Abstract
In this paper, a new technique is proposed for automatic segmentation of multiple sclerosis (MS) lesions from brain magnetic resonance imaging (MRI) data. The technique uses a trained support vector machine (SVM) to discriminate between the blocks in regions of MS lesions and the blocks in non-MS lesion regions mainly based on the textural features with aid of the other features. The classification is done on each of the axial, sagittal and coronal sectional brain view independently and the resultant segmentations are aggregated to provide more accurate output segmentation. The main contribution of the proposed technique described in this paper is the use of textural features to detect MS lesions in a fully automated approach that does not rely on manually delineating the MS lesions. In addition, the technique introduces the concept of the multi-sectional view segmentation to produce verified segmentation. The proposed textural-based SVM technique was evaluated using three simulated datasets and more than fifty real MRI datasets. The results were compared with state of the art methods. The obtained results indicate that the proposed method would be viable for use in clinical practice for the detection of MS lesions in MRI.
Keywords: MRI, Texture Analysis, Brain Segmentation, Multiple Sclerosis, SVM, ROI, Sectional View, Multi-Channels.
1. INTRODUCTION
Multiple Sclerosis (MS) is an autoimmune disease of central nervous system. It may result in a variety of symptoms from blurred vision to severe muscle weakness and degradation, depending on the affected regions in brain [1-4]. To better understand this disease and to quantify its evolution, magnetic resonance imaging (MRI) is increasingly used nowadays [5]. Manual delineation of MS lesions in MR images by human expert is time-consuming, subjective and prone to inter-expert variability. Therefore, automatic segmentation is needed as an alternative to manual segmentation. However, the progression of the MS lesions shows considerable variability and MS lesions present temporal changes in shape, location, and area between patients and even for the same patient [6-9], which renders the automatic segmentation of MS lesions a challenging problem.
A variety of methods have been proposed for the automatic segmentation of MS Lesions [10-18]. Texture analysis in MRI has been used with some success in neuroimaging to detect lesions and abnormalities. Textural analysis refers to a set of processes applied to characterize special variation patterns of voxels grayscale in an image. Segmentation based on texture properties is promising in cases of lesions that are inhomogeneous, unsharp, and faint, but show an intensity pattern that is different from the adjacent healthy tissue [18]. Textural features have been used in [19] to differentiate between lesion white matter, normal white matter and normal appearing white matter. Texture classification was also used for the analysis of multiple sclerosis lesions [20]. Ghazel et al., [21] proposed textural features extraction method based on optimal filter designed to detect classify region of interests (ROIs) selected manually by an expert that enclose potential MS lesions into MS lesions and healthy tissues background.
However, to the best of our knowledge, texture based MS segmentation approaches that have been previously reported were applied to ROIs that are manually selected by an expert to indicate potential regions including MS lesions, which makes the segmentation process semi-automated. Therefore, efforts are needed to automate the use of textural features in the detection of MS lesions.
The use of textural features is a promising approach for providing accurate segmentation of MS lesions, especially when taking advantage of various MRI sequences to benefit from their relevant and complementary information for MS segmentation. Utilizing textural features without the need for manual labeling of ROIs, enabling segmentation using multi-channels MRI data and taking into account the different sectional views of the lesion volume are the three pillars of the proposed technique.
In this paper, we propose a technique that uses textural features to describe the blocks of each MRI slice along with other features. The technique applies the classification process on slices of each sectional view of the brain MRI independently. For each sectional view, a trained classifier is used to discriminate between the blocks and detect the blocks that potentially include MS lesions mainly based on the textural features with aid of the other features. The blocks classification is used to provide an initial coarse segmentation of the MRI slices. The textural-based classifier is built using Support Vector Machine (SVM), one of the widely used supervised learning algorithms that have be utilized successfully in many applications [22, 23]. The resultant segmentations of the three sectional views segmentations are aggregated to generate the final MS lesion segmentation.
Multi-channels MRI (T1-weighted, T2-weighted and FLAIR) are used in the task of MS segmentation. Due to the higher accuracy of the FLAIR imaging sequence in revealing MS lesions and assessing the total lesion load [24, 25], FLAIR MRI was used in this paper as the source of textural features. The other channels are used to provide additional enabling features in the segmentation. Meanwhile, T1-weighted imaging data is used for registration with probabilistic atlas to provide tissues prior probabilities.
The paper consists of five sections including this introduction section. In section 2, the details of the proposed segmentation technique are illustrated. The experimental results are presented in section 3 and discussed in section 4. The paper conclusion is presented in section 5. For completeness, appendix A provides the details for calculating the textural features.
2. MATERIALS AND METHODS
2.1. Datasets
2.1.1. Synthetic Data
The simulated MRI datasets generated using the McGill University BrainWeb MRI Simulator [26-30] include three brain MRI datasets with mild, moderate and severe levels of multiple sclerosis lesions. We will refer to these templates in this paper, including the results, as MSLES 1, MSLES 2 and MSLES 3 for the mild, moderate and severe levels, respectively. The MRI data was generated using T1, T2 and Inversion Recovery (IR) channels. Isotropic voxel size of 1mm x 1mm x 1mm and spatial in-homogeneity of 0% are used in this paper. For each channel, the images are available at six different noise levels (0%, 1%, 3%, 5%, 7% and 9%).
2.1.2. Real Data
Datasets of 61 cases are used to verify the segmentation technique proposed by this paper. The sources of these datasets are the workshop of MS Lesion Segmentation Challenge 2008 [31, 32] and real MRI studies for MS subjects acquired at the University of Miami.
2.1.2.1. MS Lesion Segmentation Challenge 2008
Datasets used for evaluation in this paper include 51 cases which are publicly available from the MS Lesion Segmentation Challenge 2008 website [32]. For each case, three MR channels are made available (T1-, T2-weighted and Flair). The datasets are divided into labeled cases used for training (20 cases) and non-labeled cases used for testing (31 cases). The MRI datasets are from two separate sources: 28 datasets (10 for training and 18 for testing) from Children’s Hospital Boston (CHB) and 23 datasets (10 for training and 13 for testing) from University of North Carolina (UNC). The UNC cases were acquired on a Siemens 3T Allegra MRI scanner with slice thickness of 1 mm and in-plane resolution of 0.5 mm. No scanner information was provided about the CHB cases. All subjects MRI are re-sliced to be 512x512x512 with resolution 0.5mm x 0.5 mm x 0.5mm. We will refer to training datasets of CHB and UNC as CHB_train_CaseXX and UNC_train_CaseXX respectively (with XX refers to study number in two digits). Similarly the test datasets of CHB and UNC are referred to by CHB_test1_CaseXX and UNC_test1_CaseXX respectively.
2.1.2.2. MS Subjects MRI Data Acquired at University of Miami
The acquired MRI datasets for MS subjects are composed of multi-channel MRI, including T1, T2 and FLAIR, for 10 subjects (4 males, age range: 50-72 and 6 females, age range: 30-59). The corresponding volumes in the different sequences are co-centered and have the same field of view of 175x220 mm. The slice thickness and spacing between slices for T2, PD and FLAIR sequences are 3mm and 3.9 mm, respectively, while both of the slice thickness and spacing between slices for the T1 sequence is 1mm, respectively, for the same field of view. On average, each T2 and FLAIR MRI sequence consists of thirty seven slices while the T1 MRI sequence consists of one hundred and sixty slices that cover the whole brain. The axial FLAIR sequences used in this paper were acquired using the following imaging parameters: 9000/103/2500/256×20-4/17/123 (repetition time ms/echo time ms/inversion time/matrix size/echo train length/ imaging frequency). The parameters for axial T1 sequences: 2150/3.4/256×208/1/123 (repetition time ms/echo time ms /matrix size/echo train length/ imaging frequency), while the parameters for axial T2 sequences: 6530/120/256×20-4/11/123 (repetition time ms/echo time ms /matrix size/echo train length/ imaging frequency). All the subjects were referred for brain MRI studies based on an earlier diagnosis of MS. The MS lesions were manually labeled on the FLAIR sequences by a neuroradiologist. The ten MRI studies were acquired using a 3.0T MR scanner under a human subject’s protocol approved by our institutional review board. We will refer to these subjects as MSX (with X is an integer number from 2 to 11).
