Abstract
Background
Thlaspi caerulescens is a natural selected heavy metal hyperaccumulator that can not only tolerate but also accumulate extremely high levels of heavy metals in the shoots. Thus, to identify the transportors involved in metal long-distance transportation is very important for understanding the mechanism of heavy metal accumulation in this hyperaccumulator.
Methodology/Principal Findings
We cloned and characterized a novel gene TcOPT3 of OPT family from T. caerulescens. TcOPT3 was pronouncedly expressed in aerial parts, including stem and leaf. Moreover, in situ hybridization analyses showed that TcOPT3 expressed in the plant vascular systems, especially in the pericycle cells that may be involved in the long-distance transportation. The expression of TcOPT3 was highly induced by iron (Fe) and zinc (Zn) deficiency, especially in the stem and leaf. Sub-cellular localization showed that TcOPT3 was a plasma membrane-localized protein. Furthermore, heterogonous expression of TcOPT3 by mutant yeast (Saccharomyces cerevisiae) complementation experiments demonstrated that TcOPT3 could transport Fe2+ and Zn2+. Moreover, expression of TcOPT3 in yeast increased metal (Fe, Zn, Cu and Cd) accumulation and resulted in an increased sensitivity to cadmium (Cd) and copper (Cu).
Conclusions
Our data demonstrated that TcOPT3 might encode an Fe/Zn/Cd/Cu influx transporter with broad-substrate. This is the first report showing that TcOPT3 may be involved in metal long-distance transportation and contribute to the heavy metal hyperaccumulation.
Introduction
Metal hyperaccumulators can not only tolerate high concentration of heavy metals in the soils but also take them up actively and accumulate and distribute them to appropriate tissues at extreme high levels, thus make them very attractive for the remediation of heavy metal polluted soils [1]. A large amount of different metal hyperaccumulators have been recognized in different regions all over the world [2], among them, Thlaspi caerulescens, a Cd/Zn/Ni hyperaccumulator, has been used as a model plant to study the physiological and molecular mechanisms of heavy metal hyperaccumulation [3], [4].
The efficient metal loading and unloading ability in the vascular systems was regarded as the key step of hyperaccumulating process for hyperaccumulators [5]. Nonetheless, knowledge about the mechanisms and proteins involved in transporting heavy metals from soil via the roots and stems into their storage sites is much limited. Transporter families, including ZIP (ZRT/IRT like Protein), Nramp (Natural Resistance and Macrophage Protein), MTP (Metal Tolerance Protein), HMA (Heavy Metal transporting P-type ATPase), ABC-type (ATP-binding cassette), COPT family CTR/COPT (high-affinity Cu transporters) as well as YSL (Yellow strip1-Like transporters) families have been porved to involve in transit metal movements, they either act at the plasma membrane (PM) to transport metals into cytoplasm or redistribute metals from intracellular compartments into the cytoplasm [6], [7], [8], [9]. Besides the above transporters families, novel transporters related to metal homeostasis are continuously under discovery, oligopeptide transporter (OPT) family is among those being focused recently [10], [11], [12]. OPT family is one of three families responsible for peptide transport; the other two are the ATP-binding cassette (ABC-type) transporters superfamily and the peptide transporter (PTR) superfamily [10]. Peptides, existing abundantly in xylem and phloem saps, are important for plant growth, development and signaling. Transport of peptides is taken as a more efficient means of nitrogen distribution than transport of individual amino acids, implying the important role of peptide transporters in long-distance transport of nutrients [10]. Besides the peptides, inorganic nutrients and transit metals have been reported as the substrates of peptide transporters in the recent studies. For intances, the ABC transporters AtMRP3 [13], AtATM3 [14] and AtPDR8 [15] are proposed to involve in heavy metal (Cd, Pb) resistance and movement.
The first OPT family is identified in the pathogenic yeast Candida albicans [16] and subsequently in Schizosaccharomyces pombe [17] and Saccharomyces cerevisiae [18]. Later, it is also found in bacteria, plants, and archaea [11], but still not in animals. OPT family is grouped into two distinct subfamilies, the yellow stripe (YS) clade and the peptide transport (PT) clade [11], [12]. The function of YS1-like (YSL) transporter on transportation of metal-complex has been documented well in planta. For example, YSLs mediate long-distance transport of specific Fe species [19], [20]. ZmYS1 cloned from Zea mays is the founding member of YSL family, which functions in root Fe-phytosiderophore (Fe-PS) uptake from the soil [21]. HvYS1 encoding a YSL transporter is a specific transporter for iron (III)-PS in barley roots [22]. Since non-Poaceae species do not synthesize phytosiderophores, the substrates of YSL transporters in dicotyledonous plant are suggested to be Fe(II)/Fe(III)-nicotianamine (Fe-NA) functioning in the long-distance transporting system [23], [24], [25]. NA is a chelator for several micronutrient metals and involves in the movement of micronutrients and heavy metals throughout the plant [26]. Three YSL genes cloned from T. caerulescens express at higher levels as compared with their Arabidopsis thaliana homologs with distinct patterns. TcYSL3 was further confirmed as a Fe/Ni-NA influx transporter [19]. Functional analysis in yeast demonstrated that TcYSL3 can transport both Ni-NA and Fe-NA complexes into yeast, indicating that TcYSL3 may be involved in the long-distance translocation of Ni in T. caerulescens.
In Arabidopsis, nine putative OPT orthologues (AtOPT1 to AtOPT9) have been identified [27], [28]. OPT transporters predominantly recognize tetrapeptides and pentapeptides, thus they are also called peptide transporters, but this is challenged recently as AtOPTs have been proved to play a role in metal homeostasis and movements. Transcript of AtOPT2 is induced in root by iron and zinc deficiency, and that of AtOPT3 by iron, copper and manganese deficiency [28], [29]. AtOPT3 is further proved to involve in the movement of iron to the developing seeds [30]. Moreover, both AtOPT6 and AtOPT7 can transport cadmium or cadmium-glutathione conjugates when heterologously expressed in yeast [31], [32]. All these studies indicate that OPT family might play important roles in metal uptake and transportation.
In the present study, we isolated a member of the OPT family from a metal hyperaccumulator, Thlaspi caerulescens, named TcOPT3. We characterized its expression in different tissues, and conducted heterologous expression in yeast to investigate the role of TcOPT3 in metal uptake. We demonstrated that TcOPT3 was involved in Fe/Zn/Cd/Cu transportation and it should be a very important component in the metal long-distance transport system, and contribute to heavy metal hyperaccumulation in the shoots.
Results
Identification of TcOPT3 Gene
Degenerated primers were designed from the most conserved regions according to the AtOPT3, BjGT1 and ZmGT mRNA sequence. We isolated the full-length cDNA from Thlaspi caerulescens by RACE PCR, and designated it as TcOPT3 (Genebank accession no. HQ69984). The coding region of TcOPT3 was 2211 bp, and a corresponding 737 amino-acid sequence was predicted.
