Abstract
Quantum-chemical computations were used to investigate the structure–antioxidant parameter relationships of α-lipoic acid and its natural metabolites bisnorlipoic acid and tetranorlipoic acid in their oxidized and reduced forms. The enantiomers of lipoic and dihydrolipoic acid were optimized using the B3LYP/6-311+G(3df,2p), B3LYP/aug-cc-pVDZ and MP2(full)/6-31+G(d,p) levels of theory as isolated molecules and in the presence of water. The geometries of the metabolites and the values of their antioxidant parameters (proton affinity, bond dissociation enthalpy, adiabatic ionization potential, spin density, and the highest occupied molecular orbital energy) were calculated at the B3LYP/6-311+G(3df,2p) level of theory. The results obtained reveal similarities between these structures: a pentatomic, nonaromatic ring is present in the oxidized forms, while an unbranched aliphatic chain (as found in saturated fatty acids) is present in both the oxidized and the reduced forms. Analysis of the spin density and the highest occupied molecular orbital energy revealed that the SH groups exhibited the greatest electron-donating activities. The values obtained for the proton affinity, bond dissociation enthalpy and adiabatic ionization potential indicate that the preferred antioxidant mechanisms for α-lipoic acid and its metabolites are sequential proton loss electron transfer in polar media and hydrogen atom transfer in vacuum.
Electronic supplementary material
The online version of this article (doi:10.1007/s00894-011-1306-y) contains supplementary material, which is available to authorized users.
Keywords: α-Lipoic acid, Antioxidant, Metabolites, DFT method, MP2 method, C-PCM model
Introduction
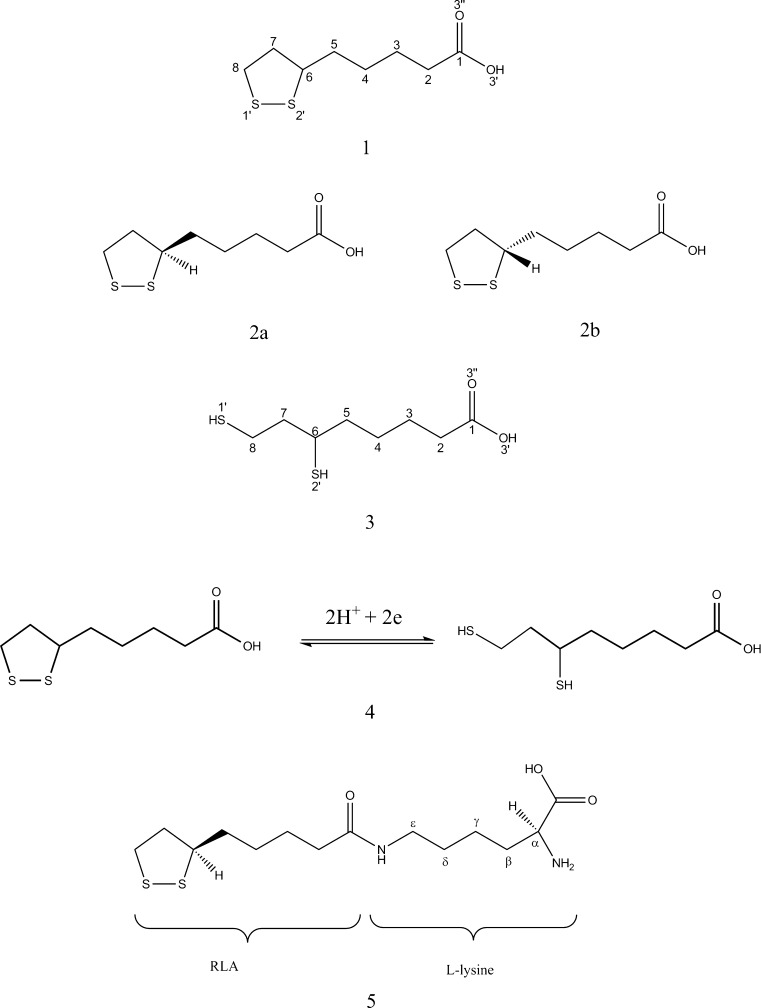
α-Lipoic acid [LA, IUPAC: 5-(1,2-dithiolan-3-yl)-pentanoic acid; 1 in Fig. 1] is an organosulfur compound that is synthesized in small amounts by microorganisms, plants, animals, and humans [1, 2]. It is one of the most important cofactors of the multi-enzyme complexes in mitochondria. In the peptide chain, LA is covalently bound to the nitrogen atom of the lysine, forming lipoyllysine (LA-Lys; 5 in Fig. 1). Physiological demand for LA is met by its synthesis in every organism. While there is currently no record of a disease caused by LA deficiency [3], the usage of LA as a dietary supplement has recently become one of the most popular topics in the scientific and medical worlds. The largest amounts of LA-Lys can be found in the kidneys, liver, heart, and in spinach and broccoli (∼1–3 μg/g) [4].
Fig. 1.
Molecular structures of lipoic acid (1), the enantiomers of lipoic acid: R-(+)-LA (2a) and S-(−)-LA (2b), dihydrolipoic acid (3), the LA/DHLA redox pair (4), and lipoyllysine (5)
α-Lipoic acid was discovered by L.J. Reed during studies aimed at isolating growth-stimulating factor for Lactobacillus casei (1949–1951; note that the prefix α has no chemical meaning, but it is helpful in distinguishing the isolated compound from structurally related forms in biological extracts [5]). The crystallographic structure of LA was determined by Stroud and Carlisle 20 years later [6]. LA is a derivative of octanoic acid with two sulfur atoms attached to carbon atoms C6 and C8. The C6 atom is the chiral center of the molecule, so LA has two optical isomers—the enantiomers R-(+)- and S-(−)- (2a and 2b in Fig. 1, respectively)—and it also exists as a racemic mixture (R/S-LA). However, only the R-(+)-enantiomer is synthesized by organisms and biologically active. In its oxidized form, LA has a characteristic pentatomic ring, as the two sulfur atoms are connected via a disulfide bond (S–S). The oxidized form can be reduced to an open-chain structure with two sulfhydryl groups: dihydrolipoic acid (DHLA; see 3 in Fig. 1). The redox pair LA/DHLA (4 in Fig. 1) interacts with different ROSs (reactive oxygen species), RNSs (reactive nitrogen species), as well as other oxidants and compounds with thiol groups, such as glutathione, cysteine, and N-acetylcysteine [7–9].