2.2. MS lesions Multi-Sectional Views Segmentation Framework
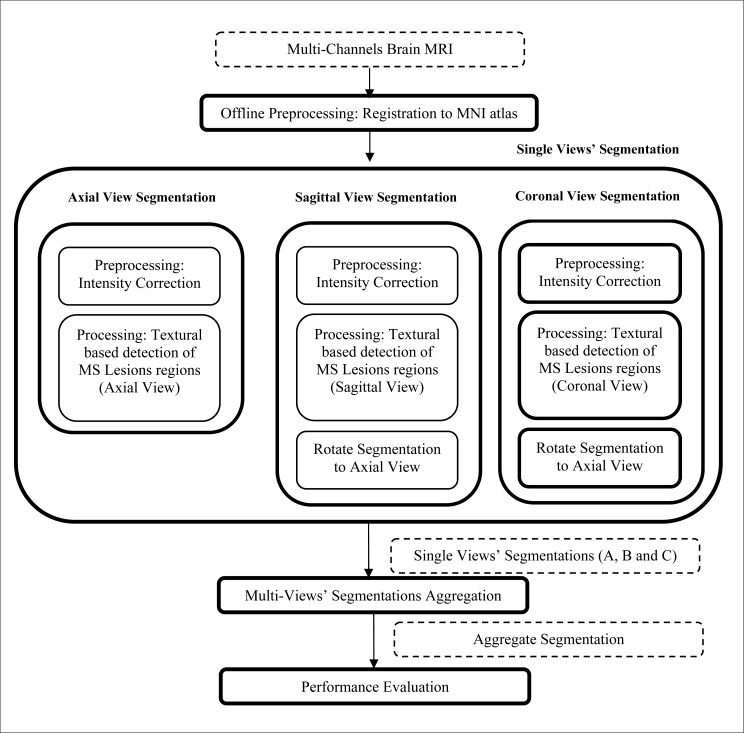
The proposed MS lesions multi-sectional views segmentation framework is described in Fig. (1). The input studies are preprocessed offline for registration of the MR images to the probabilistic MNI atlas [33] and co-registration among the different channels. The multi-sectional segmentation consists of three similar engines; each is applied to a sectional view (Axial, Sagittal and Coronal views). Through each engine, the multi-channels MRI slices of the brain are preprocessed for intensity correction to remove the effect of noise and variations in brightness and contrast among corresponding sequences of different subjects. The next step in each sectional view engine is the processing module, which is used for the detection of initial MS lesions regions based on textural features. The output segmentations of the sagittal and coronal view engines are rotated to be in the axial view. The output segmentations of the axial, sagittal and coronal sectional views engines are noted as A, B and C, respectively. The multi-sectional views’ segmentations aggregation step is applied to A, B and C to generate the final output segmentation.
Fig. (1).
MS lesions multi-sectional views segmentation framework.
2.3. Preprocessing
In this step, intensity correction and registration of the subject’s MRI data with an atlas for probabilistic tissues are performed prior to textural-based segmentation. In addition special handling for the different dataset is performed.
2.3.1. Registration
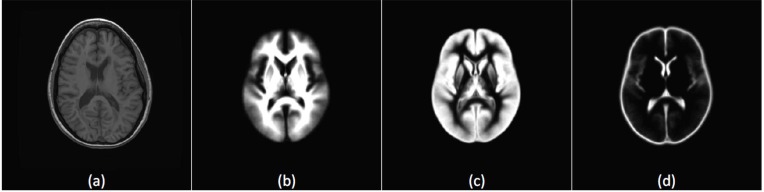
For the MS Lesion Segmentation Challenge datasets, all datasets were rigidly registered to a common reference frame and re-sliced to isotropic voxel spacing, with resolution 512x512x512, using B-spline based interpolation. The images were then aligned on the mid-sagittal plane [34]. The probabilistic MNI atlas [33] provides for each voxel the probability of belonging to the white matter (WM), the gray matter (GM) and the cerebro-spinal fluid (CSF). The template of the MNI atlas is a T1 sequence, thus the T1 sequence of the MRI datasets is used for registration with the atlas. Based on the registration step, three values are provided for each voxel representing the prior probabilities of the voxel to belong to white matter, gray matter or CSF tissues. A sample slice along with the registered priors of the probabilistic MNI atlas is shown in Fig. (2). Fig. (2a) shows a T1 slice from subject CHB_train_Case01 and figure 2(b-d) are the corresponding slice from the probabilistic tissue for the white matter (WM), gray matter (GM) and CSF tissue prior probabilities, respectively.
Fig. (2).
Registration to the tissues probabilistic atlas with T1 sequence. (a) A T1 slice from the subject CHB_train_Case01 registered to the MNI atlas. (b) Registered white matter tissue probability. (c) Registered gray matter tissue probability. (d) Registered CSF tissue probability.
As described in the datasets section 2.1, the datasets acquired at the University of Miami Miller School of Medicine, have different resolutions across the channels. All the datasets were re-sliced to be in the same resolution conditions of MS Lesion Segmentation Challenge datasets to be tested by the models trained by MS Lesion Segmentation Challenge training data. Then they were registered to the MNI atlas using Automated Image Registration (AIR) software [35].
We have created a spacial priors atlas for the BrainWeb datasets analog to the standard MNI atlas. The BrainWeb database provides twenty anatomical models for normal brain. These anatomical models consist of a set of fuzzy tissue membership volumes, one for each tissue class, i.e., white matter, gray matter, cerebrospinal fluid, fat. The voxel values in these volumes reflect the proportion of tissue present in that voxel, in the range [0,1]. Since we are interested in this paper in the white matter, gray matter and CSF tissues probabilities, the corresponding tissues membership volumes in the anatomical models are averaged over the twenty brain templates to get a spacial priors atlas to be used when testing the BrainWeb data.
2.3.2. Intensity Correction
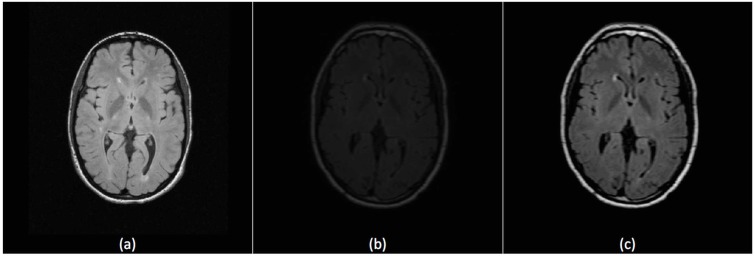
Due to different operating conditions, brightness and contrast of the imaging slices may vary among subjects. This affects performance of segmentation that is based on textural features which are calculated based on grayscale intensities. If a dataset is used for training, better histogram matching of the dataset to be segmented and the training data will lead to more accurate definition of MS lesions. We used our preprocessing technique used before in [36] that starts with applying contrast-brightness correction to maximize the intersection between the histogram of the training and segmentation datasets followed by using 3D anisotropic filter to eliminate empty histogram bins. Fig. (3) shows the effect of the preprocessing technique on improving a FLAIR slice from UNC_train_Case01 (subject dataset to be segmented) with reference to UNC_train_Case02 (subject dataset used in training).
Fig. (3).
Intensity correction for FLAIR Sequence. (a) A slice from the reference subject UNC_train_Case02 (used in training). (b) A slice from subject UNC_train_Case01 before preprocessing. (c) The same slice of UNC_train_Case01 after preprocessing.