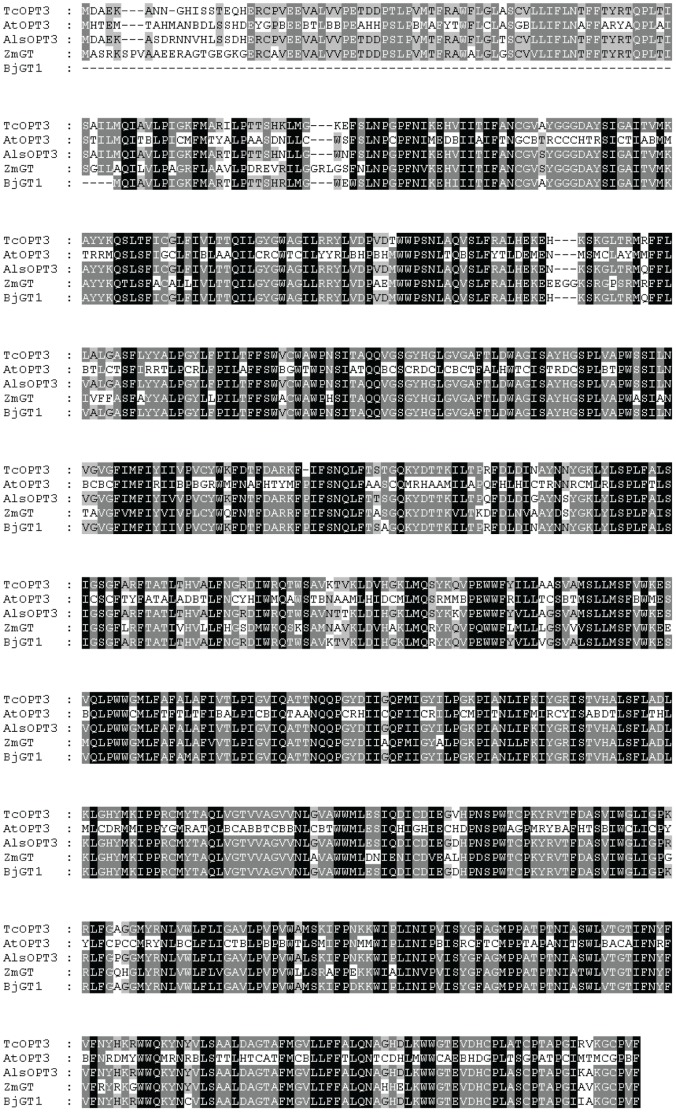
TcOPT3 exhibited 79% identity with its homologous AtOPT3, and the deduced protein TcOPT3 showed 95% identity with AtOPT3 (Fig. 1). It contained 14 putative transmembrane domains (TM I-TM XIV, Fig. 1) and two highly conserved motifs (NPG motif and KIPPR motif, Fig. 1) of OPT family, thus TcOPT3 is likely to be localized to membrane and owns the structure of a transporter protein.
Figure 1. Sequence analysis of TcOPT3.
Sequence alignment among the TcOPT3, AtOPT3 ATOPT3, ZmGT and BjGT1. CLUSTAL W (version 1.8) alignment of deduced amino acid sequences from the OPTs. Amino acids are numbered from the initiator ATG. Black-shaded areas represent the consensus, dark-gray-shaded areas represent identical amino acids, and light-gray-shaded areas represent similar amino acids. The putative transmembrane (TM) domains of the TcOPT3 were determined by the TMHMM algorithm. The predicted transmembrane membrane spanning domains are shown as lines above the sequences, and numbered TM I–TM XIV respectively. The bars under the sequence show the location of the two conserved motifs (NPG and KIPPR motifs). Tc, T. caerulescens; At, Arabidopsis thaliana; Als, Arabidopsis lyrata subsp.; Zm, Zea mays; Bj, Brassica juncea. Accession numbers are: HQ699884 (TcOPT3), NP_567493 (AtOPT3), XP_002868139 (AlsOPT3), ACL82964 (ZmGT), CAD91127.1 (BjGT1).
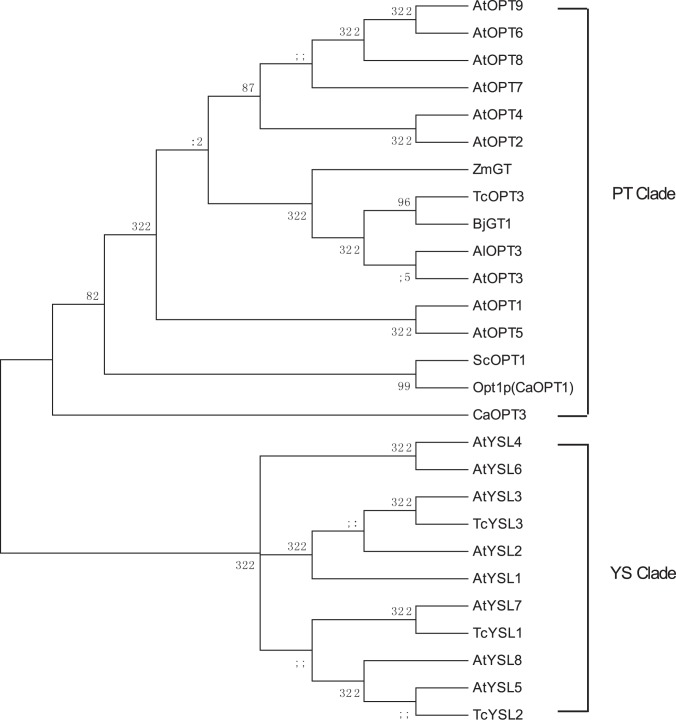
From the rooted phylogenetic tree to compare TcOPTs with OPTs and YSLs in A. thaliana and the related species. We can see that TcOPT3 belongs to the PT clade, moreover, it clusters together in one branch with the OPTs from T.caerulescens, B. juncea, A. lyrata and A. thaliana and is most closely related to BjGT1 (Fig. 2).
Figure 2. Phylogenetic tree of OPT gene transporters based on the amino acid sequences.
Dendogram showing sequence comparisons of several known members of the PT family from different species. Analysis was performed using the CLUSTAL X method in MEGA (4.0) using Neighbor-Joining method (Tamura K et al., 2007). Accession numbers are as follows: AtOPT1, NP_200404.1 GI: 15241078; AtOPT2, NP_172464.1 GI15218331; AtOPT3, NP_567493.5 GI:240255930; AtOPT4, NP_201246.1 GI:15237689; AtOPT5, NP_194389.1 GI:15236800; AtOPT6, NP_194503.1 GI:15234254; AtOPT7, NP_192815.1 GI:15236912; AtOPT8, NP_564525.1 GI:18402162; AtOPT9, NP_200163.1 GI:15238761; AlOPT3, XP_002868139.1 GI:297800510; AtYSL1, NP_567694.2 GI:79484897; AtYSL2, NP_197826.2 GI:79518939; AtYSL3, NP_200167.2 GI:145359208; AtYSL4, NP_198916.2 GI:42568235; AtYSL5, NP_566584.1 GI:18401590; AtYSL6, NP_566806.1 GI:18405202; AtYSL7, NP_176750.1 GI:15218799; AtYSL8, NP_564525.1 GI:18402162; TcYSL1, ABB76761.1 GI:82468791; TcYSL2, ABB76762.1 GI:82468793; TcYSL3, ABB76763.1 GI:82468795; CaOPT1, AAB69628.1 GI:2367386; CaOPT3, ABD17824.1 GI:87045965; ScOPT1, NP_012323.1 GI:6322249; ZmGT, ACL82964.1 GI:220901863 BjGT1, CAD91127.1 GI:30722286.
Tissue- and Organ- Specific Analysis of TcOPT3
The expression of TcOPT3 was investigated by real-time RT-PCR, where ubiquitin-conjugating enzyme gene (UBQ10) was used as a control. The transcript level of TcOPT3 mRNA was 2-fold more in the leaf and stem tissues than that in the root (Fig. 3).
Figure 3. Tissue-specific analysis of the TcOPT3.
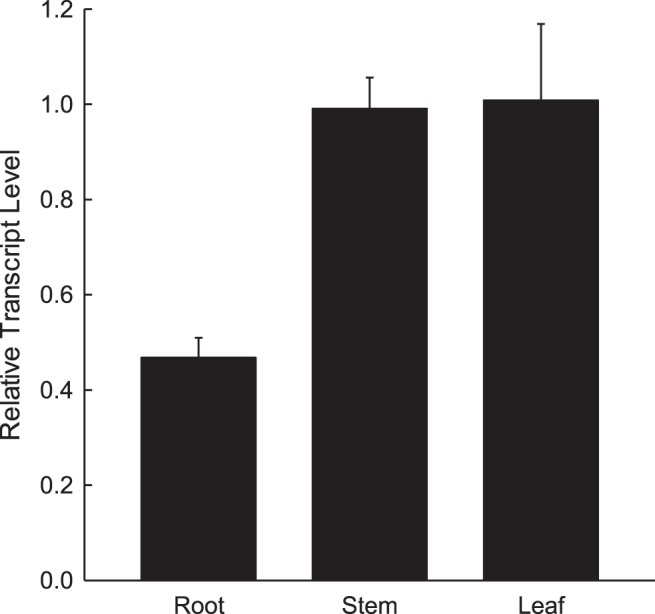
Real-time RT-PCR expression analysis of the TcOPT3 gene expression in roots (R), leaves (L) and stems (S). The ΔCp values were calculated as follows: CP of target gene (TcOPT3) – CP of constitutive control gene (ubiquitin-conjugating enzyme), where the CP value is the fractional cycle number of crossing point (CP). The ΔCP values represent the mean of three technical replicates (±SD) of one experiment representative of three independent experiments. Relative transcript levels (RTL) were calculated as follows: RTL = 2−ΔCP.