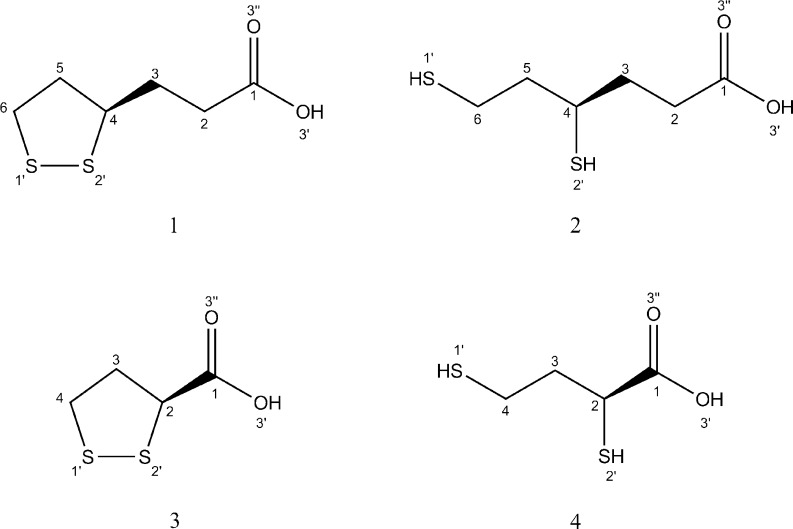
RLA is a multifunctional therapeutic compound which has shown activity in many experimental in vitro and in vivo studies. RLA/RDHLA is one of the strongest natural antioxidants, and is capable of deactivating various free radicals such as the superoxide anion (O2 • –), the hydroxyl radical (•OH), singlet oxygen (1O2), peroxynitrite (ONOO–), and hypochlorous acid (HClO) [10–12]. It is also a very important regenerating factor for other antioxidants: thioredoxin, vitamin E, vitamin C, and glutathione (GSH) [13–16]. It has been proven that α-lipoic acid can induce GSH synthesis [17] and act as a metal-chelating agent [18–22] and intercellular signal-transducing factor [7, 23]. RLA is viewed as a very promising therapeutic and nutritional supplement. On the basis of clinical studies, it was shown that adding RLA to the therapy reduces the symptoms of rheumatoid arthritis [7], cardiovascular diseases [24–31], diabetic polyneuropathy [32], multiple sclerosis [33–36], and Alzheimer’s disease [7]. The biological and therapeutical properties of RLA are dependent on its bioavailability, tissue accumulation, and the process of metabolization [7]. RLA and RDHLA can be β-oxidized and/or S-methylated in cells [37, 38]. The most common RLA/RDHLA metabolites are presented in Fig. 2.
Fig. 2.
Molecular structures of RLA/RDHLA metabolites: bisnorlipoic acid (RBLA, 1), 4,6-bisthiohexanoic acid (RDHBLA, 2), tetranorlipoic acid (STLA, 3), and 2,4-bisthiobutanoic acid (SDHTLA, 4)
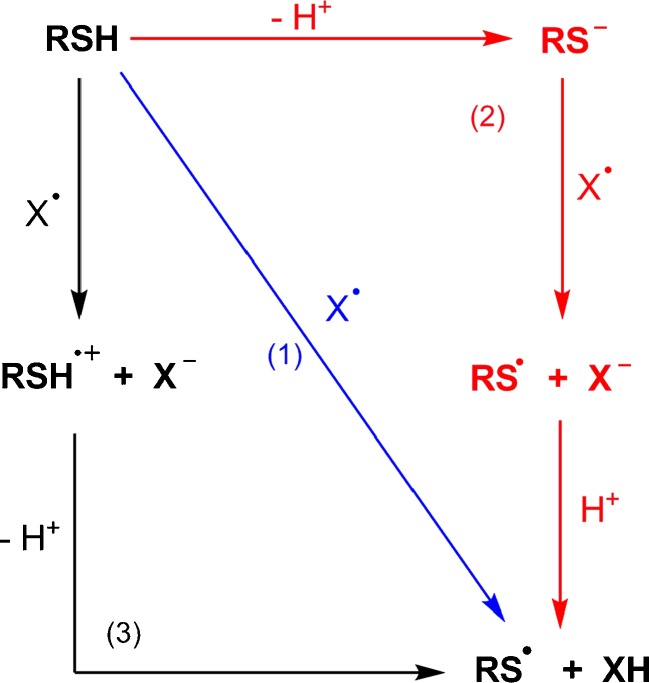
It is well known that free radicals can be deactivated in interactions with phenolic antioxidants via three mechanisms [39]. However, due to the similarity of O–H and S–H bonds, these processes can also be used to describe the antioxidant action of organosulfur compounds such as α-lipoic acid and its metabolites (Fig. 3). We can therefore distinguish the following mechanisms of free-radical scavenging:
HAT (hydrogen atom transfer) mechanism:
 |
1.1 |
-
2.
SPLET (sequential proton loss electron transfer) mechanism:
 |
1.2 |
 |
1.3 |
-
3.
SET-PT (single electron transfer followed by proton transfer) mechanism:
 |
1.4 |
 |
1.5 |
Fig. 3.
Antioxidant mechanisms of organosulfur compounds: HAT (1), SPLET (2), SET-PT (3). RSH organosulfur antioxidant, X • free radical. From [39]
There have been numerous publications describing experimental studies of the antioxidant properties of LA/DHLA, but this compound has not yet been investigated theoretically. In this paper, we report our detailed analysis of several of the molecular properties of LA/DHLA and RLA/RDHLA metabolites (RBLA/RDHBLA and STLA/SDHTLA), such as their molecular geometries and total molecular energies. We also describe the antioxidant properties of the compounds studied, which were derived by calculating physical antioxidant descriptors such as the adiabatic ionization potential (AIP), the bond dissociation enthalpy (BDE), the proton affinity (PA), the highest occupied molecular orbital energy (E HOMO), and the spin density (SD) [40–44]. Because the free-radical scavenging action occurs in aqueous solution, we account for the solvating effect of water in our quantum-mechanical computations. Moreover, based on the calculations performed, we predict which of the three antioxidant mechanisms specified above is preferred for each of the compounds studied.
Computational methods
The quantum-chemical computations were performed using the Gaussian 03 W software package [45]. Full optimization of the geometries and energies for all of the molecules studied was carried out in the gas phase and in water without symmetry constraints. The geometries of the neutral compounds in their ground states were optimized using DFT with the restricted rB3LYP hybrid functional [46–48] combined with the 6-311 + G(3df,2p) and aug-cc-pVDZ basis sets. The geometry optimization of mono-, di-, and trianions was carried out with the rB3LYP/6-311 + G(df,2p) method. To optimize the radicals and cation radicals, the unrestricted uB3LYP/6-311 + G(3df,2p) level of theory was applied, since it gives the best results for open-shell molecular systems. Additionally, the neutral forms of LA and DHLA enantiomers were optimized using second-order Møller–Plesset perturbation theory [49] combined with the 6-31 + G(d,p) basis set in vacuum and water. rMP2(full)/6-31 + G(d,p) was applied to achieve a better description of electron correlation effects in the compounds studied. Aside from analyzing the geometries, the single point energies for RLA and SLA were calculated using DFT and MP2(full) methods combined with a wide spectrum of basis sets [6-31G(d,p), 6-31+G(d,p), 6-31++G(d,p), 6-311G(d,p), 6-311+G(d,p), 6-311++G(d,p), 6-31G(3df,2p), 6-31+G(3df,2p), 6-31++G(3df,2p), 6-311G(3df,2p), 6-311+G(3df,2p), 6-311++G(3df,2p), cc-pVDZ, cc-pVTZ, cc-pVQZ, aug-cc-pVDZ, and aug-cc-pVTZ] in order to compare the energies of stereoisomers.
To establish the starting geometries of the compounds studied and the locations of the structures with minimum energy, a conformational analysis was performed. Rotational potential energy profiles at rHF/6-31 + G(d,p) for RLA, RDHLA, SLA, and SDHLA were constructed by scanning the internal dihedral angles (α = C1–C2–C3–C4 and β = C3–C4–C5–C6 for LA, α = C1–C2–C3–C4 and γ = C6–C7–C8–S1′ for DHLA) over the ranges 180° ≤ α ≤ 360°, 180° ≤ β ≤ 360°, 180° ≤ γ ≤ 360°, with no constraint placed on any other geometric parameter (see the “Electronic supplementary material,” ESM). In the next step, the most stable structures obtained from the scan were fully optimized around each potential minimum without symmetry constraints. For all of the optimized structures, harmonic vibrational frequencies were calculated in order to verify the stationary points on the potential energy surfaces (PES).