2.4. Multi-Sectional Views Textural based SVM for MS Lesion Segmentation
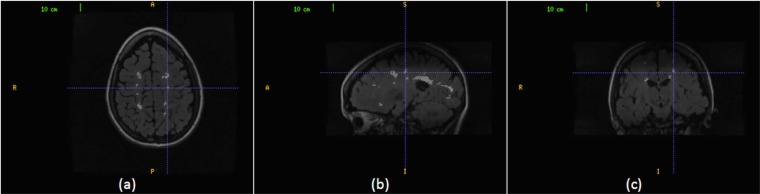
The MS lesion volumes can be viewed from the different sectional views of the brain MRIs. Fig. (4) shows an MS lesion volume in the three sectional views. A lesion region in the axial view is pointed to by a cursor in Fig. (4a), its corresponding lesion region in the sagittal view in Fig. (4b) and its corresponding lesion region in the coronal view in Fig. (4c). The proposed technique aims to benefit from the multiple views in improvement the quality of MS lesions segmentation. Our segmentation technique strategy is to superimpose the segmentations, in which slice segmentation are performed on axial, sagittal and coronal views and the resultant segmentations are aggregated to generate the final segmentation.
Fig. (4).
An MS lesion volume in the three sectional views. (a) The cursor points to an MS lesion region in the axial view. (b) The cursor points to the corresponding MS lesion region in the sagittal view. (c) The cursor points to the corresponding MS lesion region in the coronal view.
In the following two subsections, the segmentation engine used for each of the sectional views’ segmentation is explained and the aggregation of the multi-sectional views’ segmentations is explored.
2.4.1. Single Sectional View Segmentation
For each sectional brain view (axial, sagittal or coronal), a trained classifier is designed to segment the MS lesions in the corresponding view. The details discussed in this subsection apply to the three classifiers. The single view classifiers are identical in the structure, criteria and algorithm while the difference is only in the slices view from which the features are extracted.
Each preprocessed MRI slice is processed through a trained detector engine to obtain initial MS lesions regions. The detector engine in our method is implemented using support vector machine. Training the detector engine is done by processing the training dataset and dividing its slices into square blocks and assigning a binary class for each block. If the block contains at least one pixel manually labeled as MS, it is classified as MS block (class 1). Otherwise, the block is classified as non-MS block (class 0) if all of its pixels are labeled as non-MS pixels. Each block is described by a feature vector which mainly represents textural features of the block. During segmentation, the slice to be segmented is divided into square non-overlapping blocks and each block is classified by the trained engine as MS block or non-MS block.
2.4.1.1. Block Size
Statistics were previously made to measure the size of the multiple sclerosis lesions. The common values for the diameter are between 3.5 mm and 13.5mm [37]. For input MRI studies, the size of the square blocks wxw pixel2 is selected automatically to be within the range of (4x4 mm2) which tightly covers the smallest possible MS lesion diameter based on the input dataset pixel size which is determined from the input resolution and field of view.
2.4.1.2. Features Vector
In order to describe each square block of the MRI slice, a features vector of 39 features is calculated. The block features are classified into five categories: twenty four textural features, two position features, two co-registered intensities, three tissues priors and eight neighboring blocks features. The textural features are calculated for the FLAIR sequence. Textural features include histogram-based features (mean and Variance), gradient-based features (gradient mean and gradient Variance), run length-based features (gray level non-uniformity, run length non-uniformity) and co-occurrence matrix-based features (contrast, entropy and absolute value). Run length-based features are calculated 4 times for horizontal, vertical, 45 degrees and 135 degrees directions. Co-occurrence matrix-based features are calculated using a pixel distance d=1 and for the same angles as the run length-based features. The details for calculating the textural-based features are provided in Appendix A. The position features are the slice relative location with reference to the bottom slice and the radial Euclidean distance between the block’s top left pixel and the center of the slice normalized by dividing it by the longest diameter of the slice. The center and the longest diameter of the slice are parameters that are geometrically calculated in the preprocessing step. The other channels features include intensity means for the corresponding block in the T1and T2 channels. The atlas spacial prior probabilities features include the means of the priors extracted from the probabilistic atlas (White Matter, Gray Matter and CSF probabilities) for the block. The neighboring blocks features are the difference between the mean intensity of the current block and the mean intensity of each of the eight neighboring blocks in the same slice.
These categories of features are selected to have analogy with the features used non-intentionally by the expert in the task of manual labeling of MS areas. When the expert labels MS lesions in the FLAIR slice, the hyper intense areas are the potential areas to have the lesion. This is emulated in our technique by using the textural features. The candidate areas are filtered by the expert based on the positions of these areas. This is emulated by using the position based features. The expert takes into account the neighboring areas intensities and we emulate this by using the neighboring features. The expert also tests intensities of the other channels (if available) for the same areas to verify them and we emulate this by taking the multi-channels grayscales into account. The expert can specify for each area the corresponding tissue, which helps in taking decision about the candidate areas. This act is emulated by using probabilistic atlas to get spacial probabilistic tissues features.
2.4.1.3. SVM Training and Segmentation
Support Vector Machine (SVM) is a supervised learning algorithm, which has at its core a method for creating a predictor function from a set of training data where the function itself can be a binary, a multi-category, or even a general regression predictor. To accomplish this mathematical feat, SVMs find a hypersurface which attempts to split the positive and negative examples with the largest possible margin on all sides of the hyperplane. It uses a kernel function to transform data from input space into a high dimensional feature space in which it searches for a separating hyperplane. The radial basis function (RBF) kernel is selected to be the kernel of the SVM. This kernel nonlinearly maps samples into a higher dimensional space so it can handle the case when the relation between class labels and attributes is nonlinear. The library libsvm 2.9 [38] includes all the methods needed to do the implementation, training and prediction tasks of the SVM. It is incorporated in our method to handle all the SVM operations.
2.4.1.4. Training
The dataset of one subject (or more) is used to generate the SVM training set. In the selected brain view, the slices of this training dataset are divided into n square blocks of size wxw pixels. SVM training set (T) is composed of training entries ti (xi, yi) where xi is the features vector of the block bi, yi is the class label of this block for i =1 to n (n is the number of blocks included in the training set). The segmentation of MS lesions amounts to a binary classification problem, i.e., yi is either 0 or 1. The training entry is said to be positive entry if yi is 1 and negative in the other case.
For each slice of the training dataset, each group of connected pixels labeled manually as MS pixels forms a lesion region. Blocks involved in the set of positive training entries (TP) are generated by localizing all the lesion regions and for each of them, the smallest rectangle that encloses the lesion region is divided into non-overlapping square blocks of size wxw pixels. Each block bi of these blocks is labeled by yi=1 if any of the w2 pixels inside this block is manually labeled as MS pixel. Any block that contains at least 1 MS pixel is referred to in our method as MS block.
Similarly, the blocks involved in the set of negative training entries (TN) are generated by localizing the non-background lesion regions whose pixels are not manually labeled as MS pixels and dividing them into non-overlapping square blocks of size wxw pixels. These blocks are referred to in our method as non-MS blocks. Each block bi of these blocks is labeled by yi=0. Feature vector xi is calculated for each block of both positive and negative training entries. The positive training entries Tp contain blocks that contain 1 to w2 MS pixels. This helps the SVM engine to learn the features of the blocks that either partially or completely contain MS pixels. The training set T is composed of the positive training entries and the negative training entries: Y = Tp ∪ TN. The training set entries were fed to the SVM engine to generate a MS classifier which is able to classify any square wxw block of a brain MRI slice as MS block (y=1) or non-MS block (y=0) based on its features vector (x).
Training the classifier with more than one subject dataset allow the classifier to learn broad ranges for the features of real MS-blocks. However, due to computation time, training using many datasets is supported with each contributing only with a specified percentage to the overall training set. Usually the amount of MS blocks is much lower than the non-MS blocks. Hence, the share of blocks from each single dataset in the training set includes all MS blocks and the share is completed by selecting randomly non-MS blocks that cover all brain areas. One of the datasets will be the reference for intensity correction, while the other datasets are intensity corrected according to the reference dataset before adding their shares to the training set to maintain one intensity reference that can be used for the datasets to be segmented.