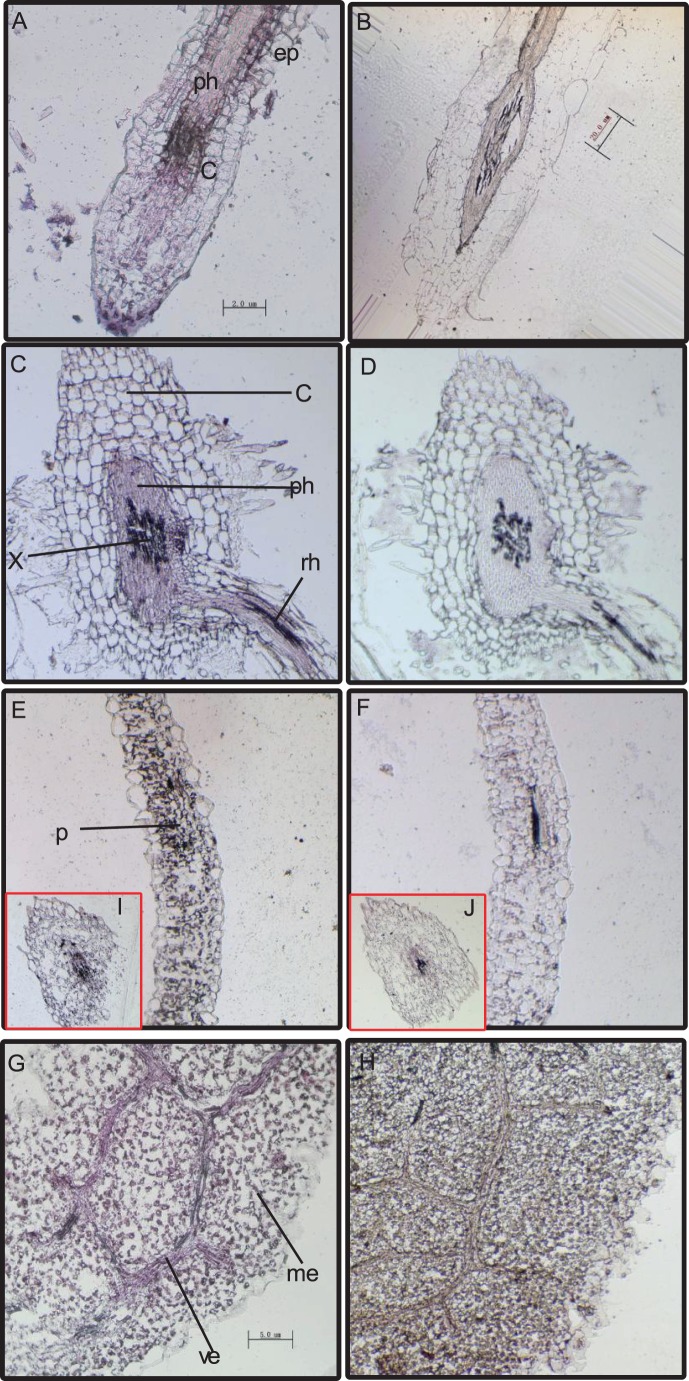
To further identify the spatiotemporal expression of TcOPT3 in vivo, we performed in situ hybridization on the longitudinal and transverse sections of root, stem and leaf using DIG-labeled TcOPT3 antisense and sense RNA as probes (Fig. 4). Sense-oriented probe was used as the negative control (Fig. 4 B, D, F, H, J). In roots, more intense signals were detected in the pericycle and the vascular tissues, including xylem and phloem (Fig. 4A). This pattern of localization was further confirmed in the cross-section (Fig. 4C). In the stem, TcOPT3 transcripts were strong in almost all the central cylinder cells (Fig. 4E, I). Additionally, in the leaf, extremely strong signals were detected in the vein of the mesophyll (Fig. 4G). All these suggest that as a transporter protein, TcOPT3 most probably plays a role in the long-distance transporting system.
Figure 4. Localization of TcOPT3 expression by in situ hybridization.
(A,C,E,G,I) represents hybridization with TcOPT3 antisense probe. (B,D,F,H,J) shows hybridization with the sense probe (negative control). In situ hybridization of the sense and antisense TcOPT3 probes to sections of Thlaspi caerulescens root tissues (A) to (D), stem tissues(E,F,I,J) and leaf tissues(G,H). Abbreviation: c, cortex; ep, epidermis; p, pericycle; ph, phloem; rh, root hair; x, xylem; cb, cambium; ve, vein; me, mesophyll; vc, vascular cambium.
TcOPT3 Expression in Response to Metal Deficiency
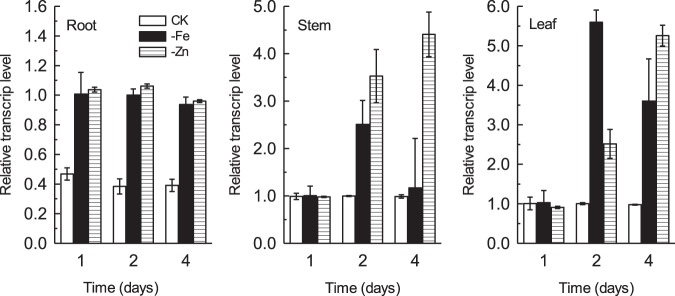
To see whether the gene responses to metal deficiency, we measured the expression of TcOPT3 in root, stem and leaf tissues (Fig. 5) and found it was also highly induced by Fe and Zn deficiency, especially in the stem and leaf. But the expression of TcOPT3 responded to Fe and Zn deficiency faster in the root as the enhancement was detected in 1-day treatment, while 2-day in the aerial parts.
Figure 5. Effect of element deficiency on mRNA expression of the TcOPT3 gene.
Real-time RT-PCR expression analysis of the TcOPT3 gene expression in roots (R), leaves (L) and stems (S) with the treatment of Fe or Zn deficient for 1, 2, 4 days. The ΔCp values were calculated as follows: CP of target gene (TcOPT3) – CP of constitutive control gene (ubiquitin-conjugating enzyme), where the CP value is the fractional cycle number of crossing point (CP). The ΔCP values represent the mean of three technical replicates (±SD) of one experiment representative of three independent experiments. Relative transcript levels (RTL) were calculated as follows: RTL = 2−ΔCP.
Sub-Cellular Localization of the TcOPT3 Protein
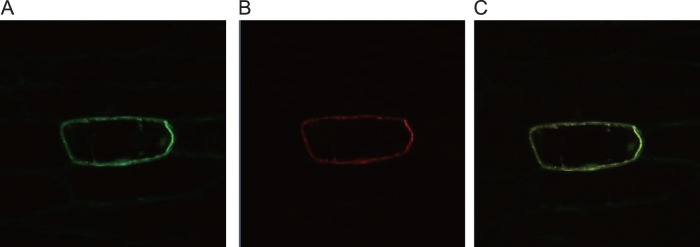
To investigate the sub-cellular localization, TcOPT3 fused with a green fluorescent protein (GFP) under the control of the cauliflower mosaic virus 35 S promoter and pm-rk (Plasma membrane marker) were co-expressed in the onion epidermis cells. The confocal microscopy observation showed that the green fluorescence and red pm-rk were both confined to the plasma membrane (Fig. 6) and can be merged perfectly, indicating that TcOPT3 encoded a plasma membrane-localized protein.
Figure 6. Sub-cellular localization of TcOPT3 protein.
Onion epidermal cells transiently co-transformed with TcOPT3::GFP and pm-rk (Plasma membrane marker). (A) Fluorescence image of epidermal cell expressing the p35S::EGFP fusion protein. (B) Fluorescence image of epidermal cell expressing the pm-rk. (C) Merged fluorescence image of epidermal cell expressing the p35S-TcOPT3::EGFP fusion protein and pm-rk marker.