The antioxidant properties of the molecules studied were described by calculating the numerical parameters (at 298 K) associated with their antioxidant mechanisms (described above), such as their proton affinities (PA; the SPLET mechanism), bond dissociation enthalpies (BDE; the HAT mechanism), and adiabatic ionization potentials (AIP; SET-PT mechanism) [50–55]:
 |
1.6. |
where H a is the enthalpy of the anion, H p is the enthalpy of the proton, and H m is the enthalpy of the parent molecule;
 |
1.7. |
where H r is the enthalpy of the radical, H H is the enthalpy of the H atom, and H m is the enthalpy of the parent molecule; and
 |
1.8. |
where E cr is the total energy of the cation radical, and E m is the total energy of the parent molecule.
In the calculations, the following enthalpy values were used: H(H•)vacuum = −0.49764 Ha (i.e. hartrees), [42], Δhydr H(H•) = −0.00152 Ha [56, 57]; H(H+)vacuum = 0.00236 Ha [40]; Δhydr H(H+) = −0.41516 Ha [58].
Additional parameters describing the electron-donating properties of the compounds studied (the HOMO orbital distribution and the spin density distribution) were calculated at the B3LYP/6-311 + G(3df,2p) level of theory in vacuum and in water.
The optimizations of all forms of the compounds investigated and the calculations of the antioxidant parameters in water were performed using the C-PCM solvation model (conductor-like polarizable continuum model), [59]. In the calculations, a dielectric constant of 78.39 was used for water. This approach was employed and described in our previous works on the antioxidant properties of phenolic compounds (trans-resveratrol and its derivatives) [41, 60–62].
Results and discussion
The optimized geometries of LA and DHLA enantiomers
Optimizing all of the compounds’ geometries at the rB3LYP/6-311 + G(3df,2p), rB3LYP/aug-cc-pVDZ, and rMP2(full)/6-31 + (d,p) levels of theory leads to the absolute energy minima on the potential energy surface, which is confirmed by the absence of imaginary frequencies. The bond lengths (in Å) and the angles (in degrees) of the equilibrium optimized neutral structure of RLA in comparison and the corresponding experimental crystallographic data are summarized in Table 1 and Table S1 of the ESM. It should be pointed out, that regardless of the method and basis set used, the equilibrium values of geometrical parameters are almost identical and very similar to the results obtained by Stroud and Carlisle (1972) for the crystallographic structure of LA [6]. Additionally, the results presented in Tables 2 and 3 and Tables S2 and S3 of the ESM reveal that there are no geometric differences between enantiomers (RLA and SLA; RDHLA and SDHLA, see Fig. 4), which is in good agreement with the statement that optical isomers are like mirror images of each other. The analysis of ΔE total for RLA and SLA, as calculated using DFT and MP2(full) employing a wide range of basis sets (Table 4 and Table S4 of the ESM), confirms that the enantiomers are isoenergetic structures.
Table 1.
The bond lengths (in Å) and angles (in degrees) of RLA [obtained using rMP2(full)/6-31 + (d,p) and optimized in vacuum and water] along with the experimental values from the crystallographic structure of RLA [6]
| Bond length or angle | rMP2(full)/6-31 + G(d,p) | Experiment | |
|---|---|---|---|
| Vacuum | Water | ||
| C1–C2 | 1.50 | 1.50 | 1.50 |
| C2–C3 | 1.52 | 1.52 | 1.55 |
| C3–C4 | 1.52 | 1.52 | 1.53 |
| C4–C5 | 1.53 | 1.53 | 1.51 |
| C5–C6 | 1.52 | 1.52 | 1.53 |
| C6–C7 | 1.54 | 1.54 | 1.55 |
| C7–C8 | 1.54 | 1.54 | 1.51 |
| S1′–C8 | 1.82 | 1.82 | 1.79 |
| S2′–C6 | 1.82 | 1.83 | 1.83 |
| S1′–S2′ | 2.07 | 2.08 | 2.05 |
| O3′–C1 | 1.36 | 1.36 | 1.31 |
| O3″–C1 | 1.22 | 1.22 | 1.20 |
| C1–C2–C3 | 113.02 | 113.46 | 112.99 |
| C2–C3–C4 | 111.92 | 111.70 | 110.39 |
| C3–C4–C5 | 112.17 | 111.93 | 109.28 |
| C4–C5–C6 | 112.46 | 112.18 | 115.38 |
| C5–C6–C7 | 113.48 | 113.56 | 112.89 |
| C6–C7–C8 | 111.86 | 112.29 | 112.61 |
| C7–C8–S1′ | 109.26 | 109.36 | 112.61 |
| C8–S1′–S2′ | 90.22 | 109.36 | 95.56 |
| C6–S2′–S1′ | 90.63 | 90.97 | 92.84 |
| C5–C6–S2′ | 110.46 | 110.27 | 111.37 |
| C7–C6–S2′ | 108.17 | 108.28 | 106.76 |
| O3′–C1–C2 | 111.24 | 111.50 | 113.39 |
| O3″–C1–C2 | 126.16 | 126.09 | 124.89 |
| O3′–C1–O3″ | 122.60 | 122.41 | 121.89 |
Table 2.
The bond lengths (in Å) and angles (in degrees) of RLA and SLA [obtained using rMP2(full)/6-31 + (d,p) and optimized in vacuum and water]
| Bond length or angle | rMP2(full)/6-31 + G(d,p) | |||
|---|---|---|---|---|
| Vacuum | Water | |||
| RLA | SLA | RLA | SLA | |
| C6–C7 | 1.54 | 1.54 | 1.54 | 1.54 |
| C7–C8 | 1.54 | 1.54 | 1.54 | 1.54 |
| S1′–C8 | 1.82 | 1.82 | 1.82 | 1.82 |
| S2′–C6 | 1.82 | 1.83 | 1.83 | 1.83 |
| S1′–S2′ | 2.07 | 2.07 | 2.08 | 2.08 |
| O3′–C1 | 1.36 | 1.36 | 1.36 | 1.36 |
| O3″–C1 | 1.22 | 1.22 | 1.22 | 1.22 |
| C6–C7–C8 | 111.86 | 111.95 | 112.29 | 112.38 |
| C7–C8–S1′ | 109.26 | 109.41 | 109.36 | 109.72 |
| C8–S1′–S2′ | 90.22 | 90.75 | 90.60 | 91.65 |
| S1′–S2′–C6 | 90.63 | 90.25 | 90.97 | 90.29 |
| S2′–C6–C7 | 108.17 | 107.91 | 108.28 | 107.59 |
| C1–C2–C3–C4 | 179.47 | 179.99 | 179.97 | 179.77 |
| C3–C4–C5–C6 | 177.65 | 172.96 | 177.37 | 172.53 |
| C8–S1′–S2′–C6 | 46.99 | 46.76 | 46.02 | 45.35 |
Table 3.