2.4.1.5. Segmentation
In the selected brain view, each of the slices of the subject dataset to be segmented is divided into non-overlapping square blocks of size wxw pixels. The features vector for each block is calculated. The trained SVM for the corresponding brain view is used to predict the class labels for all the non-overlapping blocks. The block division is done in a non-overlapping manner to reduce computation time taking in consideration the high resolution of each slice and performing the segmentation three times, once for each sectional view. For any block classified as MS block, assuming true positive classification, this does not mean that all pixels of the block should be classified as MS pixels because the SVM engine is trained to detect the blocks that contains MS pixels completely or partially. However, during segmentation, if any block is classified as MS block (y=1), all pixels inside the block are marked as MS voxel while the final decision for the individual pixels taken by the aggregate segmentation step. For the sagittal and coronal sectional-view segmentations, the output segmentation is rotated to be in the axial view. The output segmentations of the axial, sagittal and coronal sectional views segmentation engines are noted as A, B and C, respectively.
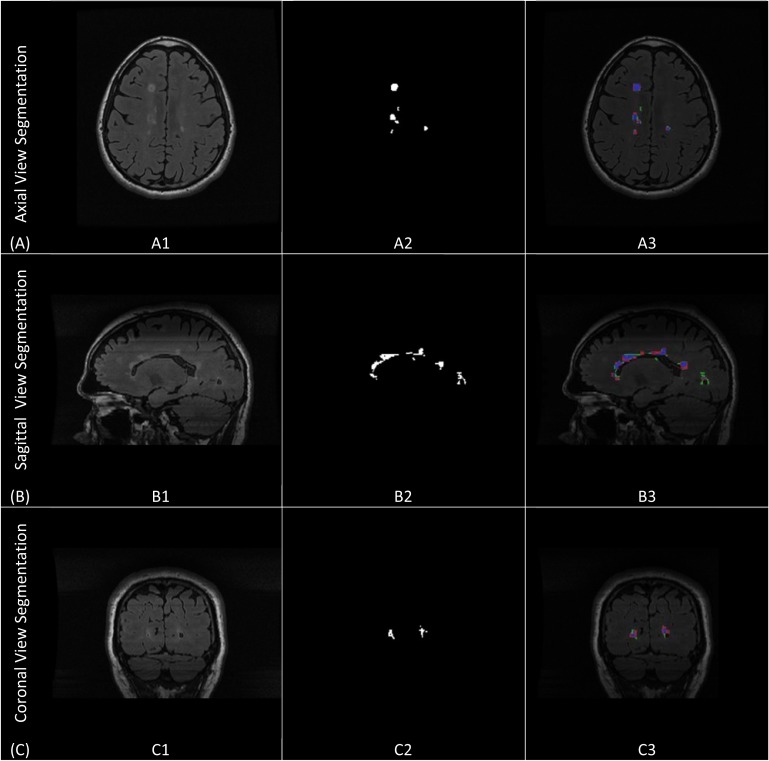
Fig. (5) shows the single sectional view segmentation done in each of the three views. It shows segmentation of sample slices (axial, sagittal and coronal) from subject (CHB_train_Case01). Figs. (5A, A1, A2 and A3) demonstrate axial sectional view segmentation. Fig. (5A1) shows the A sample FLAIR slice in axial view. Fig. (5A2) shows the ground truth for the lesions generated through manual segmentation. Fig. (5A3) provides the colored evaluation of the automatic segmentation generated by the axial view segmentation engine where the true positive pixels are marked by blue, false positive are marked by red, false negatives are marked by green and true negatives are the background pixels. Similarly, Figs. (5B, B1, B2 and B3) and Figs. (5C, C1, C2 and C3) demonstrates sagittal and coronal views segmentations.
Fig. (5).
Single sectional-view segmentation. (A) Axial sectional-view segmentation. (A1) A sample slice from CHB_train_Case07 in the axial view. (A2) The ground truth for the sample slice. (A3) The colored evaluation of the axial view automatic segmentation. (B) Sagittal sectional-view segmentation. (B1) A sample slice from CHB_train_Case07 in the sagittal view. (B2) The ground truth for the sample slice. (B3) The colored evaluation of the sagittal view automatic segmentation.(C) Coronal sectional-view segmentation. (C1) A sample slice from CHB_train_Case07 in the coronal view. (C2) The ground truth for the sample slice. (C3) The colored evaluation of the coronal view automatic segmentation.
2.4.2. Aggregate Segmentation
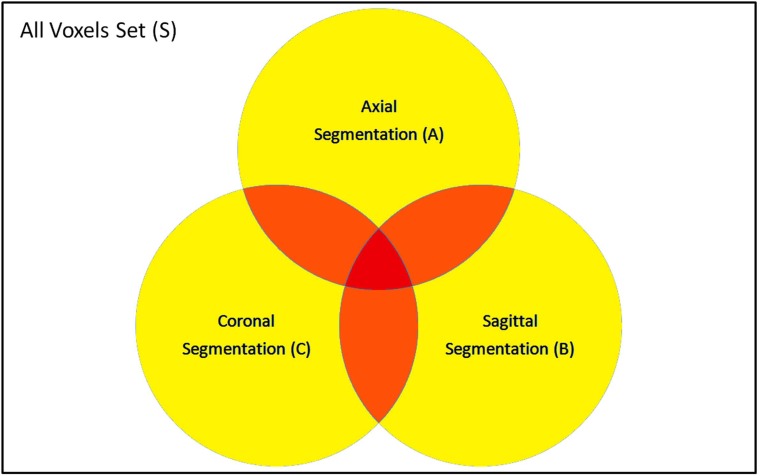
The set S includes all voxels in the dataset to be segmented. The resultant segmentations from the single view classification can be represented by sets A, B and C for the axial, sagittal and coronal segmentations, respectively. Each set includes the voxels that are classified as MS-voxel in the corresponding view segmentation. In the best case scenario, all the positive and negative classes are true, and A, B and C should be identical. However, reality is that the three sets intersect with each other as shown in Fig. (6). As the number of segmentation sets where a certain voxel is included increases, the higher the probability of this voxel to be true positives.
Fig. (6).
Segmentation sets of single views: Axial (A), Sagittal (B) and Coronal (C).
The aggregate segmentation function decides for each voxel v Є S if it should be included in the aggregate segmentation set (G) according to the relation between υ and the three sets of the sectional views segmentations A, B and C. We model the aggregation as a binary classification problem that assigns for each voxel v Є S either the class MS1 (included in G) or the class MS0 (not included in G).
For each voxel v, we define a discrete variable Xv that represents the number of segmentation sets where the voxel υ is included. The variable Xv can take discrete symbolic values X0, X1, X2 and X3 according to the following definition:
| (1) |
According to the above definition and the demonstration in Fig. (6), Xν= X0 if ν falls inside the white area (ν is not included in any of A,B or C), Xν= X1 if ν falls inside the yellow area (ν is inside either A,B or C), Xν= X2 if ν falls inside the orange area (ν is included by any two sets of A,B and C) and Xν= X3 if v falls inside the red area (ν is included in A,B and C).
The Bayesian decision rule is applied to obtain the posterior probability P(MS1| Xν). This involves the calculation of the classes priors P(MSi) and the likelihood functions P(Xν|MSi) for (i=0 to 1). In order to obtain the prior probabilities and the likelihoods, statistical analysis was performed on the training datasets (the 20 training datasets from the MS Lesion Challenge). The prior probability P(MS1) is the percentage of voxels that are manually labeled as MS-voxels in the datasets ground truth while the prior probability P(MS0) is the percentage of voxels that are manually labeled as normal voxels. Then the training datasets were segmented using the three engines of the single view (Axial, Sagittal and Coronal) segmentations to get the three sets A, B and C respectively for the training datasets. The likelihood function P(Xν=Xi|MS1) is the percentage of voxels that are manually labeled as MS-voxels in the datasets ground truth and in the same time Xv=Xi according to the automatic segmentation for (i=0 to 3). In the same manner, P(Xν=Xi|MS0) is calculated for (i=0 to 3).