Functional Characterization of TcOPT3 by Heterologous Expression in Yeast
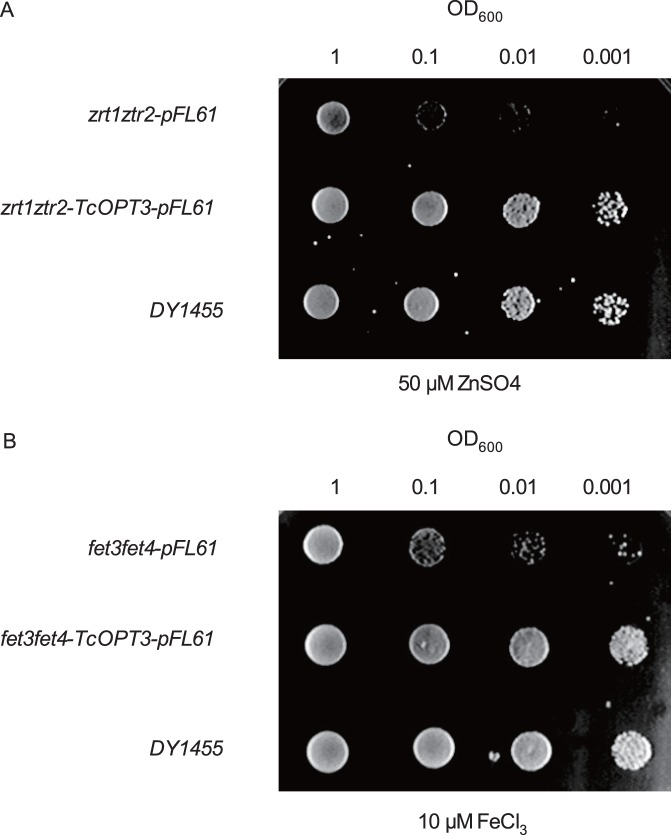
To study the function of TcOPT3 in Fe and Zn transport, we tested whether expression of a TcOPT3 cDNA could restore the growth of yeast double mutants fet3fet4 (strain DEY1453) and zrt1zrt2 (strain ZHY3), which can not grow on the Fe- and Zn-limited medium respectively [33], [34], [35]. As expected, expression of TcOPT3 complemented the growth of mutants fet3fet4 and zrt1zrt2 (Fig. 7) when both metal sources in the medium were provided at a low concentration (10 µM Fe2+ or 50 µM Zn2+), suggesting that TcOPT3 can transport both Fe2+ and Zn2+.
Figure 7. Complementation of the fet3fet4 and ZHY3 (zrt1ztr2) yeasts mutant by T. caerulescens cDNAs.
Yeast strains defective in iron uptake (fet3fet4) and zinc uptake (zrt1ztr2) were transformed with pFL61 (empty vector) and pFL61-TcOPT3. Serial dilutions of yeast cells were dropped onto a low-zinc medium (LZM) supplemented with 50 µM ZnSO4 (A) and a low-iron medium (LIM) supplemented with 10 µM FeCl3 (B) assayed for growth on SD-ura plates. The entire experiment was performed twice.
TcOPT3 Contributes to Fe/Zn/Cu/Cd Accumulation
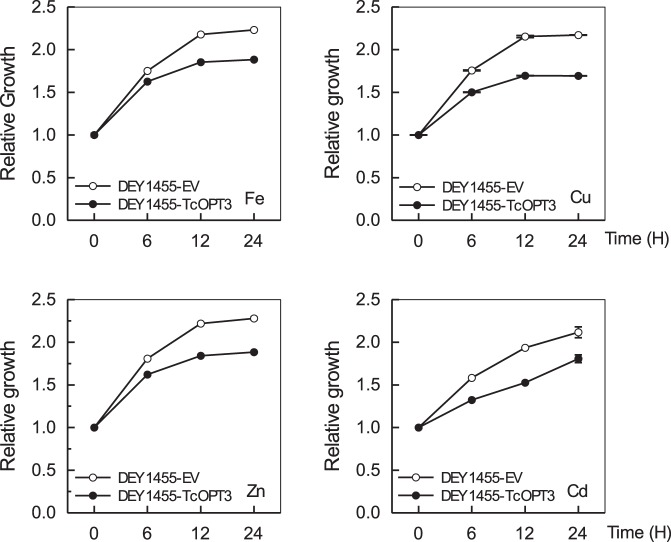
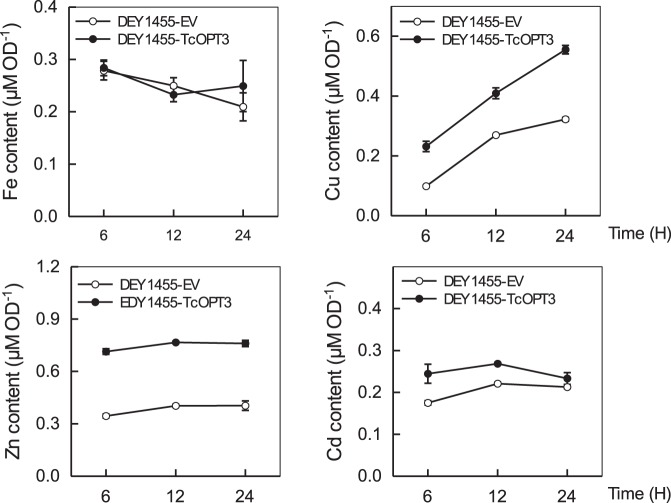
The role of TcOPT3 in the metal accumulation was further investigated by expressing TcOPT3 and empty vector in the wild-type yeast line (stain DY1455) cultured in the solutions containing 50 µM FeCl3, 50 µM ZnSO4, 50 µM CuSO4 or 20 µM CdSO4, respectively. Compared with those empty vector-expressing strains, the TcOPT3-expressing trains grew worse (Fig. 8), meanwhile the corresponding contents of these metal ions were significantly higher as well, suggesting that TcOPT3 can transport Fe/Zn/Cd/Cu into yeast cells (Fig. 9).
Figure 8. Growth of the wild-type (DY1455) and TcOPT3-transformed yeast cells under different metal supplies.
Yeast cells were grown to an OD600 of 1.0, then supplemented with 50 µM FeCl3, 50 µM ZnSO4, 20 µM CdCl2 or 50 µM CuSO4 respectively. Data are the means ± SE per experiment (n = 3), P<0.05 by Student’s t-test.
Figure 9. Heavy metal accumulation of wild-type (DY1455) and TcOPT3-transformed yeast cells.
Zn, Fe, Ni, Cd and Cu accumulation in yeast transformants. Metal accumulation was conducted in liquid SD media supplemented with 50 µM FeCl3, 50 µM ZnSO4, 20 µM CdCl2, or 50 µM CuSO4 respectively. Data are the means ± SE per experiment (n = 3), P<0.05 by Student’s t-test.
Discussion
The Oligopeptide transporters (OPTs) were initially characterized as small-peptide transporters. However, this has been challenged by their new functions in metal trafficking recently. The first report is from Wintz et al who found that heterologous expression of AtOPT3 in yeast can transport Cu, Mn and Fe [28]. Later, AtOPT6 and AtOPT7 were also proved to transport Cd or Cd-glutathione chelate in yeast [31]. Recently, AtOPT3 was further demonstrated to be involved in Fe homeostasis [30]. It was showed that heterologously expressed AtOPT3 increased Cd sensitivity of S. cerevisiae opt2 mutants and contributed to Cd accumulation [36]. All the above reports indicate the potential role of OPTs in transit metal transportantion. Thlaspi caerulescens, a Cd/Zn/Ni hyperaccumulator, ecotype of Ganges, from southern France, can accumulate 10 000 mg Cd kg−1 at shoot dry weight base [3], [37]. In this study, we cloned and elucidated the function of a novel member of OPT family, TcOPT3 from T. caerulescens.