The bond lengths (in Å) and angles (in degrees) of RDHLA and SDHLA [obtained using rMP2(full)/6-31 + (d,p) and optimized in vacuum and water]
| Bond length or angle | rMP2(full)/6-31 + G(d,p) | |||
|---|---|---|---|---|
| Vacuum | Water | |||
| RDHLA | SDHLA | RDHLA | SDHLA | |
| C6–C7 | 1.53 | 1.53 | 1.53 | 1.53 |
| C7–C8 | 1.52 | 1.52 | 1.52 | 1.52 |
| S1′–C8 | 1.83 | 1.82 | 1.83 | 1.82 |
| S1′–H1′ | 1.33 | 1.33 | 1.33 | 1.33 |
| S2′–C6 | 1.83 | 1.83 | 1.83 | 1.83 |
| S2′–H2′ | 1.33 | 1.33 | 1.33 | 1.33 |
| O3′–C1 | 1.36 | 1.36 | 1.36 | 1.36 |
| O3″–C1 | 1.22 | 1.22 | 1.22 | 1.22 |
| C7–C8–S1′ | 110.06 | 108.79 | 110.37 | 108.93 |
| C7–C6–S2′ | 112.73 | 112.94 | 112.74 | 112.78 |
| C6–C7–C8 | 116.05 | 114.00 | 116.34 | 113.88 |
| C1–C2–C3–C4 | 179.97 | 179.86 | 179.90 | 179.71 |
| C5–C6–C7–C8 | 176.62 | 175.94 | 177.79 | 174.01 |
Fig. 4.
The geometries of RLA (1), RDHLA (2), SLA (3), and SDHLA (4) [obtained using rMP2(full)/6-31 + (d,p) and optimized in water]
Table 4.
The total energy values [in Ha] of RLA and SLA [obtained using rMP2(full) and optimized in vacuum]. ΔE total (in kcal mol−1) of RLA and SLA
| Basis set | MP2(full) | ||
|---|---|---|---|
| RLA | SLA | ΔE total | |
| 6-31G(3df,2p) | −1258.253 | −1258.253 | 0.305 |
| 6-31 + G(3df,2p) | −1258.275 | −1258.275 | 0.226 |
| 6-31++G(3df,2p) | −1258.276 | −1258.277 | 0.282 |
| 6-311++G(3df,2p) | −1258.810 | −1258.811 | 0.526 |
| cc-pVDZ | −1257.728 | −1257.728 | 0.447 |
| cc-pVTZ | −1258.507 | −1258.507 | 0.498 |
| cc-pVQZ | −1258.877 | −1258.878 | 0.543 |
| aug-cc-pVDZ | −1257.861 | −1257.861 | 0.369 |
| aug-cc-pVTZ | −1258.597 | −1258.597 | 0.474 |
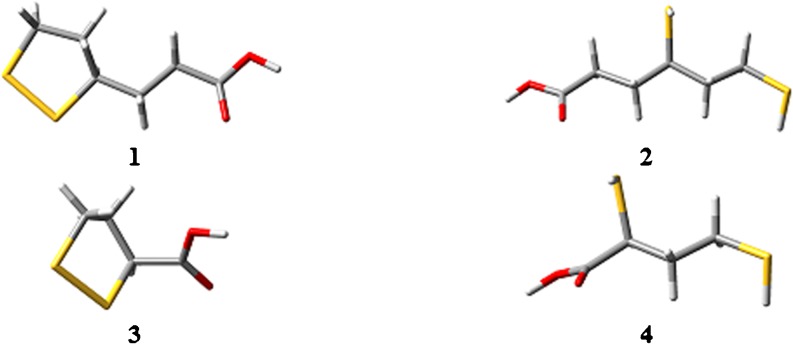
The optimized geometries of RLA/RDHLA metabolites
Optimization of the geometries of the metabolites was performed at the rB3LYP/6-311 + G(3df,2p) level of theory in vacuum and water. The equilibrium geometries (Fig. 5) are at the absolute energy minima on the potential energy surface, as confirmed by the absence of imaginary frequencies. A comparative analysis of the geometrical parameters in vacuum (Table S5 of the ESM) and in water (Table S6 of the ESM) reveals that there is good agreement between the molecular structures of RLA, RDHLA, and their metabolites. All of the compounds studied demonstrate similarities in the geometry of the aliphatic chain. The values of the dihedral angle C*–S1′–S2′–C* in the cyclic forms (where C* has a different number in each cyclic form) indicate that the pentatomic ring is not a planar system but has the conformation of an envelope. The 1′-S–H and 2′-S–H bond lengths of the reduced forms are practically identical in both vacuum and water, which indicates a lack of influence of solvation effects.
Fig. 5.
The geometries of RLA/RDHLA metabolites [obtained using rB3LYP/6-311 + G(3df,2p) and optimized in water]: RBLA (1), RDHBLA (2), STLA (3), and SDHTLA (4)
AIP, BDE, PA, spin density, and HOMO orbital distribution
AIP is a very important physical property that describes the process of single electron donation by the antioxidant. The smaller the AIP value, the greater the chance that ionization will occur. Molecules with low AIP values are considered very good antioxidants. Calculations (Table 5) indicate that the lowest values of AIP in both vacuum and water media are associated with oxidized forms: RLA, RBLA, STLA (in order of increasing AIP) in vacuum and STLA, RLA, RBLA (in order of increasing AIP) in water. In vacuum, the same order can be ascribed to the reduced forms: RDHLA, RDHBLA, SDHTLA, but in water, we observe the following growing order: RDHBLA, RDHLA, SDHTLA. The AIP values in water are lower than in vacuum for all of the compounds studied. This is connected to the strong electrostatic interaction of the polar medium with cation radical forms. Because AIP represents the SET-PT mechanism, it is possible that this mechanism is more preferable in water than in vacuum.
Table 5.
The AIP (in kcal mol−1) and ΔAIP (in kcal mol−1) values [calculated at the B3LYP/6-311 + G(3df,2p) level in vacuum and water] of LA/DHLA enantiomers and RLA/RDHLA metabolites
| Compound | Vacuum | Water | ||
|---|---|---|---|---|
| AIP (kcal/mol) | ΔAIP (kcal/mol) | AIP (kcal/mol) | ΔAIP (kcal/mol) | |
| RLA | 171 | 0 | 131 | 0 |
| SLA | 171 | 0 | 131 | 0 |
| RDHLA | 193 | 22 | 159 | 28 |
| SDHLA | 191 | 21 | 157 | 26 |
| RBLA | 173 | 2 | 132 | 1 |
| RDHBLA | 195 | 24 | 158 | 27 |
| STLA | 193 | 22 | 128 | −3 |
| SDHTLA | 199 | 28 | 162 | 31 |
The stability of the S–H bond can be evaluated using a thermodynamic parameter, the homolytic BDE. Hydrogen atom transfer from antioxidant to radical is a possible antioxidant mechanism. Because the reduced forms of the compounds studied have two S–H bonds, we have taken into consideration all possible reactions. The calculated values of the BDE are presented in Table 6. Based on the results obtained, it is clear that RDHLA and its metabolites have similar S–H bond stabilities. Just as for the AIP results (Table 5), there are no differences between the DHLA stereoisomers in vacuum and in water. In water, the S–H bond is less prone to homolytic fission than in vacuum, because of higher BDE values. This means that the HAT mechanism is not preferable in a polar medium for the compounds studied. Hydrogen atom transfer is hindered because a polar medium increases the stability of S–H bonds.
Table 6.