The posterior probability P(MS1| Xν) is calculated using the Bayesian decision rule:
| (2) |
where the evidence P(Xv) is given by:
| (3) |
Finally, the aggregate segmentation function uses the posterior probability P(MS1| Xν) to select voxels from the the S to be included in the aggregate set G according the following rule:
| (4) |
with τ is a threshold that converts the posterior into a binary decision. The value 0.5 is selected for τ.
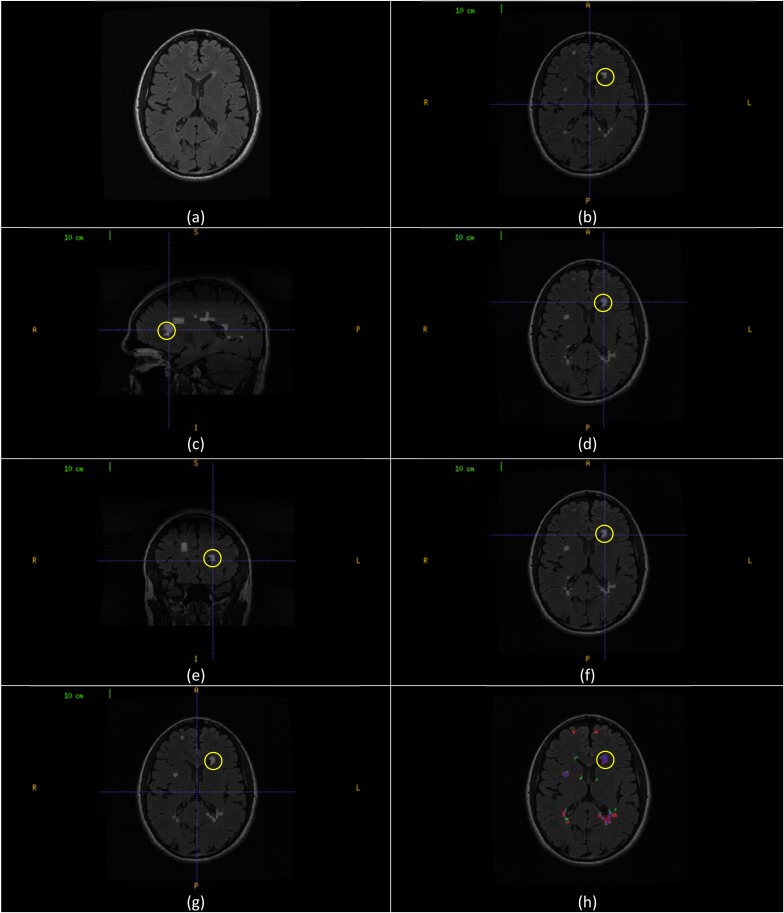
The aggregate segmentation of the multi-sectional views’ segmentations is demonstrated in Fig. (7). Fig. (7a) shows a sample axial slice (257) from CHB_train_Case07. Fig. (7b) shows the axial view segmentation for this slice referred to by A257 (A stands to the axial segmentation set and the subscript 257 stands to the slice number). A segmented lesion region (RA) is highlighted by yellow circle. Fig. (7c) shows sagittal sectional-view segmentation performed in a sagittal slice with the highlighted area intersects with (RA). After performing sagittal view segmentation for all sagittal slices, the resultant segmentation is rotated to the axial view (B). The result of this segmentation that corresponds to the original sample slice, referred to by B257 (B stands to the set of sagittal segmentation rotated to axial view and the subscript 257 stands to the slice number in axial view) is shown in Fig. (7d) with the highlighted area is a new segmentation for RA namely (RB). Similarly, Fig. (7e) shows coronal sectional-view segmentation performed in a coronal slice with the highlighted area intersects with (RA). After performing coronal view segmentation for all coronal slices, the resultant segmentation is rotated to the axial view (C). The result of this segmentation that corresponds to the original sample slice, referred to by C257 (C stands to the set of coronal segmentation rotated to axial view and the subscript 257 stands to the slice number in axial view) is shown in Fig. (7f) with the highlighted area is a new segmentation for RA namely (RC). Fig. (7g) gives the aggregate segmentation of A257, B257 and C257 referred to by G257 (G stands to the aggregate segmentation set and the subscript 257 stands to the slice number in axial view). The highlighted lesion region (RG) is the aggregate of the (RA), (RB) and (RC). Fig. (7h) shows the colored evaluation of the aggregate segmentation with the highlighted lesion region (RG) is colored mostly with blue (true positive voxels) along with small portions colored with red and green (false positive and false negative voxels respectively).
Fig. (7).
Aggregate segmentation of the multi-sectional views’ segmentations. (a) Axial slice (257) from CHB_train_Case07. (b) Axial sectional-view segmentation of the slice (A257) with a lesion region (RA) highlighted in yellow. (c) Sagittal sectional-view Segmentation. The highlighted lesion region intersects with (RA). (d) Segmentation of the original slice (B257) in sagittal sectional-siew segmentation rotated to axial view with lesion region (RB) highlighted in yellow. (e) Coronal sectional-view segmentation. The highlighted lesion region intersects with (RA). (f) Segmentation of the original slice (C257) in coronal sectional-siew segmentation rotated to axial view with lesion region (RC) highlighted in yellow. (g) Segmentation of the original slice (G257) by aggregates segmentation of (A257), (B257) and (D257). The marked lesion region (RG) is the aggregate of (RA), (RB) and (RC). (h) Colored evaluation of the aggregate segmentation (G257) and the highlighted area is the colored evaluation if (RG).
2.5. Evaluation of the Proposed Method
To evaluate the performance of the proposed segmentation method, comparisons are performed with state of the art methods. Different metrics are used to evaluate the performance for these publications, therefore, according to the comparison needed; difference metrics will be calculated in the next section of this paper that shows the results of the segmentation. Along with evaluation of the segmentation with reference to the ground truth and comparing results with other methods in a manual scheme, the proposed method have been tested on the test cases provided by the MS Lesion Segmentation Challenge [32] and results were obtained automatically and compared to other techniques on the same test cases. In the following, the different metrics used in comparisons are defined.
2.5.1. Dice Similarity (DS)
The dice similarity (DS) is a measure of the similarity between the manual segmentation (X) and the automatic segmentation (Y). The equation for the calculation can be written as:
| (5) |
As stated in [39, 40], a DS score above 0.7 is generally considered as very good. For each slice, each group of connected pixels automatically or manually labeled as MS pixels forms an automatic or manual lesion region respectively. In our evaluation, dice similarity is calculated based on the similarity of lesion regions. In equation (5), the term |X∪Y| is substituted by the number of common MS lesion regions between manual and automatic segmentation. Also, |X| and |Y| are substituted by the number of MS lesion regions of manual and automatic segmentation respectively. In this context, the automatically segmented lesion region that shares at least one pixel with a manually segmented lesion region is considered as a common MS lesion region [10].
2.5.2. Detected Lesion Load (DLL)
We introduce the Detected Lesion Load (DLL) metric as a percentage of detected lesion volume with reference to the original lesion volume. The detected lesion volume takes into account all the positive lesions whether true or false. Having a DLL close to 1.0 is clinically satisfactory since it provides a relatively accurate measure of the MS lesions volume.
