BALB/c and C57BL/6 gamma 2a genes originate from two distinct isotypes. EMBO J. 1989;8:3245–3251. doi: 10.1002/j.1460-2075.1989.tb08484.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bruggemann M. Evolution of the rat immunoglobulin gamma heavy-chain gene family. Gene. 1988;74:473–482. doi: 10.1016/0378-1119(88)90180-1. [DOI] [PubMed] [Google Scholar]
- 66.Bensmana M, Huck S, Lefranc G, Lefranc MP. The human immunoglobulin pseudo-gamma IGHGP gene shows no major structural defect. Nucleic Acids Res. 1988;16:3108. doi: 10.1093/nar/16.7.3108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Das S, Nozawa M, Klein J, Nei M. Evolutionary dynamics of the immunoglobulin heavy chain variable region genes in vertebrates. Immunogenetics. 2008;60:47–55. doi: 10.1007/s00251-007-0270-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nei M. The new mutation theory of phenotypic evolution. Proc Natl Acad Sci U S A. 2007;104:12235–12242. doi: 10.1073/pnas.0703349104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bao Y, Guo Y, Xiao S, Zhao Z. Molecular characterization of the VH repertoire in Canis familiaris. Vet Immunol Immunopathol. 2010;137:64–75. doi: 10.1016/j.vetimm.2010.04.011. [DOI] [PubMed] [Google Scholar]
- 70.Berman JE, Mellis SJ, Pollock R, Smith CL, Suh H, et al. Content and organization of the human Ig VH locus: definition of three new VH families and linkage to the Ig CH locus. EMBO J. 1988;7:727–738. doi: 10.1002/j.1460-2075.1988.tb02869.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.van Dijk KW, Mortari F, Kirkham PM, Schroeder HW, Jr, Milner EC. The human immunoglobulin VH7 gene family consists of a small, polymorphic group of six to eight gene segments dispersed throughout the VH locus. Eur J Immunol. 1993;23:832–839. doi: 10.1002/eji.1830230410. [DOI] [PubMed] [Google Scholar]
- 72.Sinclair MC, Gilchrist J, Aitken R. Bovine IgG repertoire is dominated by a single diversified VH gene family. J Immunol. 1997;159:3883–3889. [PubMed] [Google Scholar]
- 73.Johnston CM, Wood AL, Bolland DJ, Corcoran AE. Complete sequence assembly and characterization of the C57BL/6 mouse Ig heavy chain V region. J Immunol. 2006;176:4221–4234. doi: 10.4049/jimmunol.176.7.4221. [DOI] [PubMed] [Google Scholar]
- 74.Currier SJ, Gallarda JL, Knight KL. Partial molecular genetic map of the rabbit VH chromosomal region. J Immunol. 1988;140:1651–1659. [PubMed] [Google Scholar]
- 75.Nguyen VK, Muyldermans S, Hamers R. The specific variable domain of camel heavy-chain antibodies is encoded in the germline. J Mol Biol. 1998;275:413–418. doi: 10.1006/jmbi.1997.1477. [DOI] [PubMed] [Google Scholar]
- 76.Sun J, Kacskovics I, Brown WR, Butler JE. Expressed swine VH genes belong to a small VH gene family homologous to human VHIII. J Immunol. 1994;153:5618–5627. [PubMed] [Google Scholar]
- 77.Butler JE. Immunoglobulin gene organization and the mechanism of repertoire development. Scand J Immunol. 1997;45:455–462. doi: 10.1046/j.1365-3083.1997.d01-423.x. [DOI] [PubMed] [Google Scholar]
- 78.Charlton KA, Moyle S, Porter AJ, Harris WJ. Analysis of the diversity of a sheep antibody repertoire as revealed from a bacteriophage display library. J Immunol. 2000;164:6221–6229. doi: 10.4049/jimmunol.164.12.6221. [DOI] [PubMed] [Google Scholar]
- 79.Baker ML, Tachedjian M, Wang LF. Immunoglobulin heavy chain diversity in Pteropid bats: evidence for a diverse and highly specific antigen binding repertoire. Immunogenetics. 2010;62:173–184. doi: 10.1007/s00251-010-0425-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tutter A, Riblet R. Conservation of an immunoglobulin variable-region gene family indicates a specific, noncoding function. Proc Natl Acad Sci U S A. 1989;86:7460–7464. doi: 10.1073/pnas.86.19.7460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Johansson J, Aveskogh M, Munday B, Hellman L. Heavy chain V region diversity in the duck-billed platypus (Ornithorhynchus anatinus): long and highly variable complementarity-determining region 3 compensates for limited germline diversity. J Immunol. 2002;168:5155–5162. doi: 10.4049/jimmunol.168.10.5155. [DOI] [PubMed] [Google Scholar]