The BDE (in kcal mol−1) and ΔBDE (in kcal mol−1) values [calculated at the B3LYP/6-311 + G(3df,2p) level in vacuum and water] of RDHLA, SDHLA, RDHBLA, and SDHTLA
| Bond | Vacuum | Water | ||
|---|---|---|---|---|
| BDE (in kcal/mol) | ΔBDE (in kcal/mol) | BDE (in kcal/mol) | ΔBDE (in kcal/mol) | |
| 1′-SH–RDHBLA | 86 | 1 | 96 | 1 |
| 1′-SH–RDHLA | 85 | 0 | 96 | 0 |
| 1′-SH–SDHLA | 85 | 0 | 95 | 0 |
| 1′-SH–SDHTLA | 85 | 1 | 96 | 1 |
| 2′-SH–RDHBLA | 85 | 1 | 97 | 1 |
| 2′-SH–RDHLA | 86 | 1 | 96 | 1 |
| 2′-SH–SDHLA | 86 | 2 | 97 | 2 |
| 2′-SH–SDHTLA | 87 | 2 | 97 | 2 |
The PA values were calculated for the neutral forms of RDHLA, RDHBLA, and SDHTLA, as well as for their 3′-O-monoanions, 1′-S,3′-O-dianions, and 2′-S,3′-O-dianions. The results obtained are presented in Table 7. For all of the compounds studied, the PA values in water are much smaller than those in vacuum, which means that in a polar medium the SPLET mechanism is greatly preferred. Turning our attention to the ΔPA values, we notice that S–H bonds in the monoanions and dianions are more prone to heterolytic fission than those in neutral forms. The higher PA values observed for the 2′-S–H bond than for the 1′-S–H bond suggest that the former break more easily.
Table 7.
The PA (in kcal mol−1) and ΔPA (in kcal mol−1) values [calculated at the rB3LYP/6-311 + G(3df,2p) level in vacuum and water] of the neutral, mono-, and dianionic forms of RDHLA and its metabolites
| Compound | 1′-S–H | 2′-S–H | ||||||
|---|---|---|---|---|---|---|---|---|
| Vacuum | Water | Vacuum | Water | |||||
| PA | ΔPA | PA | ΔPA | PA | ΔPA | PA | ΔPA | |
| RDHLA | 344 | 0 | 34 | 0 | 342 | 0 | 44 | 0 |
| RDHBLA | 350 | 6 | 23 | −11 | 341 | −1 | 32 | −11 |
| SDHTLA | 349 | 4 | 33 | 0 | 339 | −3 | 29 | −15 |
| 3′-O-mRDHLA | 384 | 40 | 34 | 0 | 388 | 46 | 34 | −10 |
| 3′-O-mRDHBLA | 398 | 53 | 25 | −8 | 408 | 66 | 26 | −17 |
| 3′-O-mSDHTLA | 414 | 70 | 27 | −7 | 434 | 92 | 28 | −16 |
| 2′-S,3′-O-dRDHLA | 458 | 113 | 36 | 2 | − | − | − | − |
| 1′-S,3′-O-dRDHLA | − | − | − | − | 461 | 120 | 37 | −7 |
| 2′-S,3′-O-dRDHBLA | 466 | 122 | 37 | 3 | − | − | − | − |
| 1′-S,3′-O-dRDHBLA | − | − | − | − | 476 | 135 | 38 | −6 |
| 2′-S,3′-O-dSDHTLA | 481 | 137 | 37 | 3 | − | − | − | − |
| 1′-S,3′-O-dSDHTLA | − | − | − | − | 501 | 160 | 38 | −6 |
Upon comparing the AIP, PA, and BDE values for all of the compounds studied, it is clear that the preferred antioxidant pathway in a polar medium for lipoic acid and its metabolites is the SPLET mechanism. The HAT mechanism is preferred in nonpolar media.
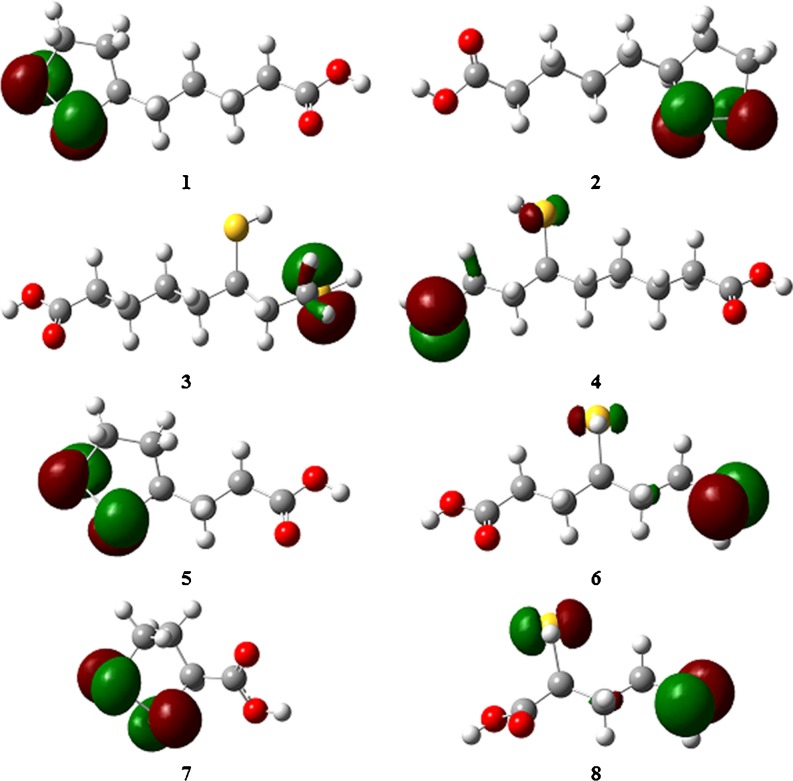
When describing the free-radical scavenging activities of molecules, parameters such as the spin density (SD) distribution in free radicals and the energy of the HOMO orbital are also useful. The SD has been calculated for the 1′-S- and 2′-S-radicals of RDHLA, SDHLA, RDHBLA, and SDHTLA. According to these calculations (Table 8), the highest concentration of SD is observed for the S1′ atom in 1′-S-radicals and for S2′ in 2′-S-radicals. In comparison, the SD values for the O3′ and O3″ atoms in these radicals are very small, which proves the absence of electron delocalization through the whole molecule. The SD values in water are slightly higher than those in vacuum for all of the radicals studied (Table 8). We can therefore conclude that the antioxidant mechanisms involving radical action are not preferred in polar media. The energy of the HOMO orbital is another important molecular parameter associated with the scavenging of free radicals. Molecules with higher E HOMO values have stronger electron-donating abilities. All of the compounds studied have similar values of E HOMO (Table 9). The electronic density distribution in these orbitals is concentrated on the sulfur atoms (Fig. 6). Similar to the SD distribution, there is no delocalization of HOMO orbitals. It can be predicted that S–S and S–H bonds are the most probable sites of free-radical attack in all of the molecules studied.
Table 8.