Role of TcOPT3 in Metal Transport
AtOPT3 has been proved to play a role in Fe homeostasis in A. thaliana [30]. Here, we also found that heterologous expression of TcOPT3 in Saccharomyces cerevisiae rescued the growth of Fe-depleted mutant fet3fet4 (Fig. 7), suggesting that TcOPT3 can transport Fe too. As T. caerulescens is also a Zn hyperaccumulator, we tested the possible role of TcOPT3 in transporting Zn. As expected, TcOPT3 restored the growth of Zn-uptake-defective mutant zrt1zrt2. Furthermore, heterologous expression of TcOPT3 negatively affected the growth of S. cerevisiae when treated with elevated Fe, Zn, Cd, or Cu (Fig. 8) as more Fe, Zn, Cd or Cu was accumulated in the transformed yeast (Fig. 9), suggesting a broader substrates specificity for TcOPT3. The broad substrate specificity of cytoplasmic transition metal importers may ensure their universal potential to fulfill different developmental stages, without perpetuating the consequences of limited metal transporter specificity throughout the plant [7], while the specificity of transition metal export from cytoplasm may serve to establish specificity through the differential storage of transition metals in specific tissues of cell types. Therefore, the specificity of the efflux transporters appears to be more pounced than that of the influx transporters. For example, the P1B-type ATPases HMA3 from T.caerulescens showed high specificity for Cd, that serves to efflux Cd into vacoule [38]. As T. caerulescens can also accumulate Ni, we tested the possibility of transporting nickel (Ni), and found that altough the growth was negatively affected by Ni addtion (Fig. S1), the content in the yeast did not change much (Fig. S2), suggeting that TcOPT3 does not transport Ni, and there must be other transporters responsible for Ni hyperaccumulation, such as TcYSL3 [19]. Futhermore, we still found that TcOPT3 did not transport Pb into yeast too (Fig. S1, S2).
Roles of membrane transporters that are responsible for hyperaccumulation include uptake, efflux, translocation and sequestration of transit metals. Metal transporters herein identified include ZIP (ZRT/IRT like Protein), Nramp (Natural Resistance and Macrophage Protein), MTP (Metal Resistance Protein), and HMA (Heavy metal/CPX-type ATPases) families in A.thaliana and hyeperaccmulators such as T. caerulescens and A. halleri [4], [38], [39], [40], [41]. Transporters of the HMA family and the MTP family that are involved in metal efflux from the cytoplasm, either by movement across the plasma membrane (PM) or into organelles, whereas metal uptake transporters include the NRAMP family and the ZIP families that are transporters either act at the PM to move metals into the cytoplasm or remobilize metals from intracellular compartments into the cytoplasm [6]. In this study, heterologous expression of TcOPT3 in S. cerevisiae increased the sensitivity of yeast by accumulating more metals, indicating that TcOPT3 function as an uptake transporter for heavy metals (Fig. 8, 9).
Potential Role of TcOPT3 in Plant Long-distance Metal Transport and Hyperaccumulation
In this study, qRT-PCR results showed that TcOPT3 was preferentially expressed in the aerial parts (stem and leaf) than roots both under normal condition or Fe/Zn deficient conditions (Fig. 3, 5). While in Arabidopsis, AtOPT3 expressed stronger in flower, leaf and root, but relatively lower in the stem [42]. It may be due to the different growth stage tested, but most probably, the different expression patterns between AtOPT3 and TcOPT3 may suggest their different roles in trafficking heavy metals within plants. It was reported that reduced expression of AtOPT3 in the Arabidopsis mutant opt3-2 resulted in the accumulation of very high levels of Fe in tissues except seeds [30]. Recently, it was showed that heterologously expressed AtOPT3 increased Cd sensitivity of S. cerevisiae opt2 mutants and contributed to Cd accumulation [36]. Consistent with the role of OPT3 in Cd detoxification, the early-stage seedlings of A. thaliana opt3-3 knockdown allele were extremely sensitive to Cd. In contrast, leaves of hydroponically-grown opt3-3 mature plants were more tolerant to Cd compared to the wild type [36]. We also found that heterologously expressed TcOPT3 increased heavy metal (Cu/Cd) sensitivity of S. cerevisiae and contributed to metal accumulation, suggesting a similar role of TcOPT3 in heavy metal trafficking in plant. However, little evidence from overexpression OPTs in metal distribution is reported to date. Given the different expression patterns between TcOPT3 and AtOPT3, it is interesting to investigate the potential role of TcOPT3 in hyperaccmulation in T. caerulescens. The tissue specific overexperssion of TcOPT3 is highly desirable in order to verify its potential role in creating metal hyperaccumulators for the purpose of phytoremediation of heavy metal polluted environments in future.
As proposed by Milner and Kochian [4], Zn hyperaccumulation of T.caerulescens appears to include at least five physiological events: (1) Increased Zn2+ influx across the root-cell PM; (2) Reduced Zn sequestration in the root-cell vacuole; (3) Increased Zn transport into the xylem and via the xylem to the shoot; (4) Increased Zn2+ influx into leaf mesophyll cells; and (5) Zn and Cd stored primarily in leaf epidermal cells. In situ hybridization results showed that, TcOPT3 was highly expression in vascular cells both in roots and shoots (Fig. 4). This expression pattern is similar to the previous report on the expression of AtOPT3 in A. thaliana, which is also highly expressed in the vascular tissues both in light-grown seedlings and adult plants [42]. As efficient translocation of metals from root to shoot is one of the most important hallmarks of hyperaccumulators. Thus, efficient transporters expressed in loading and unloading tissues are fundamental for hyperaccumulation. For example, HMA4 is characterized metal transporter primarily localized in the root stele. It plays a critical role in loading heavy metals into xylem for long-distance transportation from root to shoot not only in T.caerulescens but also in another hyperaccumulating plant A.halleri [43], [44], [45]. TcHMA3, another member of HMA family was proved to be a tonoplast-localized transporter highly specific for Cd, which is responsible for sequestration of Cd into the leaf vacuoles, and that a higher expression of this gene is required for Cd hypertolerance in the Cd-hyperaccumulating ecotype of T. caerulescens [38]. Uptake transporters responsible for hyperaccumulation, TcIRT1, TcIRT2, TcZNT1 and TcZNT5 of ZIP families in T.caerulescens, for instance, were expressed only in roots but not in leaves [46]. The yellow-stripe 1-like (YSL) subfamily is included in the OPT superfamily, some of which was proved to be involved in loading and unloading of nicotianamine-metal chelates from the vascular tissues. TcYSLs of YSL transporters from T. caerulescens, especially for TcYSL3 and TcYSL7, were expressed in xylem parenchyma and phloem [19]. Furthermore, TcYSL3 was shown to transport Ni-NA chelates. Here we demonstrated that TcOPT3 is a plasma membrane-localized protein, and can transport metals into yeast cells (Fig. 6, 8). TcOPT3 was expressed primarily in shoots, especially under nutrient deficiency, indicating their specificity in metal uptake and transport in shoots (Fig. 3, 5). As it is highly and constructively expressed in vascular system, including xylem, phloem and vein, it is reasonable to assume that it plays important roles in unloading of nutrients and heavy metals in those tissues, thus consists of an important component in the long-distance transportation system. However, it needs further investigation.
In this study, ecotype Ganges instead of ecotype Prayon was studied. Considered that heavy metal hyperaccumulation varies greatly among different ecotypes, it is interesting to further confirm the expression and functional analysis of TcOPT3 in two contrasting ecotypes of the hyperaccumulator Thlaspi caerulescens. We have analyzed the amino acids sequence and expression of TcOPT3-p preliminarily; however, there is no significant difference between TcOPT3-g and TcOPT3-p expression pattern. Further studies need to be conducted in the future.
In conclusion, we demonstrated that TcOPT3 in a metal hyperaccumulator, Thlaspi caerulescens, was an Fe/Zn/Cu/Cd influx transporter with non-specificity substrate. This is the first report showing that TcOPTs gene may be involved in metal long-distance transport systems that contribute to heavy metal hyperaccumulation.
Materials and Methods
Plant Growth
Seeds of Thlaspi caerulescens J. & C. Presl, ‘Ganges’ ecotype, were surface sterilized by 75% alcohol and 10% NaClO4, then stored at 4°C for 3 days. Seeds were germinated on agar with modified MS medium (with addition 270 µM ZnSO4 in Murashige and Skoog, Sigma, U.S.) for two weeks at 14-h/25°C d and a 10-h/18°C night regime, a light intensity of 100 µmol photons m−2 s−1. Then the seedlings were transferred to modified Hoagland nutrient solution (2 mM Ca(NO3)2, 0.1 mM KH2PO4, 0.5 mM MgSO4, 0.1 mM KCl, 0.7 mM K2SO4, 0.1 mM Fe-EDTA, 10 µM H3BO3, 0.5 µM MnSO4, 10 µM ZnSO4, 0.2 µM CuSO4 and 0.01 µM (NH4)6Mo7O2, pH 5.5).
