Abstract
In previous studies of a genetic isolate, we identified significant linkage of attention deficit hyperactivity disorder (ADHD) to 4q, 5q, 8q, 11q and 17p. The existence of unique large size families linked to multiple regions, and the fact that these families came from an isolated population, we hypothesized that two-locus interaction contributions to ADHD were plausible. Several analytical models converged to show significant interaction between 4q and 11q (P<1 × 10−8) and 11q and 17p (P<1 × 10−6). As we have identified that common variants of the LPHN3 gene were responsible for the 4q linkage signal, we focused on 4q–11q interaction to determine that single-nucleotide polymorphisms (SNPs) harbored in the LPHN3 gene interact with SNPs spanning the 11q region that contains DRD2 and NCAM1 genes, to double the risk of developing ADHD. This interaction not only explains genetic effects much better than taking each of these loci effects by separated but also differences in brain metabolism as depicted by proton magnetic resonance spectroscopy data and pharmacogenetic response to stimulant medication. These findings not only add information about how high order genetic interactions might be implicated in conferring susceptibility to develop ADHD but also show that future studies of the effects of genetic interactions on ADHD clinical information will help to shape predictive models of individual outcome.
Keywords: ADHD, genetic interaction, LPHN3, NCAM1, DRD2
Introduction
Using multigenerational and extended pedigrees from a genetic isolate our group identified several loci significantly linked to attention deficit hyperactivity disorder (ADHD), namely on 4q, 5q, 8q, 11q and 17p.1 Regions on 11q and 17p replicated linkage described by others.2 Minimal critical regions for each one of these regions, defined by recombination events in affected individuals as well as the 1-LOD-support criterion, have been reported elsewhere.1 Given the significant linkage to different genomic regions, the existence of unique large size families linked to multiple regions, and the fact that these families came from an isolated population, we hypothesized that two-locus interaction contributions to ADHD were plausible.
Methods and Results
To screen for possible interacting regions, we initially performed a correlation subset analysis3 between the linked regions. We used 134 nuclear families from the Paisa genetic isolate and single-nucleotide polymorphisms (SNPs) spanning each minimal critical region with a resolution of 200 kb (mapping resolution as well as SNPs selection criteria have been published in detail elsewhere4). We calculated the non-parametric linkage (NPL) score using GENEHUNTER5 with the weight function weight1−0, where families that demonstrated nominal NPL (NPL>1.00) to a region were included in the analysis (that is, were coded as 1 for weight1−0) and those families not demonstrating linkage were excluded (that is, were coded as 0 for weight1−0).3 To measure heterogeneity the weight function weight0−1 (defined analogously) was used, where only those families that did not demonstrate linkage to a region were included in the analysis.3 Of the entire possible set of pair-wise comparisons, two gave increases in the NPL statistic greater than two units when compared with linkage analyses using all families. Selecting families linked to 17p increased the nonparametric linkage statistic from 0.55 to 3.88 on 11q (17p–11q interaction) and selecting families linked to 4q increased the NPL statistic from 0.55 to 3.24 in an overlapping region on 11q (4q–11q interaction; Figure 1a, Supplementary Information). Using the method implemented in GENEHUNTER-TWOLOCUS,6, 7 we found a maximal non-parametric score of 6.08 (P<1 × 10−8) produced by SNPs rs1038426 (4q) and rs1293344 (11q; 4q–11q interaction; Figure 1b, Supplementary Information) and a maximal non-parametric score of 5.51 (P<1 × 10−6), produced by SNPs rs9227 (17p) and rs1293344 (11q; 17p–11q interaction; Figure 1c, Supplementary Information). Convergent evidence of interaction was detected while using two-locus parametric analysis. Furthermore, power analyses to detect two interacting loci while considering ADHD as a binary trait and also to detect interacting loci underlying quantitative traits, shows that, in general, our discovery sample exhibits exceptional power to detect two-locus interactions (see procedures attached to the Supplementary Information).
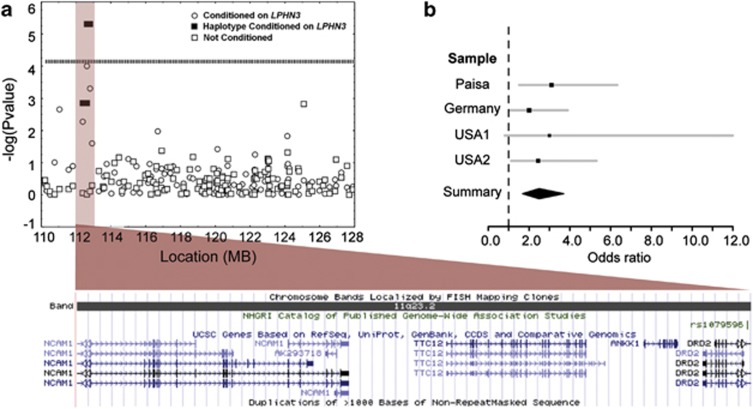
Figure 1.