The spin density (SD) values [calculated at the uB3LYP/6-311 + G(3df,2p) level in vacuum and water] of RDHLA, SDHLA, RDHBLA, and SDHTLA radicals
| Radical | Vacuum | Water | ||||||
|---|---|---|---|---|---|---|---|---|
| S1′ | S2′ | O3′ | O3″ | S1′ | S2′ | O3′ | O3″ | |
| 1′-S-RDHBLA | 1.0662 | 0.0137 | −0.0002 | 0.0000 | 1.0700 | 0.0149 | −0.0002 | 0.0000 |
| 1′-S-RDHLA | 1.0487 | 0.0033 | 0.0000 | 0.0000 | 1.0642 | 0.0045 | 0.0000 | 0.0000 |
| 1′-S-SDHLA | 1.0796 | 0.0084 | 0.0000 | 0.0000 | 1.0830 | 0.0080 | 0.0000 | 0.0000 |
| 1′-S-SDHTLA | 1.0522 | 0.0116 | −0.0001 | 0.0001 | 1.0569 | 0.0110 | −0.0002 | 0.0000 |
| 2′-S-RDHBLA | −0.0042 | 1.0496 | 0.0002 | 0.0000 | −0.0064 | 1.0540 | 0.0002 | −0.0001 |
| 2′-S-RDHLA | 0.0041 | 1.0611 | 0.0000 | 0.0001 | 0.0032 | 1.0693 | 0.0001 | 0.0001 |
| 2′-S-SDHLA | −0.0088 | 1.0560 | 0.0000 | 0.0001 | 0.0163 | 1.0397 | 0.0001 | 0.0003 |
| 2′-S-SDHTLA | 0.0011 | 1.0313 | 0.0031 | 0.0147 | 0.0005 | 1.0349 | 0.0024 | 0.0130 |
Table 9.
The HOMO orbital distributions (E HOMO and ΔE HOMO) of LA/DHLA enantiomers and RLA/RDHLA metabolites [obtained using rB3LYP/6-311 + G(3df,2p) and optimized in vacuum and water]
| Compounds | Vacuum | Water | ||||
|---|---|---|---|---|---|---|
| E HOMO (Ha) | E HOMO (eV) | ΔE HOMO (eV) | E HOMO (Ha) | E HOMO (eV) | ΔE HOMO (eV) | |
| RLA | −0.215 | −5.9 | 0.0 | −0.211 | −5.7 | 0.0 |
| SLA | −0.214 | −5.8 | 0.0 | −0.212 | −5.8 | 0.0 |
| RDHLA | −0.245 | −6.7 | 0.8 | −0.240 | −6.5 | 0.8 |
| SDHLA | −0.244 | −6.6 | 0.8 | −0.241 | −6.5 | 0.8 |
| RBLA | −0.218 | −5.9 | 0.1 | −0.212 | −5.8 | 0.0 |
| RDHBLA | −0.246 | −6.7 | 0.8 | −0.247 | −6.7 | 1.0 |
| STLA | −0.220 | −6.0 | 0.1 | −0.220 | −6.0 | 0.3 |
| SDHTLA | −0.249 | −6.8 | 0.9 | −0.246 | −6.7 | 0.9 |
Fig. 6.
The HOMO orbital distributions in RLA (1), SLA (2), RDHLA (3), SDHLA (4), RBLA (5), RDHBLA (6), STLA (7), and SDHTLA (8). The calculations were accomplished in vacuum with the isovalue 0.07
Conclusions
Structure–antioxidant parameter relationships of α-lipoic acid and its natural metabolites bisnorlipoic acid and tetranorlipoic acid in their oxidized and reduced forms have been investigated using quantum-chemical methods: DFT/B3LYP and MP2(full). On the basis of the results obtained, we can confirm that the SH groups of the reduced forms are responsible for their antioxidant properties. More importantly, we propose that the most preferable antioxidant mechanism in polar media is SPLET. However, in nonpolar media, the HAT mechanism is preferred, as shown by calculations performed for isolated molecules. α-Lipoic acid and its natural metabolites are not the only available organosulfur acids that act as antioxidants; LA/DHLA derivatives should also be included in quantum-chemical investigations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
(DOC 197719 kb)
Acknowledgments
The calculations were performed at the Poznań Supercomputing and Networking Center (PCSS). The authors are grateful for grant no. 76, “Theoretical study of lipoic acid and its biological activity.”
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- 1.Carreau JP. Biosynthesis of lipoic acid via unsaturated fatty acids. Methods Enzymol. 1979;62:152–158. doi: 10.1016/0076-6879(79)62212-7. [DOI] [PubMed] [Google Scholar]
- 2.Reed LJ. A trail of research from lipoic acid to alpha-keto acid dehydrogenase complexes. J Biol Chem. 2001;276:38329–38336. doi: 10.1074/jbc.R100026200. [DOI] [PubMed] [Google Scholar]
- 3.Biewenga GP, Haenen GR, Bast A. The pharmacology of the antioxidant lipoic acid. Gen Pharmacol. 1997;29:315–331. doi: 10.1016/S0306-3623(96)00474-0. [DOI] [PubMed] [Google Scholar]
- 4.Lodge JK, Youn HD, Handelman GJ, Konishi T, Matsugo S, Mathur VV, Packer L. Natural sources of lipoic acid: determination of lipoyllysine released from protease-digested tissues by high performance liquid chromatography incorporating electrochemical detection. J Appl Nutr. 1997;49:3–11. [Google Scholar]
- 5.Reed LJ, DeBusk BG, Gunsalus IC, Hornberger CS. Crystalline α-lipoic acid: a catalytic agent associated with pyruvate dehydrogenase. Science. 1951;114:93–94. doi: 10.1126/science.114.2952.93. [DOI] [PubMed] [Google Scholar]
- 6.Stroud RM, Carlisle CH (1972) Single-crystal structure determination of DL-6-thioctic acid, C8H14O2S2. Acta Crystallogr B 28:304–307
- 7.Shay KP, Moreau RF, Smith EJ, Smith AR, Hagen TM (2009) Alpha-lipoic acid as a dietary supplement: molecular mechanisms and therapeutic potential. Biochim Biophys Acta 1790:1149–1160 [DOI] [PMC free article] [PubMed]
- 8.Aruoma OI, Halliwell B, Hoey BM, Butler J. The antioxidant action of N-acetylcysteine: its reaction with hydrogen peroxide, hydroxyl radical, superoxide, and hypochlorous acid. Free Rad Biol Med. 1989;6:593–597. doi: 10.1016/0891-5849(89)90066-X. [DOI] [PubMed] [Google Scholar]
- 9.Agnihotri N, Mishra PC (2009) Mechanism of scavenging action of N-acetylcysteine for the OH radical: a quantum computational study. J Phys Chem B 113:12096–12104 [DOI] [PubMed]
- 10.Suzuki YJ, Tsuchiya M, Packer L. Thioctic acid and dihydrolipoic acid are novel antioxidants which interact with reactive oxygen species. Free Radic Res Commun. 1991;15:255–263. doi: 10.3109/10715769109105221. [DOI] [PubMed] [Google Scholar]
- 11.Devasagayam TP, Di Mascio P, Kaiser S, Sies H. Singlet oxygen induced singlestrand breaks in plasmid pBR322 DNA: the enhancing effect of thiols. Biochim Biophys Acta. 1991;1088:409–412. doi: 10.1016/0167-4781(91)90133-7. [DOI] [PubMed] [Google Scholar]
- 12.Haenen GR, Bast A. Scavenging of hypochlorous acid by lipoic acid. Biochem Pharmacol. 1991;42:2244–2246. doi: 10.1016/0006-2952(91)90363-A. [DOI] [PubMed] [Google Scholar]
- 13.Xu DP, Wells WW. alpha-Lipoic acid dependent regeneration of ascorbic acid from dehydroascorbic acid in rat liver mitochondria. J Bioenerg Biomembranes. 1996;28:77–85. [PubMed] [Google Scholar]