Gene and cDNA Cloning and Sequencing
Total RNA was extracted from T. caerulescens roots or leaves by the Trizol reagent (Invitrogen, USA) and purified by DNase I (RNase Free) Kit (TaKaRa, China). First strand cDNA of T. caerulescens synthesis was performed using M-MLV reverse transcriptase (TaKaRa, China) and PCR-amplified with degenerated primers designed from conserved AtOPT3 sequence. One fragment was cloned into the pMD19-T Vector (TaKaRa, China), sequenced and identified as AtOPT3 orthologs by BLAST using the NCBI database. RACE PCR (5′ and 3′) were performed in order to obtain full-length sequences from uncomplete cDNA fragments (see Table 1 for primer sequences). The 5′ and 3′ regions of cDNA fragments were cloned with a SMART™ RACE cDNA Amplification Kit (Clontech, TaKaRa Bio, USA) according to the manufacturer’s protocol. The PCR products were subcloned to pMD19-T Vector and sequenced. By sequencing alignment with its AtOPT3 ortholog from A. thaliana, it was observed that the 5′ region of the isolated TcOPT3 cDNA contained the first initiating ATG codon and the 3′ region containing the stop codon and the 3′ UTR. The gene we cloned has been deposited in Genbank with the accession number of HQ699884.
Table 1. Primers used for PCR amplification of in TcOPT3 cDNAs, 5′ and 3′ RACE, qRealTime-PCR, and plasmid constructions in order of their first mention in “Materials and Methods”.
| Purpose | Name | Sequence (5′-3′) |
| RACE PCR | TcOPT3-GSP-F | CAGTAACAGACAGGGACGATG |
| TcOPT3-GSP-R | CAGCAGGTTGGCTCAGGGTA | |
| TcOPT3-5′R-1-R | ACCTGCTGAGCCGTGATGCTATTA | |
| TcOPT3-3′R-1-F | ACATCATCGTCCCTGTCTGTTACTG | |
| TcOPT3-5′R-2-R | CGACGATTGAGTGCGAAAAGAG | |
| TcOPT3-3′R-2-F | CTTCAAATCTCGCTCAAGTTTCTCT | |
| Quantified-RealTime PCR | qRT-TcOPT3-F | CCGAGCAGCACGAGCGTTGT |
| qRT-TcOPT3-R | GCGGTTGCGTGCGGTATGTG | |
| TcUBQ-F | GGAGCCCCGCTTGGAC | |
| TcUBQ-R | CGGCGAGGCGAGTGTA | |
| in situ hybridization | TcOPT3-Probe-S | CCGGAATTCGGTTATCTCCTACGGCTTTG |
| TcOPT3-Probe-A | CCCAAGCTTTAGGGCAAGCAGTGGTATCAA | |
| Plasmid of GFP fusion | TcOPT3-EGFP-F | AAAAGTACTATGGACGCAGAGAAGGCTAACAAT |
| TcOPT3-EGFP-R | TCCCCCGGGGAAAACAGGACAGCCTTTAACTCT | |
| Plasmid of yeast transformation | TcOPT3-pFL61-F | ATAAGAATGCGGCCGCATGGACGCAGAGAAGGCTAA |
| TcOPT3-pFL61-R | ATAAGAATGCGGCCGCAATCTTAGAAAACAGGACAGC |
F, Forword; R, Reverse. S, Sense; A, Anti-sense.
Sequence Comparisons
The predicted amino-acid sequence from TcOPT3 cDNA and other selected OPT protein sequences were aligned using the CLUSTAL W program, version 1.8 (Thompson et al., 1994). The putative trans-membrane domains were predicted by the TMHMM (version 2.0; http://www.cbs.dtu.dk/services/TMHMM/). The putative amino acid sequences were aligned with the program Clustal X Version 1.8 [47] and viewed by GeneDoc version 2.6 [48]. Phylogenetic trees were constructed with the neighbor-joining algorithm using the program with MEGA 4 (http://www.megasoftware.net) [49]. Hydrophilicity plots for TcOPT3 was generated based on the Kyte and Doolittle (1982) method using Protean sequence analysis software (DNASTAR) under default parameters.
Quantitative PCR Analysis of TcOPT3 Expression
Total RNA was isolated from roots, stems and leaves of one-month-old T. caerulescens and treated by DNase I (RNase free) as described above. RT-PCR with oligo d(T)-anchor primer were performed with PrimeScript RT (Perfect Real Time) reagent kit (TaKaRa, China). For real-time RT-PCR, 2 µl of the diluted (1∶5) cDNA products were used as templates with 5 µl SYBR Pre mix (Takara, China), 0.5 µl primers (10 µM each) and 2.5 µl water. Calculation of the ΔCp values was performed as described [50]. Primers were list in Table 1.
In Situ Hybridization
One-month-old seedlings grown on hydroponic culture were obtained. Plant materials were fixed in potassium phosphate buffer (0.1 M, pH 7.4) containing 4% paraformaldehyde overnight at 4°C, then dehydrated in ethanol series, and embedded in paraffin. A gene-specific fragment containing the 416 bp fragment cross 3′UTR of TcOPT3 was amplified by PCR (primers were shown in Table 1) and cloned into pSPT19 vectors. Sense and antisense probes were synthesized using SP6 and T7 primers with DIG RNA Labeling kit according to the manufacturer’s instructions (Roche, USA). In-situ hybridization was performed as described previously [19], [51].
GFP Fusion and Subcellular Localization
To construct the TcOPT3-GFP fusion protein, the ORF without the stop codon of TcOPT3 cDNA fragment containing SmaI and SacI restriction sites (see Table 1 for primer sequences) was cloned into the modified pEGFP-N2 vector under the control of 35 S promoter. The final construct p35S::TcOPT3-EGFP and the pm-rk were transiently expressed in onion epidermal cells using a particle gun–mediated system (PDS-1000/He; Bio-Rad), where pm-rk was used as the plasma membrane-localizated maker [52]. Onion epidermal cells were bombarded with 1 µm gold particles coated with plasmid DNA and then incubated in the dark at 25°C for 20 h. Fluorescence was observed by confocal laser scanning microscopy (LSM700; Carl Zeiss).
Yeast Complementation
For yeast transformation and functional complementation, the Saccharomyces cerevisiae (Meyen) E.C. Hansen knock-out strain fet3fet4 (DEY1453; MATa/MATa ade2/+ can1 his3 leu2 trp1 ura fet3-2::HIS3 fet4-1::LEU2) [33], zrt1zrt2 (ZHY3; MATa ade6 can1 his3 leu2 trp1 ura3 zrt1::LEU2 zrt2::HIS3) [34], [35]; and its parent strain DY1455 (MATα ade2–1oc can1 his3 leu2 trp1 ura3) were used, which is defective in low- and high-affinity iron or zinc uptake, respectively. The ORF of TcOPT3 was amplified by PCR with primers (see Table 1 for primer sequences), which was then cloned into the pGEM-T Easy (Promega) vector, digested with NotI, and subsequently cloned into yeast binary vector pFL61 [53] to form TcOPT3-pFL61. Constructs were sequenced to ensure the correct orientations of the inserts and correct sequences. fet3fet4 transformants were selected on SD medium lacking uracil (SD-ura) and supplemented with 10 µM FeCl3. zrt1zrt2 transformants were selected on SD-ura and supplemented with 50 µM ZnSO4.
For complement test, fet3fet4 and zrt1zrt2 transformants were grown overnight in liquid SD-ura and supplemented with 10 µM FeCl3 or 50 µM ZnSO4, respectively. Yeast cells were recovered by centrifugation, resuspended in SD-ura (without added Fe or Zn) at an OD600 of 1, 0.1, 0.01 and 0.001. Then 10 µL of each culture was spotted on SD-ura plates; plates were incubated at 30°C for 2 d and then photographed.
Heavy Metal Tolerance in Yeast
Wild-type yeast DY1455 was transformed with TcOPT3-pFL61, or with pFL61, which served as a negative control. DY1455 transformants were grown on liquid SD-ura medium overnight at an OD600 of 1. After washed by CaCl2 and centrifugation, resuspended in SD-ura with 50 µM FeCl3, 50 µM ZnSO4, 20 µM CdCl2, or 50 µM CuSO4 were grown for another 0, 6, 12, 24 hours. After measured the OD values and washed by CaCl2 to remove adsorbed transit metals in apoplast cells, aliquots of yeast cells were taken for measurement of metal accumulation.