- 82.Kullander K, Carlson B, Hallbook F. Molecular phylogeny and evolution of the neurotrophins from monotremes and marsupials. J Mol Evol. 1997;45:311–321. doi: 10.1007/pl00006235. [DOI] [PubMed] [Google Scholar]
- 83.Janke A, Xu X, Arnason U. The complete mitochondrial genome of the wallaroo (Macropus robustus) and the phylogenetic relationship among Monotremata, Marsupialia, and Eutheria. Proc Natl Acad Sci U S A. 1997;94:1276–1281. doi: 10.1073/pnas.94.4.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Air GM, Thompson EO. Studies on marsupial proteins. IV. Amino acid sequence of myoglobin from the red kangaroo, Megaleia rufa. Aust J Biol Sci. 1971;24:75–95. doi: 10.1071/bi9710075. [DOI] [PubMed] [Google Scholar]
- 85.Retief JD, Winkfein RJ, Dixon GH. Evolution of the monotremes. The sequences of the protamine P1 genes of platypus and echidna. Eur J Biochem. 1993;218:457–461. doi: 10.1111/j.1432-1033.1993.tb18396.x. [DOI] [PubMed] [Google Scholar]
- 86.Janke A, Gemmell NJ, Feldmaier-Fuchs G, von Haeseler A, Paabo S. The mitochondrial genome of a monotreme–the platypus (Ornithorhynchus anatinus). J Mol Evol. 1996;42:153–159. doi: 10.1007/BF02198841. [DOI] [PubMed] [Google Scholar]
- 87.Almagro JC, Hernandez I, Ramirez MC, Vargas-Madrazo E. Structural differences between the repertoires of mouse and human germline genes and their evolutionary implications. Immunogenetics. 1998;47:355–363. doi: 10.1007/s002510050370. [DOI] [PubMed] [Google Scholar]
- 88.Lucier MR, Thompson RE, Waire J, Lin AW, Osborne BA, et al. Multiple sites of V lambda diversification in cattle. J Immunol. 1998;161:5438–5444. [PubMed] [Google Scholar]
- 89.Haino M, Hayashida H, Miyata T, Shin EK, Matsuda F, et al. Comparison and evolution of human immunoglobulin VH segments located in the 3′ 0.8-megabase region. Evidence for unidirectional transfer of segmental gene sequences. J Biol Chem. 1994;269:2619–2626. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Multiple sequence alignments of guinea pig VH genes.
(DOC)
Multiple sequence alignments of guinea pig Vκ genes.
(DOC)
Multiple sequence alignments of guinea pig Vλ genes.
(DOC)
Guinea pig immunoglobulin heavy chain constant region encoding gene amino acid sequences. Four guinea pig encoding gene amino acid sequences of constant region (IgM, IgG, IgE and IgA) are cloned by 3′RACE.
(TIF)
Analysis results of the guinea pig Ig μ, γ, ε and α genes in scaffold 54. Four guinea pig constant region encoding genes of tansmembrane type for IgM, IgG, IgE and IgA were identified by bioinformatics analysis.
(TIF)
Alignment of the IgG amino acid sequences of the guinea pig. Alignment of IgG1, IgG2 and IgG squence of guinea pig. Dots indicate similar residues as in G1 and G, whereas dashes indicate gaps introduced for optimal alignment. The diference is represented by grey background between G and G1.
(TIF)
Southern blotting analysis of guinea pig genomic DNA. Southern blotting analysis of guinea pig heavy chain constant region IgG genes. The genomic DNA was digested with BamH I (B), EcoR I (E), Hind III (H) and Xba I (X), and hybridized with probes for IgG-CH1 sequence.
(TIF)
Guinea pig germline DH segments. The nonamers (9-mer) and heptamers (7-mer) are displayed. Heptamer components that are different from the consensus (5′: CACTGTG and 3′: CACAGTG) are shadowed. The deduced amino acids of all three reading frames of the coding region of D segments are shown, all the DH segments were attached by 12 bp spacer.
(RAR)
Primers were designed for amplify guinea pig four classes immunoglobulin M, E, A, and G genes.
(DOC)
GenBank accession numbers or references of the gene sequences from other species.
(RAR)
The guinea pig immunoglobulin heavy chain DNA segments location in scaffolds 54 and 75.
(RAR)
The guinea pig immunoglobulin κ light chain DNA segments location in scaffolds 37 and 24.
(RAR)
The guinea pig immunoglobulin λ light chain DNA segments location in scaffold 4.
(RAR)