2.5.3. True Positive Rate (TPR) and Positive Predictive Value (PPV)
For the two sets manual segmentation (X) and automatic segmentation (Y), true positives (TP), false positives (FP) and false negatives (FN) can be calculated as:
| (6) |
| (7) |
| (8) |
The True positive rate (TPR) and Positive Predictive Value (PPV) are defined as:
| (9) |
| (10) |
2.5.4. MS Lesion Segmentation Challenge Metrics and Score
The automated evaluation system used by the MS Lesion Segmentation Challenge uses the volume difference (Volume diff.), average distance (Avg. Dist.), true positive rate (True Pos.) and false positive rate (False Pos.) to evaluate the segmentation. These metrics are defined in [31] as follows:
Volume Difference, in percent:
The total absolute volume difference of the segmentation to the reference is divided by the total volume of the reference, in percent.
Average Distance, in millimeters:
The border voxels of segmentation and reference are determined. These are defined as those voxels in the object that have at least one neighbor (of their 18 nearest neighbors) that does not belong to the object. For each voxel along one border, the closest voxel along the other border is determined (using unsigned Euclidean distance in real world distances, thus taking into account the different resolutions in the different scan directions). All these distances are stored, for border voxels from both reference and segmentation. The average of all these distances gives the averages symmetric absolute surface distance.
True Positive Rate, in percent:
This is measured by dividing the number of lesions in the segmentation that overlap with a lesion in the reference segmentation with the number of overall lesions in the reference segmentation. This evaluates whether all lesions have been detected that are also in the reference segmentation.
False Positive Rate, in percent:
This is measured by dividing the number of lesions in the segmentation that do not overlap with any lesion in the reference segmentation with the number of overall lesions in the segmentation. This rate represents whether any lesions are detected that are not in the reference.
All measures have been scored in relation to how the expert raters compare against each other. A score of 90 for any of the metric indicate a comparable performance with an expert rater. The overall score for each test case is an average of the score of the above four metrics calculated for two different raters. An overall score is an average for the scores of each individual test case.
3. RESULTS
Evaluation was performed using both synthetic data and real MRI data containing varying levels of MS lesion load at different locations in the brain. The following subsections provide details about the evaluation settings, metrics, and result in comparison to other methods. Comments on the results are provided in the discussion section.
3.1. Synthetic Data
The BrainWeb database provides three simulated subjects brains involving three levels of MS lesion; mild, moderate and severe referred to in this paper as MSLES 1, MSLES2 and MSLES3, respectively. The features vector used in the proposed method uses multi-channels image intensities with textural features from the FLAIR sequence. However, the FLAIR sequence is not provided in the BrainWeb MRI data, but the Inversion recovery sequence (IR) provided in the BrainWeb MRI data is used instead since it is the closest sequence to FLAIR. Besides, the feature vector depends on spatial priors’ atlas to provide white matter, gray matter and CSF tissues probability for each voxel. For this purpose, a simulated atlas was generated using anatomical models provided for the twenty normal simulated brains in the BrainWeb database as described in preprocessing section.
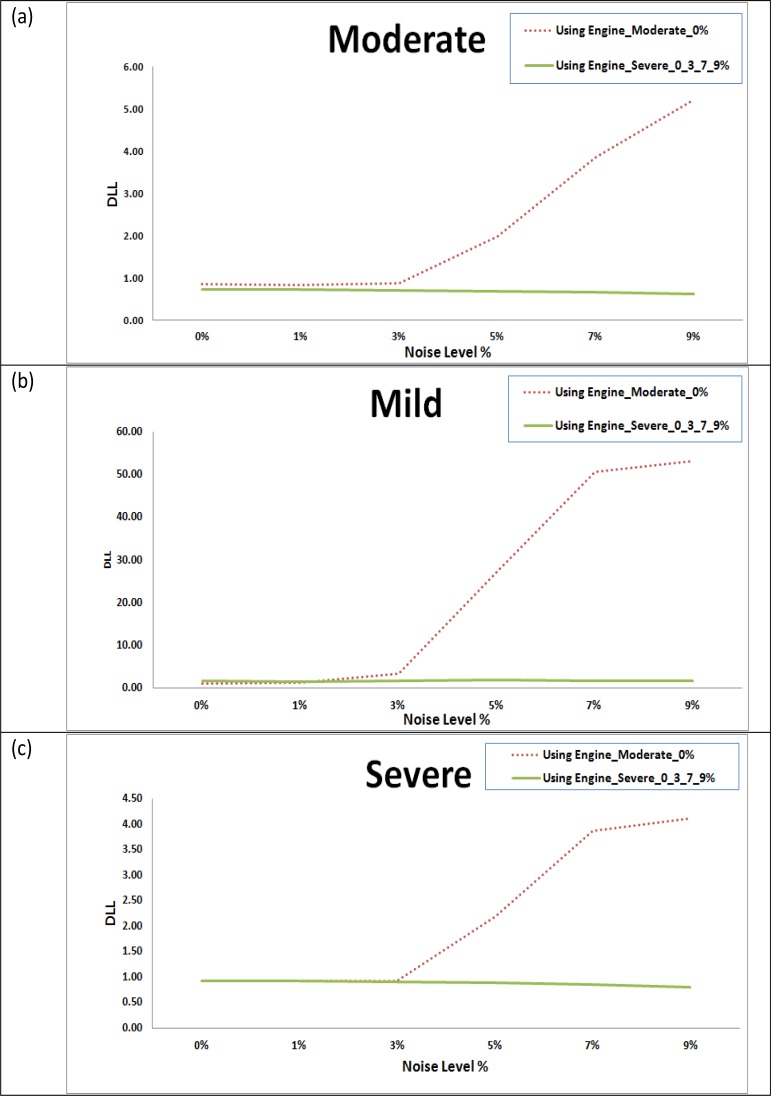
In the training phase, two different engines were trained. The first engine (Engine_Moderate_0%) was trained using moderate level lesion subject MSLES2 with 0% of noise level. The second engine (Engine_Severe_0_3_7_9%) was trained using severe level lesion subject MSLES3 at noise levels 0%, 3%, 7% and 9% with each noise level sharing 25% of the training set. In segmentation phase, all the three subjects were tested with all noise levels. For each case, the segmentation is evaluated using detected lesion load (DLL) and dice similarity (DS) for both trained engines. Fig. (8) shows the DLL for segmentation results using the two engines for the mild, moderate and severe cases in Fig. (8a), (8b) and (8c) respectively for all noise levels.
Fig. (8).
Detected Lesion Load (DLL) for testing the two trained engines on BrainWeb datasets for different noise levels based on dice similarities for different subject templates. (a) Mild lesion level subject. (b) Moderate lesion level subject. (c) Severe lesion level subject.