Elemental Analysis
For metal accumulation, yeast cells were concentrated at 5,000 rpm for 5 min and washed by 0.5 mM CaCl2 for 5 min twice to remove adsorbed transit metals from the yeast cell walls. Then cell sediments were dried at 70°C for 2 days, and digested in HNO3. Metal concentration in the digested solution was determined by inductively coupled plasma-atomic emission spectrometry (ICP-AES; IRIS/AP optical emission spectrometer).
Statistics
Data were statistically analyzed using analysis of variance (ANOVA) in Origin 8, and tested for significant (P≤0.05) treatment differences using Student’s t-test.
Supporting Information
Growth of the wild-type (DY1455) and TcOPT3-transformed yeast cells under different metal supplies. Yeast cells were grown to an OD600 of 1.0, and then supplemented with 100 µM PbNO3 or 100 µM NiSO4 respectively. Data are the means ± SE per experiment (n = 3), P<0.05 by Student’s t-test.
(EPS)
Heavy metal accumulation of wild-type (DY1455) and TcOPT3-transformed yeast cells. Pb and Ni accumulation in yeast transformants. Metal accumulation was conducted in liquid SD media supplemented with 100 µM PbNO3 or 100 µM NiSO4 respectively. Data are the means ± SE per experiment (n = 3), P<0.05 by Student’s t-test.
(EPS)
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by Changjiang Scholarship, NSFC(30625026) and the Fundamental Research Funds for Central Universities. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Salt DE, Smith RD, Raskin I. PHYTOREMEDIATION. Annu Rev Plant Biol. 1998;49:643–668. doi: 10.1146/annurev.arplant.49.1.643. [DOI] [PubMed] [Google Scholar]
- 2.Macnair MR. The hyperaccumulation of metals by plants. Adv Bot Res J. A. Callow, Academic Press. 2003;40:63–105. [Google Scholar]
- 3.Ingrouille MJ, Smirnoff N. Thlaspi caerulescens J. & C. Presl. (T. alpestre L.) in Britain. New Phytol. 1986;102:219–233. doi: 10.1111/j.1469-8137.1986.tb00812.x. [DOI] [PubMed] [Google Scholar]
- 4.Milner MJ, Kochian LV. Investigating heavy-metal hyperaccumulation using Thlaspi caerulescens as a model system. Ann Bot. 2008;102:3–13. doi: 10.1093/aob/mcn063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clemens S. Toxic metal accumulation, responses to exposure and mechanisms of tolerance in plants. Biochimie. 2006;88:1707–1719. doi: 10.1016/j.biochi.2006.07.003. [DOI] [PubMed] [Google Scholar]
- 6.Colangelo EP, Guerinot ML. Put the metal to the petal: metal uptake and transport throughout plants. Curr Opin Plant Biol. 2006;9:322–330. doi: 10.1016/j.pbi.2006.03.015. [DOI] [PubMed] [Google Scholar]
- 7.Krämer U, Talke IN, Hanikenne M. Transition metal transport. FEBS Lett. 2007;581:2263–2272. doi: 10.1016/j.febslet.2007.04.010. [DOI] [PubMed] [Google Scholar]
- 8.Puig S, Peñarrubia L. Placing metal micronutrients in context: transport and distribution in plants. Curr Opin Plant Biol. 2009;12:299–306. doi: 10.1016/j.pbi.2009.04.008. [DOI] [PubMed] [Google Scholar]
- 9.Pilon M, Cohu CM, Ravet K, Ghany SE, Gaymard F. Essential transition metal homeostasis in plants. Curr Opin Plant Biol. 2009;12:347–357. doi: 10.1016/j.pbi.2009.04.011. [DOI] [PubMed] [Google Scholar]
- 10.Stacey G, Koh S, Granger C, Becker JM. Peptide transport in plants. Trends Plant Sci. 2002;7:257–263. doi: 10.1016/s1360-1385(02)02249-5. [DOI] [PubMed] [Google Scholar]
- 11.Lubkowitz M. Setlow J, editor. The OPT family functions in long-distance peptide and metal transport in plants. 2006. pp. 35–55. editor. Genetic Engineering: Springer US. [DOI] [PubMed]
- 12.Lubkowitz M. The Oligopeptide Transporters: A small gene family with a diverse group of substrates and functions. Mol Plant. 2011;4:407–415. doi: 10.1093/mp/ssr004. [DOI] [PubMed] [Google Scholar]
- 13.Bovet L, Eggmann T, Meylan-Bettex M, Poloer J, Kammer P, et al. Transcript levels of AtMRPs after cadmium treatment: induction of AtMRP3. Plant Cell Environ. 2003;26:371–381. [Google Scholar]
- 14.Kim DY, Bovet L, Kushnir S, Noh EW, Martinoia E, et al. AtATM3 is involved in heavy metal resistance in Arabidopsis. Plant Physiol. 2006;140:922–932. doi: 10.1104/pp.105.074146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim DY, Bovet L, Maeshima M, Martinoia E, Lee Y. The ABC transporter AtPDR8 is a cadmium extrusion pump conferring heavy metal resistance. Plant J. 2007;50:207–218. doi: 10.1111/j.1365-313X.2007.03044.x. [DOI] [PubMed] [Google Scholar]
- 16.Lubkowitz MA, Hauser L, Breslav M, Naider F, Bercker JM. An oligopeptide transport gene from Candida albicans. Microbiology. 1997;143:387–396. doi: 10.1099/00221287-143-2-387. [DOI] [PubMed] [Google Scholar]
- 17.Lubkowitz MA, Barnes D, Breslav M, Burchfield A, Naider F, et al. Schizosaccharomyces pombe isp4 encodes a transporter representing a novel family of oligopeptide transporters. Mol Microbio. 1998;28:729–741. doi: 10.1046/j.1365-2958.1998.00827.x. [DOI] [PubMed] [Google Scholar]
- 18.Hauser M, Donhardt AM, Barnes D, Naider F, Becker JM. Enkephalins are transported by a novel Eukaryotic peptide uptake system. J Biol Chem. 2000;275:3037–3041. doi: 10.1074/jbc.275.5.3037. [DOI] [PubMed] [Google Scholar]
- 19.Gendre D, Czernic P, Conéjéro G, Pianelli K, Briat JF, et al. TcYSL3, a member of the YSL gene family from the hyper-accumulator Thlaspi caerulescens, encodes a nicotianamine-Ni/Fe transporter. Plant J. 2007;49:1–15. doi: 10.1111/j.1365-313X.2006.02937.x. [DOI] [PubMed] [Google Scholar]
- 20.Morrissey J, Guerinot ML. Iron uptake and transport in plants: the good, the bad, and the ionome. Chem Rev. 2009;109:4553–4567. doi: 10.1021/cr900112r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Curie C, Panaviene Z, Loulergue C, Dellaporta SL, Briat JF, et al. Maize yellow stripe1 encodes a membrane protein directly involved in Fe(III) uptake. Nature. 2001;409:346–349. doi: 10.1038/35053080. [DOI] [PubMed] [Google Scholar]
- 22.Harada E, Sugase K, Namba K, Iwashita T, Murata Y. Structural element responsible for the Fe(III)-phytosiderophore specific transport by HvYS1 transporter in barley. FEBS Lett. 2007;581:4298–4302. doi: 10.1016/j.febslet.2007.08.011. [DOI] [PubMed] [Google Scholar]
- 23.Anderegg G, Ripperger H. Correlation between metal complex formation and biological activity of nicotianamine analogues. J Chem Soc Chem Commun. 1989;10:647–650. [Google Scholar]
- 24.von Wirén N, Klair S, Bansal S, Briat JF, Khodr H, et al. Nicotianamine chelates both FeIII and FeII. Implications for metal transport in plants. Plant Physiol. 1999;119:1107–1114. doi: 10.1104/pp.119.3.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chu HH, Chiecko J, Punshon T, Lanzirotti A, Lahner B, et al. Successful reproduction requires the function of Arabidopsis YELLOW STRIPE-LIKE1 and YELLOW STRIPE-LIKE3 metal-nicotianamine transporters in both vegetative and reproductive structures. Plant Physiol. 2010;154:197–210. doi: 10.1104/pp.110.159103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stephan UW, Scholz G. Nicotianamine: mediator of transport of iron and heavy metals in the phloem? Physiol Plant. 1993;88:522–529. [Google Scholar]