(a) Results of a case–control association analysis, in which one individual was selected per family based on having a susceptibility haplotype on 4q within LPHN3 (48 cases and 40 controls; circles) or irrespective of haplotype within LPHN3 (squares). The filled rectangles represent haplotypes made of the most significant markers. Results are plotted as the −log (P-value). The dashed line indicates significance as established by the Bonferroni correction (Pcorrected=0.01/139=0.000072). A single haplotype, rs677642-rs877137, is very significant in this analysis (P<0.000005; GG; 57% allele frequency cases; 23% allele frequency controls). A diagram of the region encompassed by the haplotype as depicted at the University of California, Santa Cruz genome browser is presented. (b) This figure presents the meta-analysis results for the four samples included in the study for a random effects model P<0.00001, OR=2.46 (95% 1.68–3.70).
Using a very stringent linkage disequilibrium map with a resolution of ∼68 kb (two times stronger that the resolution recommended for covering this genomic area4) we also demonstrated that throughout the scanned region there was a remarkable total absence of linkage disequilibrium gaps, meaning that our genotyped markers covered any variation inside of the 4q-linked region. With this map, we identified a unique region of association and linkage of ∼240 Kb. None other areas inside of the 4q-linked region showed positive association. The region of ∼240 Kb was harbored inside of the LPHN3 gene. Additional analysis showed that LPHN3 common variant confers susceptibility to ADHD affects brain metabolism and predicts effectiveness of stimulant medication.4 We showed that three markers harbored in LPHN3 passed the test of heterogeneity and were significant after adjusting for multiple tests: rs6551665 (odds ratio (OR)=1.23, 95% confidence interval (CI) 1.09–1.37, P=3.46 × 10−4), rs1947274 (OR=1.23, 95% CI=1.09–1.38, P=5.41 × 10−4) and rs2345039 (OR=1.21, 95% CI=1.08–1.35, P=8.97 × 10−4). The marker rs6551665 was genotyped in all the samples and showed a linkage disequilibrium r2 with these other two markers >90%.
Given the demonstrated association of ADHD susceptibility to rs6551665 (G, allele), a SNP harbored inside the genomic region coding for LPHN3, we next performed an association analysis using SNPs on 11q and 17p and conditioning on the fact of being a carrier of the G variant of susceptibility at rs6551665. Either one case or control with at least one copy of the rs6551665 LPHN3 susceptibility G variant was selected per family. Because of the rareness of individuals homozygous for the rs6551665 G allele we pooled them with G heterozygotes, when we conditioned on the fact of being a carrier of the G variant of susceptibility at rs6551665, we were able to narrow down signals at 11q and 17p. However, the signal at 11q pointed to a region containing only three genes, whereas the signal at 17p spanned a genomic region containing dozens of genes. Furthermore, given that testing three-locus interactions (LPHN3–11q–17p) will demand evaluation of thousands of models (227–2 for a dichotomic trait), we focused in this manuscript to describe results of the LPHN3–11q interaction.
LPHN3–11q interaction
Single marker and haplotype case–control association analysis revealed a single strongly associated haplotype (rs677642–rs877137, G–G; OR=4.47, CI 2.30–8.69, P<0.000005, Pcorrected<0.005; Figure 1a, Table 1a). This haplotype spans 166 kbps from intron 7 of the gene coding neural cell adhesion molecule 1 (NCAM1), encompasses the tetratricopeptide repeat domain 12 (TTC12) and the ankyrin repeat and kinase domain containing 1 (ANKK1), and is adjacent to the 5′ untranslated region of the dopamine receptor D2 (DRD2; Figure 1a). To avoid the potential effects of genetic stratification, a known cause of type I error, we performed a transmission disequilibrium test analysis in the entire set of multigenerational, nuclear and trio Paisa families. The OR for the transmission of the susceptibility variants on 4q and 11q is 3.14 (95% CI=1.49–6.62; P<0.0027) compared with transmission of neither variant (Table 1b). Both the association analysis and the transmission disequilibrium test demonstrate that the significance of the association on chromosome 11q is lost when not conditioning on the presence of the susceptibility variant within LPHN3.