- 14.Lykkesfeldt J, Hagen TM, Vinarsky V, Ames BN (1998) Age-associated decline in ascorbic acid concentration, recycling, and biosynthesis in rat hepatocytes—eversal with (R)-alpha-lipoic acid supplementation. FASEB J 12:1183–1189 [DOI] [PubMed]
- 15.Suh JH, Shigeno ET, Morrow JD, Cox B, Rocha AE, Frei B, Hagen TM (2001) Oxidative stress in the aging rat heart is reversed by dietary supplementation with (R)-(alpha)-lipoic acid. FASEB J 15:700–706 [DOI] [PMC free article] [PubMed]
- 16.Michels AJ, Joisher N, Hagen TM. Age-related decline of sodium-dependent ascorbic acid transport in isolated rat hepatocytes. Arch Biochem Biophys. 2003;410:112–120. doi: 10.1016/S0003-9861(02)00678-1. [DOI] [PubMed] [Google Scholar]
- 17.Han D, Handelman G, Marcocci L, Sen CK, Roy S, Kobuchi H, Tritschler HJ, Flohe L, Packer L. Lipoic acid increases de novo synthesis of cellular glutathione by improving cystine utilization. Biofactors. 1997;6:321–338. doi: 10.1002/biof.5520060303. [DOI] [PubMed] [Google Scholar]
- 18.Ou P, Tritschler HJ, Wolff SP (1995) Thioctic (lipoic) acid: a therapeutic metal chelating antioxidant? Biochem Pharmacol 50:123–126 [DOI] [PubMed]
- 19.Suh JH, Moreau R, Heath SH, Hagen TM (2005) Dietary supplementation with (R)-alpha-lipoic acid reverses the age-related accumulation of iron and depletion of antioxidants in the rat cerebral cortex. Redox Rep 10:52–60 [DOI] [PubMed]
- 20.Lodge JK, Traber MG, Packer L. Thiol chelation of Cu2+ by dihydrolipoic acid prevents human low density lipoprotein peroxidation. Free Radic Biol Med. 1998;25:287–297. doi: 10.1016/S0891-5849(98)00048-3. [DOI] [PubMed] [Google Scholar]
- 21.Bush AI. Metal complexing agents as therapies for Alzheimer`s disease. Neurobiol Aging. 2002;23:1031–1038. doi: 10.1016/S0197-4580(02)00120-3. [DOI] [PubMed] [Google Scholar]
- 22.Goralska M, Dackor R, Holley B, McGahan MC (2003) Alpha-lipoic acid changes iron uptake and storage in lens epithelial cells. Exp Eye Res 76:241–248 [DOI] [PubMed]
- 23.Patel MS, Packer L (2008) Lipoic acid: energy production, antioxidant activity and health effects (oxidative stress and disease). Taylor & Francis, Boca Raton
- 24.Vasdev S, Ford CA, Parai S, Longerich L, Gadag V. Dietary lipoic acid supplementation prevents fructose-induced hypertension in rats. Nutr Metab Cardiovasc Dis. 2000;10:339–346. [PubMed] [Google Scholar]
- 25.Vasdev S, Ford CA, Parai S, Longerich L, Gadag V. Dietary alpha-lipoic acid supplementation lowers blood pressure in spontaneously hypertensive rats. J Hypertens. 2000;18:567–573. doi: 10.1097/00004872-200018050-00009. [DOI] [PubMed] [Google Scholar]
- 26.Vasdev S, Gill V, Longerich L, Parai S, Gadag V. Salt-induced hypertension in WKY rats: prevention by alpha-lipoic acid supplementation. Mol Cell Biochem. 2003;254:319–326. doi: 10.1023/A:1027354005498. [DOI] [PubMed] [Google Scholar]
- 27.Vasdev S, Gill V, Parai S, Gadag V. Dietary lipoic acid supplementation attenuates hypertension in Dahl salt sensitive rats. Mol Cell Biochem. 2005;275:135–141. doi: 10.1007/s11010-005-1095-7. [DOI] [PubMed] [Google Scholar]
- 28.Louhelainen M, Merasto S, Finckenberg P, Lapatto R, Cheng ZJ, Mervaala EM. Lipoic acid supplementation prevents cyclosporine-induced hypertension and nephrotoxicity in spontaneously hypertensive rats. J Hypertens. 2006;24:947–956. doi: 10.1097/01.hjh.0000222766.37971.9f. [DOI] [PubMed] [Google Scholar]
- 29.Takaoka M, Kobayashi Y, Yuba M, Ohkita M, Matsumura Y. Effects of alpha-lipoic acid on deoxycorticosterone acetate-salt-induced hypertension in rats. Eur J Pharmacol. 2001;424:121–129. doi: 10.1016/S0014-2999(01)01120-7. [DOI] [PubMed] [Google Scholar]
- 30.Midaoui AE, de Champlain J. Prevention of hypertension, insulin resistance, and oxidative stress by alpha-lipoic acid. Hypertension. 2002;39:303–307. doi: 10.1161/hy0202.104345. [DOI] [PubMed] [Google Scholar]
- 31.Midaoui AE, Elimadi A, Wu L, Haddad PS, de Champlain J. Lipoic acid prevents hypertension, hyperglycemia, and the increase in heart mitochondrial superoxide production. Am J Hypertens. 2003;16:173–179. doi: 10.1016/S0895-7061(02)03253-3. [DOI] [PubMed] [Google Scholar]
- 32.Kramer K, Packer L (2001) R-alpha-lipoic acid. In: Kramer K, Hoppe P, Packer L (eds) Nutraceuticals in health and disease prevention. Marcel Dekker, New York, pp 129–164
- 33.Marracci GH, Jones RE, McKeon GP, Bourdette DN (2002) Alpha-lipoic acid inhibits T cell migration into the spinal cord and suppresses and treats experimental autoimmune encephalomyelitis. J Neuroimmunol 131:104–114 [DOI] [PubMed]
- 34.Morini M, Roccatagliata L, Dell’Eva R, Pedemonte E, Furlan R, Minghelli S, Giunti D, Pfeffer U, Marchese M, Noonan D, Mancardi G, Albini A, Ucelli A (2004) Alpha-lipoic acid is effective in prevention and treatment of experimental autoimmune encephalomyelitis. J Neuroimmunol 148:146–153 [DOI] [PubMed]
- 35.Yadav V, Marracci G, Lovera J, Woodward W, Bogardus K, Marquardt W, Shinto L, Morris C, Bourdette D. Lipoic acid in multiple sclerosis: a pilot study. Mult Scler. 2005;11:159–165. doi: 10.1191/1352458505ms1143oa. [DOI] [PubMed] [Google Scholar]
- 36.Yadav V, Marracci GH, Munar MY, Cherala G, Stuber LE, Alvarez L, Shinto L, Koop DR, Bourdette DN. Pharmacokinetic study of lipoic acid in multiple sclerosis: comparing mice and human pharmacokinetic parameters. Mult Scler. 2010;16:387–397. doi: 10.1177/1352458509359722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schupke H, Hempel R, Peter G, Hermann R, Wessel K, Engel J, Kronbach T. New metabolic pathways of alpha-lipoic acid. Drug Metab Dispos. 2001;29:855–862. [PubMed] [Google Scholar]
- 38.Harrison EE, McCormick DB (1974) The metabolizm of dl-(1,6-14C)lipoic acid in the rat. Arch Biochem Biophys 160:514–522 [DOI] [PubMed]