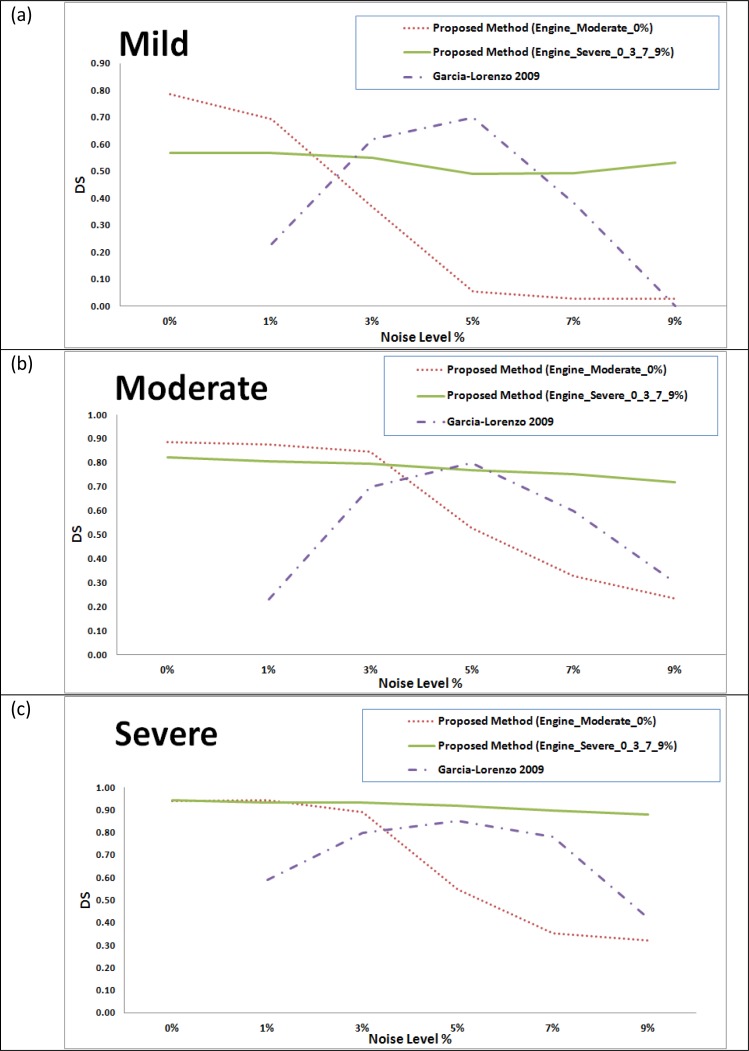
The results were compared to those obtained by Garcia-Lorenzo et al., [41] for the same dataset. The comparison for the mild, moderate and severe cases are provided in Figs. (9a), (9b) and (9c) respectively. In [41], the training was made using the 0% noise level template for each case and the segmentation is tested using the other noise levels, excluding the 0% noise level, in contrast to our testing that involves intra subject evaluation (training with a subject template and testing another subject). Therefore, the 0% will have no results in the charts for the Garcia Lorenzo [41] data in Fig. (9).
Fig. (9).
Comparison between the proposed method and a state of the art method [41] on BrainWeb datasets for different noise levels based on dice similarities for different subject templates. (a) Mild lesion level subject. (b) Moderate lesion level subject. (c) Severe lesion level subject.
Besides, the results were compared to Leemput et al., [16] and Freifeld et al., [42] for the moderate dataset, which was the only MS lesion level provided by the BrainWeb at the time of publication of these two methods. The comparison for the different noise levels is shown in Table 1. In [16] and [42], the training was made using the 0% and 1% noise levels for the moderate case template and the segmentation is tested using the other noise levels excluding the 0% and 1% noise levels for the same case where our testing involves segmentation of all moderate case noise levels using the engine trained by severe case (Engine_Severe_0_3_7_9%). Therefore, the 0% and 1% will have no results for [16, 42] data in Table 1.
Table 1.
Comparison Between the Proposed Method and State of the Art Methods [16, 42] on BrainWeb Moderate Lesion Level Dataset for Different Noise Levels Based on of Dice Similarities
3.2. Real Data
Datasets of 61 cases were used to verify the segmentation technique proposed in this paper. The sources of these datasets are the workshop of MS Lesion Segmentation Challenge 2008 (51 subjects) and real MRI datasets acquired for MS subjects at the University of Miami Miller School of Medicine (10 subjects). The subjects datasets of the MS Lesion Segmentation Challenge are categorized as twenty subjects provided for training (10 from CHB and 10 from UNC) and thirty one subjects provided for testing (18 from CHB and 13 from UNC) and labels are not provided for the testing set.
In the training phase, two different engines were trained. The first engine (Engine_CHB) was trained using four subjects of the training dataset (CHB_train_Case01, CHB_train_Case02, CHB_train_Case06 and CHB_train_Case10) with each subject sharing 25% of the training set. The second engine (Engine_UNC) was trained using four subjects of the training dataset (UNC_train_Case02, UNC_train_Case03, UNC_train_Case09 and UNC_train_Case10) with each subject sharing 25% of the training set. As recommended by Anbeek et al., [12], CHB_train_Case04, CHB_train_Case05, CHB_train_Case09, UNC_train_Case01, UNC_train_Case05 and UN-C_train_Case06 were avoided due to image and manual segmentation quality. For the UNC training set, only the manual segmentations of the CHB rater were used.
In the segmentation phase, three groups of subjects were tested. The first group is composed of the MRI dataset acquired for MS subjects at the University of Miami Miller School of Medicine. The second group is composed of the training set of CHB provided by MS Lesion Challenge. The third group is composed of the testing cases provided by MS Lesion Challenge. For each group, the segmentation is evaluated using the metrics that match those used in other methods using the MS Lesion Segmentation Challenge datasets to facilitate the comparison with other method for the purpose of the evaluation.
3.2.1. MS Subjects MRI Data
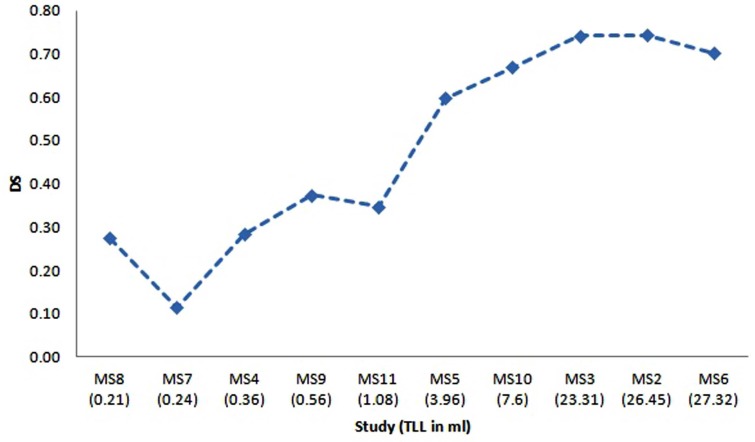
The ten MRI studies for MS subjects acquired at University of Miami, referred to as MS2 to MS11, have been re-sliced to have the same resolution of the training set of Engine_CHB and registered to the MNI atlas as described in the preprocessing section. The segmentation of these datasets was performed to check the robustness of the proposed technique when training is done using one source of data with set of conditions of acquisition and segmentation is performed using a different source of data with different set of conditions. The dice similarity for the segmentation of the ten subjects is shown in Fig. (10). The X-axis provides the subjects ordered by the total lesion load (TLL) in ml to show the effect of this parameter on the segmentation performance. An approximate total lesion load is calculated by counting the number of MS voxles in the ground truth and multiplying by the voxel volume.
Fig. (10).
Dice similarity versus the total lesion load for the segmentation of ten MRI studies for MS subjects acquired at University of Miami. X-Axis gives the study name ordered by the total lesion load in ml.
3.2.2. MS Lesion Segmentation Challenge Training Set Data
The ten CHB subjects provided by the MS Lesion Segmentation Challenge datasets are segmented by the proposed technique. For the purpose of comparison, segmentation results are evaluated using TPR and PPV in the same manner reported by Geremia et al., [5] who provides results for segmenting the training datasets. Table 2 shows a comparison between the proposed technique, Geremia et al., [11] and Souplet et al., [43] (the best result in the MS Lesion Segmentation Challenge at that time). The value marked in bold is the best metric value obtained.