Table 1. (a) Results of a case–control association analysis conditioning on individuals with the susceptibility variant within LPHN3 (P<0.000005, Pcorrected<0.005), with 48 affected cases and 39 unaffected controls; (b) results of a TDT analysis in trios derived from nuclear families from the Paisa genetic isolate, Germany and two primarily European American samples (USA1—National Human Genome Research Institute, Bethesda, MD, USA; USA2—Children's Hospital of Philadelphia, Philadelphia, PA, USA); (c) results of the fixed-effect meta-analysis on the four samples.
| (a)a | ||||||
|---|---|---|---|---|---|---|
| LPHN3b | 11q haplotypec | Cases (frequency, %) | Controls (frequency, %) | OR (95% CI) | P-value | |
| G | GG | 55 (57) | 18 (23) | 4.47 (2.30, 8.69) | P<0.000005, | |
| G | Else | 41 (43) | 60 (77) | Pcorrected<0.005 | ||
| (b)d | ||||||
|
Sample |
LPHN3b |
11q haplotypee |
Transmitted (frequency, %) |
Not transmitted (frequency, %) |
OR (95% CI) |
P-value |
| Paisa | G | GG | 31 (10.2) | 10 (3.3) | 3.14 (1.49, 6.62) | 0.0027 |
| G | Else | 39 (12.9) | 27 (8.9) | 1.46 (0.85, 2.51) | 0.1661 | |
| Else | GG | 78 (25.7) | 108 (35.8) | 0.73 (0.51, 1.06) | 0.0943 | |
| Else | Else | 155 (51.2) | 157 (52) | Reference | Reference | |
| German | G | GG | 26 (7.9) | 13 (3.9) | 1.91 (0.95, 3.84) | 0.0706 |
| G | Else | 58 (17.6) | 50 (15.2) | 1.11 (0.72, 1.71) | 0.6505 | |
| Else | GG | 75 (22.7) | 104 (31.5) | 0.69 (0.48, 0.99) | 0.0449 | |
| Else | Else | 171 (51.8) | 163 (49.4) | Reference | Reference | |
| US1 | G | GG | 9 (7.9) | 3 (2.6) | 3.1 (0.8, 12) | 0.1014 |
| G | Else | 19 (16.7) | 16 (13.9) | 1.23 (0.58, 2.59) | 0.5955 | |
| Else | GG | 23 (20.2) | 31 (27) | 0.77 (0.4, 1.45) | 0.4139 | |
| Else | Else | 63 (55.3) | 65 (56.5) | Reference | Reference | |
| US2 | G | AG | 22 (5.7) | 9 (2.4) | 2.28 (1.03, 5.08) | 0.0432 |
| G | Else | 108 (28.4) | 123 (32.4) | 0.82 (0.59, 1.13) | 0.2279 | |
| Else | AG | 40 (10.5) | 52 (13.7) | 0.72 (0.46, 1.13) | 0.1306 | |
| |
Else |
Else |
210 (55.3) |
196 (51.6) |
Reference |
Reference |
| (c) | ||||||
|
Meta-analysis |
|
|
OR (95% CI) |
|
|
P-value |
| Interaction | 2.46 (1.68, 3.70) | <0.00001 | ||||
| LPHN3 onlyb | 1.04 (0.86, 1.25) | 0.7111 | ||||
| Haplotype onlye | 0.73 (0.61, 0.87) | <0.001 | ||||
Abbreviations: CI, confidence interval; OR, odds ratio; TDT, transmission disequilibrium test.
Demonstrates an OR of 4.47 (2.30–8.69) for having both susceptibility variants compared with the variant within LPHN3 itself.
Defined by the marker rs6551665 in chromosome 4.
Defined by the markers rs677642 and rs877137 in chromosome 11.
For the Paisa sample the OR for the transmission of the susceptibility variants on 4q and 11q is 3.14 (95% CI=1.49–6.62) compared with transmission of neither variant, 2.15 (95% CI=0.9–5.1) compared with transmission solely of the susceptibility variant on 4q and 4.3 (95% CI=2.0–9.3) compared with the sole transmission of the susceptibility variant on 11q. For the German sample, the OR for the transmission of the susceptibility variants on 4q and 11q is 1.91 (95% CI=0.95–3.84) compared with transmission of neither variant, 1.72 (95% CI=0.80–3.71) compared with transmission solely of the susceptibility variant on 4q and 2.78 (95% CI=1.34–5.75) compared with the sole transmission of the susceptibility variant on 11q. For the USA1 sample, the OR for the transmission of the susceptibility variants followed the same trend as the other samples where on 4q and 11q, it is 3.10 (0.80–12) compared with transmission of neither variant, 2.53 (95% CI=0.58–10.95) compared with transmission solely of the susceptibility variant on 4q and 4.04 (95% CI=0.98–16.62) compared with the sole transmission of the susceptibility variant on 11q. For the USA2 sample, we see over-transmission of the susceptibility variants (P<0.04). The OR for variants on 4q and 11q is 2.28 (95% CI=1.03–5.08) compared with neither variant transmitted, 2.78 (95% CI=1.2–6.3) compared with transmission of only the susceptibility variant on 4q and 3.2 (95% CI=1.3–7.6) compared with transmission of only the susceptibility variant on 11q.