- 39.Shang YJ, Qian YP, Liu XD, Dai F, Shang XL, Jia WQ, Liu Q, Fang JG, Zhou B. Radical-scavenging activity and mechanism of resveratrol-oriented analogues: influence of the solvent, radical, and substitution. J Org Chem. 2009;74:5025–5031. doi: 10.1021/jo9007095. [DOI] [PubMed] [Google Scholar]
- 40.Klein E, Rimarčík J, Lukeš V (2009) DFT/B3LYP study of the O–H bond dissociation enthalpies and proton affinities of para- and meta-substituted phenols in water and benzene. Acta Chim Slovaca 2:37–51
- 41.Mikulski D, Górniak R, Molski M (2010) A theoretical study of the structure-radical scavenging activity of trans-resveratrol analogues and cis-resveratrol in gas phase and water environment. Eur J Med Chem 45:1015–1027 [DOI] [PubMed]
- 42.Wright JS, Johnson ER, DiLabio GA. Predicting the activity of phenolic antioxidants: theoretical method, analysis of substituent effects, and application to major families of antioxidants. J Am Chem Soc. 2001;123:1173–1183. doi: 10.1021/ja002455u. [DOI] [PubMed] [Google Scholar]
- 43.Leopoldini M, Marino T, Russo N, Toscano M. Antioxidant properties of phenolic compounds: H-atom versus electron transfer mechanism. J Phys Chem A. 2004;108:4916–4922. doi: 10.1021/jp037247d. [DOI] [Google Scholar]
- 44.Cao H, Pan XL, Li C, Zhou C, Deng FY, Li TH. Density functional theory calculations for resveratrol. Bioorg Med Chem Lett. 2003;13:1869–1871. doi: 10.1016/S0960-894X(03)00283-X. [DOI] [PubMed] [Google Scholar]
- 45.Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Montgomery JA Jr, Vreven T, Kudin KN, Burant JC, Millam JM, Iyengar SS, Tomasi J, Barone V, Mennucci B, Cossi M, Scalmani G, Rega N, Petersson GA, Nakatsuji H, Hada M, Ehara M, Toyota K, Fukuda R, Hasegawa J, Ishida M, Nakajima T, Honda Y, Kitao O, Nakai H, Klene M, Li X, Knox JE, Hratchian HP, Cross JB, Bakken V, Adamo C, Jaramillo J, Gomperts R, Stratmann RE, Yazyev O, Austin AJ, Cammi R, Pomelli C, Ochterski JW, Ayala PY, Morokuma K, Voth GA, Salvador P, Dannenberg JJ, Zakrzewski VG, Dapprich S, Daniels AD, Strain MC, Farkas O, Malick DK, Rabuck AD, Raghavachari K, Foresman JB, Ortiz JV, Cui Q, Baboul AG, Clifford S, Cioslowski J, Stefanov BB, Liu G, Liashenko A, Piskorz P, Komaromi I, Martin RL, Fox DJ, Keith T, Al-Laham MA, Peng CY, Nanayakkara A, Challacombe M, Gill PMW, Johnson B, Chen W, Wong MW, Gonzalez C, Pople JA (2004) Gaussian 03, revision C02. Gaussian Inc., Wallingford
- 46.Hohenberg K, Kohn W. Inhomogeneous electron gas. Phys Rev. 1964;136:B864–B871. doi: 10.1103/PhysRev.136.B864. [DOI] [Google Scholar]
- 47.Kohn W, Sham LJ (1965) Self-consistent equations including exchange and correlation effects. Phys Rev A 140:1133–1138
- 48.Lee C, Yang W, Parr RG (1988) Development of the Cole–Salvetti correlation energy formula into a functional of the electron density. Phys Rev B 37:785–789 [DOI] [PubMed]
- 49.Møller C, Plesset MP. Note on an approximation treatment for many-electron systems. Phys Rev. 1934;46:618–622. doi: 10.1103/PhysRev.46.618. [DOI] [Google Scholar]
- 50.Leopoldini M, Russo N, Toscano M. The molecular basis of working mechanism of natural polyphenolic antioxidants. Food Chem. 2011;125:288–306. doi: 10.1016/j.foodchem.2010.08.012. [DOI] [Google Scholar]
- 51.Foti MC, Daquino C, Geraci C (2004) Electron-transfer reaction of cinnamic acids and their methyl esters with the DPPH· radical in alcoholic solutions. J Org Chem 69:2309–2314 [DOI] [PubMed]
- 52.Litwinienko G, Ingold KU. Abnormal solvent effects on hydrogen atom abstraction. 2. Resolution of the curcumin antioxidant controversy. The role of sequential proton loss electron transfer. J Org Chem. 2004;69:5888–5896. doi: 10.1021/jo049254j. [DOI] [PubMed] [Google Scholar]
- 53.Musialik M, Litwinienko G (2005) Scavenging of dpph· radicals by vitamin E is accelerated by its partial ionization: the role of sequential proton loss electron transfer. Org Lett 7:4951–4954 [DOI] [PubMed]
- 54.Nakanishi I, Kawashima T, Ohkubo K, Kanazawa H, Inami K, Mochizuki M, Fukuhara K, Okuda H, Ozawa T, Itoh S, Fukuzumi S, Ikota N. Electron-transfer mechanism in radical-scavenging reactions by a vitamin E model in a protic medium. Org Biomol Chem. 2005;3:626–629. doi: 10.1039/b416572a. [DOI] [PubMed] [Google Scholar]
- 55.Litwinienko G, Ingold KU. Solvent effects on the rates and mechanisms of reaction of phenols with free radicals. Acc Chem Res. 2007;40:222–230. doi: 10.1021/ar0682029. [DOI] [PubMed] [Google Scholar]
- 56.Parker VD (1992) Homolytic bond (H–A) dissociation free energies in solution—applications of the standard potential of the (H+/H) couple. J Am Chem Soc 114:7458–7462
- 57.Bizarro MM, Cabral BJC, dos Santos RMB, Simões JAM (1999) Substituent effects on the O–H bond dissociation enthalpies in phenolic compounds: agreements and controversies. Pure Appl Chem 71:1249–1256
- 58.Atkins PW (1998) Physical chemistry, 6th edn. Oxford University Press, Oxford
- 59.Cossi M, Rega N, Scalmani G, Barone V. Energies, structures, and electronic properties of molecules in solution with the C-PCM solvation model. J Comput Chem. 2003;24:669–681. doi: 10.1002/jcc.10189. [DOI] [PubMed] [Google Scholar]
- 60.Mikulski D, Molski M (2010) Quantum chemistry study of interaction of Cu2+ cation and aqua–copper [Cu(H2O)1–4]2+ complexes with resveratrol. J Mol Struct THEOCHEM 956:66–76
- 61.Mikulski D, Szeląg M, Molski M, Górniak R (2010) Quantum-chemical study on the antioxidation mechanisms of trans-resveratrol reactions with free radicals. In the gas phase, water and ethanol environment. J Mol Struct THEOCHEM 951:37–48
- 62.Mikulski D, Szeląg M, Molski M (2011) Quantum-chemical study of interactions of trans-resveratrol with guanine-thymine dinucleotide and DNA-nucleobases. J Mol Model. doi:10.1007/s00894-011-0999-2 [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOC 197719 kb)