- 27.Koh S, Wiles AM, Sharp JS, Naider FR, Becker JM, et al. An oligopeptide transporter gene family in Arabidopsis. Plant Physiol. 2002;128:21–29. [PMC free article] [PubMed] [Google Scholar]
- 28.Wintz H, Fox T, Wu YY, Feng V, Chen W, et al. Expression profiles of Arabidopsis thaliana in mineral deficiencies reveal novel transporters involved in metal homeostasis. J Biol Chem. 2003;278:47644–47653. doi: 10.1074/jbc.M309338200. [DOI] [PubMed] [Google Scholar]
- 29.Stacey MG, Osawa H, Patel A, Gassmann W, Stacey G. Expression analyses of Arabidopsis oligopeptide transporters during seed germination, vegetative growth and reproduction. Planta. 2006;223:291–305. doi: 10.1007/s00425-005-0087-x. [DOI] [PubMed] [Google Scholar]
- 30.Stacey MG, Patel A, McClain WE, Mathieu M, Remley M, et al. The Arabidopsis AtOPT3 protein functions in metal homeostasis and movement of iron to developing seeds. Plant Physiol. 2008;146:589–601. doi: 10.1104/pp.107.108183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cagnac O, Bourbouloux A, Chakrabarty D, Zhang MY, Delrot S. AtOPT6 transports glutathione derivatives and is induced by primisulfuron. Plant Physiol. 2004;135:1378–1387. doi: 10.1104/pp.104.039859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pike S, Patel A, Stacey G, Gassmann W. Arabidopsis OPT6 is an oligopeptide transporter with exceptionally broad substrate specificity. Plant Cell Physiol. 2009;50:1923–1932. doi: 10.1093/pcp/pcp136. [DOI] [PubMed] [Google Scholar]
- 33.Eide D, Broderius M, Fett J, Guerinot ML. A novel iron-regulated metal transporter from plants identified by functional expression in yeast. Proc Natl Acad Sci U S A. 1996;93:5624–5628. doi: 10.1073/pnas.93.11.5624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao H, Eide D. The yeast ZRT1 gene encodes the zinc transporter protein of a high-affinity uptake system induced by zinc limitation. Proc Natl Acad Sci U S A. 1996;93:2454–2458. doi: 10.1073/pnas.93.6.2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao H, Eide D. The ZRT2 gene encodes the low affinity zinc transporter in Saccharomyces cerevisiae. J Biol Chem. 1996;271:23203–23210. doi: 10.1074/jbc.271.38.23203. [DOI] [PubMed] [Google Scholar]
- 36.Zhai Z, Gayomba S, Hail J, John D, Mike R, et al. "Novel" functions of "Old" transporters: COPT2 and OPT3 function in cadmium detoxification in Arabidopsis thaliana. 2010. Available: http://abstracts.aspb.org/pb2010/public/M20/M2004.html. Accessed 2010 Sep 10.
- 37.Lombi E, Zhao FJ, Dunham SJ, Mc Grath SP. Cadmium accumulation in populations of Thlaspi caerulescens and Thlaspi goesingense. New Phytol. 2000;145:11–20. [Google Scholar]
- 38.Ueno D, Milner MJ, Yamaji N, Yokosho K, Koyama E, et al. Elevated expression of TcHMA3 plays a key role in the extreme Cd tolerance in a Cd-hyperaccumulating ecotype of Thlaspi caerulescens. Plant J. 2011;66:852–862. doi: 10.1111/j.1365-313X.2011.04548.x. [DOI] [PubMed] [Google Scholar]
- 39.Weber M, Harada E, Vess C, Roepenack-Lahaye E, Clemens S. Comparative microarray analysis of Arabidopsis thaliana and Arabidopsis halleri roots identifies nicotianamine synthase, a ZIP transporter and other genes as potential metal hyperaccumulation factors. Plant J. 2004;37:269–281. doi: 10.1046/j.1365-313x.2003.01960.x. [DOI] [PubMed] [Google Scholar]
- 40.Oomen RJFJ, Wu J, Lelièvre F, Blanchet S, Richaud P, et al. Functional characterization of NRAMP3 and NRAMP4 from the metal hyperaccumulator Thlaspi caerulescens. New Phytol. 2009;181:637–650. doi: 10.1111/j.1469-8137.2008.02694.x. [DOI] [PubMed] [Google Scholar]
- 41.Roosens N, Leplae R, Bernard C, Verbruggen N. Variations in plant metallothioneins: the heavy metal hyperaccumulator Thlaspi caerulescens as a study case. Planta. 2005;222:716–729. doi: 10.1007/s00425-005-0006-1. [DOI] [PubMed] [Google Scholar]
- 42.Stacey MG, Koh S, Becker J, Stacey G. AtOPT3, a member of the oligopeptide transporter family, is essential for embryo development in Arabidopsis. Plant Cell. 2002;14:2799–2811. doi: 10.1105/tpc.005629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Papoyan A, Kochian LV. Identification of Thlaspi caerulescens genes that may be involved in heavy metal hyperaccumulation and tolerance. Characterization of a novel heavy metal transporting ATPase. Plant Physiol. 2004;136:3814–3823. doi: 10.1104/pp.104.044503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Courbot M, Willems G, Motte P, Arvidsson S, Roosens N, et al. A major quantitative trait locus for cadmium tolerance in Arabidopsis halleri colocalizes with HMA4, a gene encoding a heavy metal ATPase. Plant Physiol. 2007;144:1052–1065. doi: 10.1104/pp.106.095133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hanikenne M, Talke IN, Haydon MJ, Lanz C, Nolte A, et al. Evolution of metal hyperaccumulation required cis-regulatory changes and triplication of HMA4. Nature. 2008;453:391–395. doi: 10.1038/nature06877. [DOI] [PubMed] [Google Scholar]
- 46.Plaza S, Tearall K L, Zhao FJ, Buchner P, McGrath SP, et al. Expression and functional analysis of metal transporter genes in two contrasting ecotypes of the hyperaccumulator Thlaspi caerulescens. J Exp Bot. 2007;58:1717–1728. doi: 10.1093/jxb/erm025. [DOI] [PubMed] [Google Scholar]
- 47.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X Windows Interface: Flexible Strategies for Multiple Sequence Alignment Aided by Quality Analysis Tools. Nucl Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nicholas KB, Nicholas HB, Deerfield DW. GeneDoc: analysis and visualization of genetic variation. EMBNEW. EMBnet news. 1997;4:14. [Google Scholar]
- 49.Kumar V, Dey A, Singh A. MEGA: A Bio Computational Software for Sequence and Phylogenetic Analysis. World Congress on Engineering 2009, Vols I and Ii: 1863–1865 1895. 2009.
- 50.Pfaffl MW. A new mathematical model for relative quantification in real-time RT–PCR. Nucl Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Komminoth P, Long AA, Ray R, Wolfe HJ. In Situ polymerase chain reaction detection of Viral DNA, single-copy Genes, and gene rearrangements in cell suspensions and cytospins. Diagnostic Molecular Pathology. 1992;1:85–97. [PubMed] [Google Scholar]
- 52.Nelson BK, Cai X, Nebenführ A. A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J. 2007;51:1126–1136. doi: 10.1111/j.1365-313X.2007.03212.x. [DOI] [PubMed] [Google Scholar]
- 53.Minet M, Dufour ME, Lacroute F. Complementation of Saccharomyces cerevisiae auxotrophic mutants by Arabidopsis thaliana cDNAs. Plant J. 1992;2:417–422. doi: 10.1111/j.1365-313x.1992.00417.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Growth of the wild-type (DY1455) and TcOPT3-transformed yeast cells under different metal supplies. Yeast cells were grown to an OD600 of 1.0, and then supplemented with 100 µM PbNO3 or 100 µM NiSO4 respectively. Data are the means ± SE per experiment (n = 3), P<0.05 by Student’s t-test.
(EPS)
Heavy metal accumulation of wild-type (DY1455) and TcOPT3-transformed yeast cells. Pb and Ni accumulation in yeast transformants. Metal accumulation was conducted in liquid SD media supplemented with 100 µM PbNO3 or 100 µM NiSO4 respectively. Data are the means ± SE per experiment (n = 3), P<0.05 by Student’s t-test.
(EPS)