Defined by the markers rs677642 and rs877137 in chromosome 11 except in the US2 sample, for which it is defined by the markers rs754672 and rs965560.
Note: TDT results after a combination of 4q–11q variants originates a definitive decrease of the original sample size, reported by Arcos-Burgos et al. (2010). The decrease of the sample size occurs as a consequence of the ad hoc strategy of conditioning on the fact of being a carrier of the G variant of susceptibility at rs6551665. Furthermore, because of genotype limitations, the Norwegian and Spaniard samples were not genotyped for markers in 11q, and given that TDT was selected to evaluate interaction effects, much information contained in the original family structure sample was lost.
Looking for replication, we performed a transmission disequilibrium test analyses for three additional samples: one from Germany and two primarily European-American samples consisting of 95 trios collected at the National Human Genome Research Institute, Bethesda, MD, USA (US1) and 240 trios from a sample collected at Children's Hospital of Philadelphia, Philadelphia, PA, USA (US2; Table 1b). All these three samples were used for the replication of the LPHN3 association to ADHD.8 The US2 sample was not genotyped at identical SNPs on 11q, so we tested two tag-SNPs that fully describe the variation (r2=1, rs754672, rs965560) for the CEU sample (The International HapMap Consortium, 2005; http://hapmap.ncbi.nlm.nih.gov). Overall, all of these results show a similar pattern of interaction and suggest that the haplotype on 11q interacts cooperatively with the LPHN3 susceptibility variant to increase the risk to ADHD. A meta-analysis of the transmission disequilibrium test results from the four samples, using a random effects model, demonstrated a significant association to the transmission of both susceptibility variants on chromosome 4q and 11q (OR=2.46, 95% CI=1.63–3.70, P<0.00001; Figure 1b and Table 1c).
Effects of the LPHN3–11q interaction on brain metabolism and ADHD
To define how the above described LPHN3–11q interaction modulates the original effects of the LPHN3 susceptibility variant on brain metabolism, we next examined proton magnetic resonance spectroscopy (1H-MRS) data of 18 individuals from the Paisa genetic isolate to four metabolites, namely, N-acetylaspartate, myoinositol, choline and glutamine (for all taken as the ratio to creatine), in several brain regions making up part of the frontal–striatal–cerebellar circuit.
The full two-locus interaction model9, 10 was fit to the data by using linear regression, where y is the quantitative MRS metabolite phenotype, μ is the mean effect, A is the age at diagnosis, S is a code for gender (males=0, females=1), D describes disease status (unaffected=0, affected=1), xi, i=1, 2, is a variable modeling an additive effect (−1 for homozygote for allele 1, 0 for a heterozygote and 1 for a homozygote for allele 2), zi is a dummy variable for a dominant effect (−0.5 for homozygote for allele 1, 0.5 for a heterozygote and −0.5 for a homozygote for allele 2), ai and di refer to additive and dominant coefficients estimated for the single locus effect, and iaa, iad, ida and idd represent epistatic coefficients in the following model:
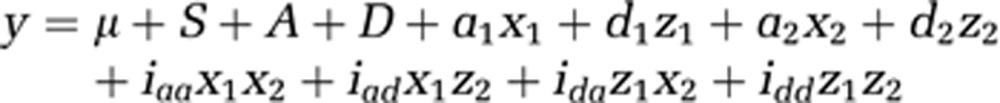 |
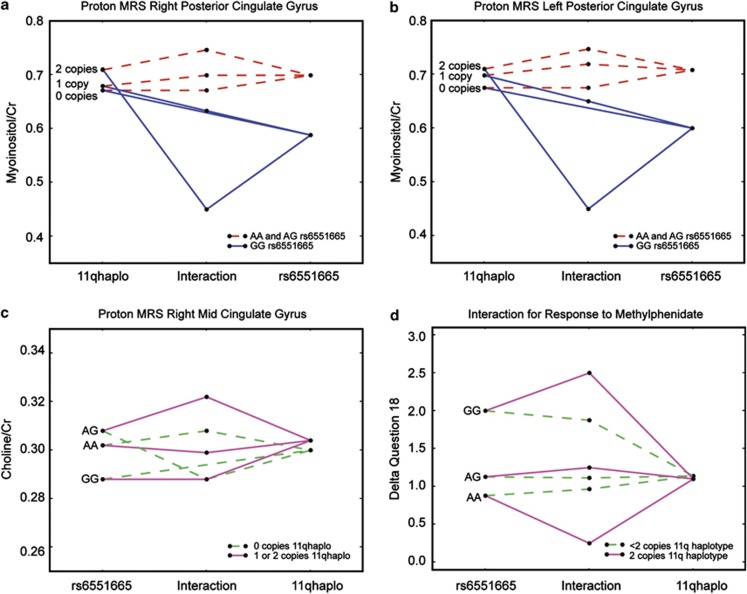
This model was compared with a nested model lacking interaction coefficients by a likelihood ratio test (LRT) that follows a χ2 distribution with four degrees of freedom. Three regions gave results that were significant after permutation (Ppermuted<0.05). They are myoinositol in the right posterior cingulate gyrus, myoinositol in the left posterior cingulate gyrus and choline in the right medial cingulate gyrus. Examining the coefficients for each disclosed that for myoinositol in the right and left posterior cingulate, the iad coefficient for an interaction between an additive effect from the haplotype on 11q and a dominant effect from rs6551665 was contributing to the better fitting model (iad left cingulate gyrus P=0.00342; iad right cingulate gyrus P=0.00298). Thus, the means were plotted for an additive effect from the 11q haplotype and a dominant effect from rs6551665 (where values from individuals that were AA or AG at rs6551665 were grouped). The results demonstrate that having two copies of the susceptibility haplotype on chromosome 11 and GG at rs6551665 correlates with a significant decrease in myoinositol in the two regions (Figure 2a and b). The results for choline in the right medial cingulate demonstrated that the iad coefficient for an interaction between an additive effect from rs6551665 and a dominant effect from the 11q haplotype was contributing to the better fitting model (P=0.00968). The results were plotted for an additive effect from rs6551665 and a dominant effect from the haplotype on 11q (where having one or two copies of the susceptibility haplotype were grouped). The results demonstrate that having AG at rs6551665 and at least one copy of the susceptibility haplotype on 11q is related to an increase in choline in the right medial cingulate and that having AG at rs6551665 and no copies of the susceptibility haplotype on 11q is related to a decreased level of choline in the right medial cingulate region (Figure 2c).
Figure 2.
(a) Fitting of myoinositol/creatinine levels in the right posterior cingulated gyrus resulted in a significant improvement when modeled with a two-loci model compared to a single, main-effects model (Ppermuted<0.05). (b) Fitting of myoinositol/creatinine levels in left posterior cingulate gyrus resulted in a significant improvement when modeled with a two-loci model compared with a single, main-effects model (Ppermuted<0.05). (c) Fitting of choline/creatinine levels in the right medial cingulate gyrus resulted in a significant improvement when modeled with a two-loci model compared with a single, main-effects model (Ppermuted<0.05). (d) Results for question 18 on the SWAN scale (hyperactive question; ‘butting into conversations') and response to methylphenidate demonstrates significant two locus model effect compared with single locus effects (Ppermuted=0.0036). In all, 82 individuals from the USA1 sample were included. The y-axis represents improvement in the symptom after taking stimulant medication. The iad coefficient for an interaction between an additive effect from chromosome 4 and a dominant effect from chromosome 11 indicated the better fitting model (P=0.027).
Effects of the LPHN3–11q interaction on ADHD pharmacogenetics
We next attempted to study the effect of the LPHN3–11q interaction with regards to the pharmacogenetic consequences that the LPHN3 ADHD-susceptibility variant had on the treatment response to stimulant medication. In all, 82 individuals with complete genotype and phenotype information from the US1 sample were included in the analysis. These individuals were sampled from the original 240 individual sample described by our group with complete demographic information of this sample presented in the Supplementary Information.8 In addition, comparative analyses regarding stimulant treatment have been also presented.8 Then we examined the relationship of SWAN scale questions individually as well as inattentive and hyperactive combined dimensions (questions 1–9 indicate inattentive symptoms and questions 10–18 indicate hyperactive symptoms before and after starting methylphenidate medication). We observed a significantly better fitting model for question 18 (hyperactive–impulsive dimension) when the iad coefficient for interaction between additive effects at LPHN3 and dominant effects at 11q are included (Pcorrected=0.0036). Overall results demonstrate that having GG at rs6551665 and two copies of the susceptibility haplotype on 11q is correlated with a significant improvement of symptoms after the treatment with stimulant medication, and that having AA at rs6551665 and fewer than two copies of the susceptibility haplotype on 11q is correlated with a poor response to stimulant medication treatment (Figure 2d).
Although statistically significant, the importance of the genetic LPHN3–11q interaction regarding the treatment response to stimulant medication requires larger studies to both replicate and assess the contribution of this interaction to ADHD symptoms and response to stimulant medication.
Discussion
Variants within LPHN3 were recently demonstrated to be associated with ADHD in samples derived from various populations.8 The demonstrated OR was ∼1.2 (95% CI=1.09–1.38, P<5.42 × 10−4) for having ADHD in individuals carrying susceptibility variants within LPHN3. In this analysis, when rs6551665 within LPHN3 is examined concomitantly with the haplotype on 11q the OR is 2.46 (95% CI=1.68–3.70, P<0.00001). The haplotype on 11q by itself does not exhibit main effects. However, the association with ADHD is demonstrated when conditioning on the susceptibility variant within LPHN3. Thus, the haplotype on 11q seems to function as a modifier of LPHN3 susceptibility.
The haplotype on chromosome 11q spans 166 kbps, from intron 7 of NCAM1, encompasses TTC12 and ANKK1 and is adjacent to DRD2. There is evidence that variation in this region is associated with human behavior. SNPs and haplotypes across this identical region were previously demonstrated to be associated with nicotine, alcohol and drug dependence. Strongest signals were located within TTC12 and/or ANKK1.11, 12, 13 The function of TTC12 is not well described, although expression has been reported in testis, prostate, lung, lymphocytes and numerous tumors.14, 15 Although there are conflicting results for replication, DRD2 is a putative candidate gene for ADHD given its involvement in dopamine metabolism.16 Of note, the commonly tested polymorphism DRD2 Taq1A was found to be within exon 8 of ANKK1, where it causes a glutamine to lysine amino acid change.17 ANKK1 was found to be expressed in placenta and spinal cord but to date has not been observed in the developing or adult brain.17 NCAM1 is a member of the immunoglobin superfamily and is thought to have a fundamental role in normal neural development, function and plasticity.18, 19 Mouse knockout models of NCAM1 have been described to mimic schizophrenia based on brain morphological changes as well as reduced prepulse inhibition of the startle response.20, 21 Furthermore, NCAM1 mouse models have also been demonstrated to have hyperactivity, aggression, anxiety and abnormal social behaviors.22 In humans, genetic variation has been suggested to be associated with neural tube defects and bipolar disorder.23, 24 Isoform variation in postmortem brain and cerebrospinal fluid has been described to be associated with schizophrenia, bipolar disorder and autism, suggesting NCAM1 has many pleiotropic effects.25 Although we have genetic evidence for locus interaction in the development of ADHD, the specific biological mechanism and gene products are not clear. Characterization of homeostatic and pathophysiological mechanisms of gene products on 11q and LPHN3 are necessary in order to determine the precise mechanistic interaction and any discussion of the mechanism until then is purely speculative.
The fact that the LPHN3–11q interaction better describes the proton MRS metabolite differences in the cingulate gyrus, with the strongest findings demonstrated for myoinositol in the posterior cingulate gyrus, might be compatible with recent findings of gray matter volume reduction in ADHD patients in the posterior cingulate26 and with reduced activation of the posterior cingulate gyrus detected by functional magnetic resonance imaging in ADHD patients during sustained tasks and inhibition failure.27 Overall this region is hypothesized to be important for the interplay of attention and motivation and in particular visuospatial attention. It is worth mentioning that the main difference regarding 1H-MRS data between this report and the original report of Arcos-Burgos et al.4 is that additional brain metabolites were known at the time of the making of this report. Analyses of association of one locus marker data to 1H-MRS data are part of a different manuscript that is currently prepared for submission. Evaluation of 1H-MRS data in the context of two-locus interaction had the main goal to show that the two-locus interaction was able of predict genetic effects on brain metabolism much better than a model of one-locus.
As exemplified by the few reproducible genes discovered to date for ADHD, there are numerous difficulties that must be overcome in mapping disorders that are thought to have complex origins, including low penetrance, heterogeneity, pleiotropy and epistasis. Assaying and accounting for these potential complexities are of extreme importance when searching for the specific genetic variations that underlie disease susceptibility. In this case, accounting for epistasis disclosed a variant that would have been missed by traditional association analysis. In summary, we demonstrate that an interaction between rs6551665 within LPHN3 and a region on chromosome 11q is involved in ADHD susceptibility and substantially increases the OR for having ADHD compared to examining the LPHN3 common variant associated with ADHD on its own.
Acknowledgments
We are grateful to the families who participated in this research endeavor. This research was supported by the Division of Intramural Research, National Human Genome Research Institute, National Institutes of Health, and in part by COLCIENCIAS, grants 1115-04-12010, 1115-04-18083, and by the Deutsche Forschungsgemeinschaft (KFO 125/1-1, SFB 581).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on the Molecular Psychiatry website (http://www.nature.com/mp)
Supplementary Material
References
- Arcos-Burgos M, Castellanos FX, Pineda D, Lopera F, Palacio JD, Palacio LG, et al. Attention-deficit/hyperactivity disorder in a population isolate: linkage to loci at 4q13.2, 5q33.3, 11q22, and 17p11. Am J Hum Genet. 2004;75:998–1014. doi: 10.1086/426154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arcos-Burgos M, Acosta MT. Tuning major gene variants conditioning human behavior: the anachronism of ADHD. Curr Opin Genet Dev. 2007;17:234–238. doi: 10.1016/j.gde.2007.04.011. [DOI] [PubMed] [Google Scholar]
- Cox NJ, Frigge M, Nicolae DL, Concannon P, Hanis CL, Bell GI, et al. Loci on chromosomes 2 (NIDDM1) and 15 interact to increase susceptibility to diabetes in Mexican Americans. Nat Genet. 1999;21:213–215. doi: 10.1038/6002. [DOI] [PubMed] [Google Scholar]
- Arcos-Burgos M, Jain M, Acosta MT, Shively S, Stanescu H, Wallis D, et al. A common variant of the latrophilin 3 gene, LPHN3, confers susceptibility to ADHD and predicts effectiveness of stimulant medication. Mol Psychiatry. 2010;15:1053–1066. doi: 10.1038/mp.2010.6. [DOI] [PubMed] [Google Scholar]
- Kruglyak L, Daly MJ, Reeve-Daly MP, Lander ES. Parametric and nonparametric linkage analysis: a unified multipoint approach. Am J Hum Genet. 1996;58:1347–1363. [PMC free article] [PubMed] [Google Scholar]
- Strauch K, Fimmers R, Kurz T, Deichmann KA, Wienker TF, Baur MP. Parametric and nonparametric multipoint linkage analysis with imprinting and two-locus-trait models: application to mite sensitization. Am J Hum Genet. 2000;66:1945–1957. doi: 10.1086/302911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietter J, Spiegel A, an Mey D, Pflug HJ, Al-Kateb H, Hoffmann K, et al. Efficient two-trait-locus linkage analysis through program optimization and parallelization: application to hypercholesterolemia. Eur J Hum Genet. 2004;12:542–550. doi: 10.1038/sj.ejhg.5201196. [DOI] [PubMed] [Google Scholar]
- Arcos-Burgos M, Jain M, Acosta MT, Shively S, Stanescu H, Wallis D, et al. A common variant of the latrophilin 3 gene, LPHN3, confers susceptibility to ADHD and predicts effectiveness of stimulant medication. Mol Psychiatry. 2010;15:1053–1066. doi: 10.1038/mp.2010.6. [DOI] [PubMed] [Google Scholar]
- Cordell HJ. Epistasis: what it means, what it doesn't mean, and statistical methods to detect it in humans. Hum Mol Genet. 2002;11:2463–2468. doi: 10.1093/hmg/11.20.2463. [DOI] [PubMed] [Google Scholar]
- Cordell HJ, Todd JA, Hill NJ, Lord CJ, Lyons PA, Peterson LB, et al. Statistical modeling of interlocus interactions in a complex disease: rejection of the multiplicative model of epistasis in type 1 diabetes. Genetics. 2001;158:357–367. doi: 10.1093/genetics/158.1.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang BZ, Kranzler HR, Zhao H, Gruen JR, Luo X, Gelernter J. Haplotypic variants in DRD2, ANKK1, TTC12, and NCAM1 are associated with comorbid alcohol and drug dependence. Alcohol Clin Exp Res. 2008;32:2117–2127. doi: 10.1111/j.1530-0277.2008.00800.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang BZ, Kranzler HR, Zhao H, Gruen JR, Luo X, Gelernter J. Association of haplotypic variants in DRD2, ANKK1, TTC12 and NCAM1 to alcohol dependence in independent case control and family samples. Hum Mol Genet. 2007;16:2844–2853. doi: 10.1093/hmg/ddm240. [DOI] [PubMed] [Google Scholar]
- Gelernter J, Yu Y, Weiss R, Brady K, Panhuysen C, Yang BZ, et al. Haplotype spanning TTC12 and ANKK1, flanked by the DRD2 and NCAM1 loci, is strongly associated to nicotine dependence in two distinct American populations. Hum Mol Genet. 2006;15:3498–3507. doi: 10.1093/hmg/ddl426. [DOI] [PubMed] [Google Scholar]
- Katoh M. Identification and characterization of human FOXN5 and rat Foxn5 genes in silico. Int J Oncol. 2004;24:1339–1344. [PubMed] [Google Scholar]
- Katoh M. Identification and characterization of TPARM gene in silico. Int J Oncol. 2003;23:1213–1217. [PubMed] [Google Scholar]
- Esposito-Smythers C, Spirito A, Rizzo C, McGeary JE, Knopik VS. Associations of the DRD2 TaqIA polymorphism with impulsivity and substance use: preliminary results from a clinical sample of adolescents. Pharmacol Biochem Behav. 2009;93:306–312. doi: 10.1016/j.pbb.2009.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neville MJ, Johnstone EC, Walton RT. Identification and characterization of ANKK1: a novel kinase gene closely linked to DRD2 on chromosome band 11q23.1. Hum Mutat. 2004;23:540–545. doi: 10.1002/humu.20039. [DOI] [PubMed] [Google Scholar]
- Ronn LC, Olsen M, Soroka V, Østergaard S, Dissing S, Poulsen FM, et al. Characterization of a novel NCAM ligand with a stimulatory effect on neurite outgrowth identified by screening a combinatorial peptide library. Eur J Neurosci. 2002;16:1720–1730. doi: 10.1046/j.1460-9568.2002.02242.x. [DOI] [PubMed] [Google Scholar]
- Ronn LC, Berezin V, Bock E. The neural cell adhesion molecule in synaptic plasticity and ageing. Int J Dev Neurosci. 2000;18:193–199. doi: 10.1016/s0736-5748(99)00088-x. [DOI] [PubMed] [Google Scholar]
- Wood SJ, Toth M. Molecular pathways of anxiety revealed by knockout mice. Mol Neurobiol. 2001;23:101–119. doi: 10.1385/MN:23:2-3:101. [DOI] [PubMed] [Google Scholar]
- Wood GK, Tomasiewicz H, Rutishauser U, Magnuson T, Quirion R, Rochford J, et al. NCAM-180 knockout mice display increased lateral ventricle size and reduced prepulse inhibition of startle. Neuroreport. 1998;9:461–466. doi: 10.1097/00001756-199802160-00019. [DOI] [PubMed] [Google Scholar]
- Stork O, Welzl H, Wolfer D, Schuster T, Mantei N, Stork S, et al. Recovery of emotional behaviour in neural cell adhesion molecule (NCAM) null mutant mice through transgenic expression of NCAM180. Eur J Neurosci. 2000;12:3291–3306. doi: 10.1046/j.1460-9568.2000.00197.x. [DOI] [PubMed] [Google Scholar]
- Arai M, Itokawa M, Yamada K, Toyota T, Haga S, Ujike H, et al. Association of neural cell adhesion molecule 1 gene polymorphisms with bipolar affective disorder in Japanese individuals. Biol Psychiatry. 2004;55:804–810. doi: 10.1016/j.biopsych.2004.01.009. [DOI] [PubMed] [Google Scholar]
- Deak KL, Boyles AL, Etchevers HC, Melvin EC, Siegel DG, Graham FL, et al. SNPs in the neural cell adhesion molecule 1 gene (NCAM1) may be associated with human neural tube defects. Hum Genet. 2005;117:133–142. doi: 10.1007/s00439-005-1299-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vawter MP. Dysregulation of the neural cell adhesion molecule and neuropsychiatric disorders. Eur J Pharmacol. 2000;405:385–395. doi: 10.1016/s0014-2999(00)00568-9. [DOI] [PubMed] [Google Scholar]
- Carmona S, Vilarroya O, Bielsa A, Tremols V, Soliva JC, Rovira M, et al. Global and regional gray matter reductions in ADHD: a voxel-based morphometric study. Neurosci Lett. 2005;389:88–93. doi: 10.1016/j.neulet.2005.07.020. [DOI] [PubMed] [Google Scholar]
- Rubia K, Smith AB, Halari R, Matsukura F, Mohammad M, Taylor E, et al. Disorder-specific dissociation of orbitofrontal dysfunction in boys with pure conduct disorder during reward and ventrolateral prefrontal dysfunction in boys with pure ADHD during sustained attention. Am J Psychiatry. 2009;166:83–94. doi: 10.1176/appi.ajp.2008.08020212. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.