Table 2.
| Study Case | TPR | PPV | ||||
|---|---|---|---|---|---|---|
| Souplet et al. 2008 [43] | Geremia et al. 2010 [11] | Proposed Method | Souplet et al. 2008 [43] | Geremia et al. 2010 [11] | Proposed Method | |
| CHB_train_Case01 | 0.22 | 0.49 | 0.73 | 0.41 | 0.64 | 0.48 |
| CHB_train_Case02 | 0.18 | 0.44 | 0.02 | 0.29 | 0.63 | 0.56 |
| CHB_train_Case03 | 0.17 | 0.22 | 0.14 | 0.21 | 0.57 | 0.06 |
| CHB_train_Case04 | 0.12 | 0.31 | 0.48 | 0.55 | 0.78 | 0.04 |
| CHB_train_Case05 | 0.22 | 0.4 | 0.44 | 0.42 | 0.52 | 0.10 |
| CHB_train_Case06 | 0.13 | 0.32 | 0.15 | 0.46 | 0.52 | 0.42 |
| CHB_train_Case07 | 0.13 | 0.4 | 0.29 | 0.39 | 0.54 | 0.54 |
| CHB_train_Case08 | 0.13 | 0.46 | 0.76 | 0.55 | 0.65 | 0.47 |
| CHB_train_Case09 | 0.03 | 0.23 | 0.18 | 0.18 | 0.28 | 0.09 |
| CHB_train_Case10 | 0.05 | 0.23 | 0.38 | 0.18 | 0.39 | 0.43 |
3.2.3. MS Lesion Segmentation Challenge Testing Set Data
After testing the technique with the available datasets that contain the ground truth, the test cases provided by MS Lesion Segmentation Challenge are segmented using the proposed method with the CHB test cases segmented using the Engine_CHB and the UNC test cases segmented using Engine_UNC for the purpose of comparison and competition with other competitors have done the same. The segmentation of the test cases were uploaded to the MS Lesion Segmentation Challenge to get the automatic evaluation of the segmentation with team name (UM-ECE_team) and the results were posted and can be accessed from the official MS Lesion Segmentation Challenge results section [44]. A snapshot of results is depicted in Table 3 with the performance metrics and scores are explained in subsection 2.5.4. For each test case, the total is the average of all scores of the different metrics with reference to both raters. The average of scores is the team score. Currently, the proposed technique segmentation score is included in the best ten scores knowing that the proposed technique is the only technique that was tested using 31 test cases where the others were tested using only 25 test cases.
Table 3.
Snapshot of the Results Table Generated Automatically by the MS Lesion Segmentation Challenge Workshop Evaluation Software for the Segmentation of the Test Cases [44]
| Ground Truth | UNC Rater | CHB Rater | Total | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| All Dataset | Volume Diff. | Avg. Dist. | TRUE Pos. | FALSE Pos. | Volume Diff. | Avg. Dist. | TRUE Pos. | FALSE Pos. | |||||||||
| [%] | Score | [mm] | Score | [%] | Score | [%] | Score | [%] | Score | [mm] | Score | [%] | Score | [%] | Score | ||
| UNC test1 Case01 | 66.2 | 90 | 14.7 | 70 | 18.6 | 62 | 63.6 | 71 | 50.6 | 93 | 10.1 | 79 | 25 | 66 | 31.8 | 90 | 78 |
| UNC test1 Case02 | -1 | 0 | -1 | 0 | -1 | 0 | -1 | 0 | -1 | 0 | -1 | 0 | -1 | 0 | -1 | 0 | 0 |
| UNC test1 Case03 | 55.3 | 92 | 3.8 | 92 | 16.2 | 61 | 26 | 94 | 42.3 | 94 | 3.2 | 93 | 19.1 | 62 | 12 | 100 | 86 |
| UNC test1 Case04 | 100 | 85 | 128 | 0 | 0 | 51 | 0 | 100 | 100 | 85 | 128 | 0 | 0 | 51 | 0 | 100 | 59 |
| UNC test1 Case05 | 86 | 87 | 11.7 | 76 | 19 | 62 | 31.2 | 91 | 68.3 | 90 | 9.3 | 81 | 30.4 | 69 | 37.5 | 87 | 80 |
| UNC test1 Case06 | 99.9 | 85 | 80.7 | 0 | 0 | 51 | 100 | 49 | 99.5 | 85 | 82 | 0 | 0 | 51 | 100 | 49 | 46 |
| UNC test1 Case07 | 96.2 | 86 | 28 | 42 | 3.3 | 53 | 20 | 97 | 91.1 | 87 | 20.9 | 57 | 10 | 57 | 20 | 97 | 72 |
| UNC test1 Case08 | 44.8 | 93 | 9.6 | 80 | 19.1 | 62 | 53.3 | 77 | 9.8 | 99 | 6 | 88 | 55.6 | 83 | 46.7 | 81 | 83 |
| UNC test1 Case09 | 256.2 | 62 | 48.2 | 1 | 0 | 51 | 100 | 49 | 402.6 | 41 | 53.4 | 0 | 0 | 51 | 100 | 49 | 38 |
| UNC test1 Case10 | 100 | 85 | 128 | 0 | 0 | 51 | 0 | 100 | 100 | 85 | 128 | 0 | 0 | 51 | 0 | 100 | 59 |
| UNC test1 Case11 | 98.2 | 86 | 15.3 | 68 | 3.2 | 53 | 25 | 94 | 98.4 | 86 | 13.2 | 73 | 4.8 | 54 | 0 | 100 | 77 |
| UNC test1 Case12 | 100 | 85 | 128 | 0 | 0 | 51 | 0 | 100 | 100 | 85 | 128 | 0 | 0 | 51 | 0 | 100 | 59 |
| UNC test1 Case13 | 143.5 | 79 | 29.3 | 40 | 0 | 51 | 100 | 49 | 153.9 | 77 | 16.1 | 67 | 33.3 | 70 | 83.3 | 59 | 62 |
| UNC test1 Case14 | 103.3 | 85 | 10.6 | 78 | 44.4 | 77 | 64.7 | 70 | 113.4 | 83 | 15.2 | 69 | 25 | 66 | 82.4 | 60 | 73 |
| CHB test1 Case01 | 243.7 | 64 | 8.1 | 83 | 45.3 | 77 | 78 | 62 | 391 | 43 | 10.5 | 78 | 77.4 | 95 | 83.9 | 59 | 70 |
| CHB test1 Case02 | 445.1 | 35 | 9.6 | 80 | 77.3 | 95 | 92.7 | 53 | 132.3 | 81 | 4.4 | 91 | 84.2 | 99 | 87.6 | 56 | 74 |
| CHB test1 Case03 | 112 | 84 | 15 | 69 | 50 | 80 | 93.5 | 53 | 2.4 | 100 | 12.6 | 74 | 53.3 | 82 | 91.9 | 54 | 74 |
| CHB test1 Case04 | 80.2 | 88 | 19 | 61 | 27.3 | 67 | 76.5 | 63 | 90.5 | 87 | 24.2 | 50 | 16.7 | 61 | 76.5 | 63 | 68 |
| CHB test1 Case04 | 80.2 | 88 | 19 | 61 | 27.3 | 67 | 76.5 | 63 | 90.5 | 87 | 24.2 | 50 | 16.7 | 61 | 76.5 | 63 | 68 |
| CHB test1 Case05 | 11422.6 | 0 | 17.4 | 64 | 70.4 | 91 | 98.5 | 50 | 2085.8 | 0 | 11.8 | 76 | 78.3 | 96 | 97.8 | 50 | 53 |
| CHB test1 Case06 | 173.6 | 75 | 3.6 | 93 | 75 | 94 | 96.9 | 51 | 185.9 | 73 | 3.8 | 92 | 45.5 | 77 | 98.3 | 50 | 75 |
| CHB test1 Case07 | 140.2 | 79 | 7.7 | 84 | 41.7 | 75 | 75 | 64 | 46.1 | 93 | 2.8 | 94 | 50 | 80 | 50 | 79 | 81 |
| CHB test1 Case08 | 19.7 | 97 | 20.4 | 58 | 18.5 | 62 | 71.4 | 66 | 46.2 | 93 | 21.4 | 56 | 11.8 | 58 | 66.7 | 69 | 70 |
| CHB test1 Case09 | 775.8 | 0 | 4.9 | 90 | 80.5 | 97 | 84.4 | 58 | 638.6 | 6 | 4.4 | 91 | 67.3 | 90 | 86.7 | 57 | 61 |
| CHB test1 Case10 | 754.2 | 0 | 10.2 | 79 | 68.4 | 90 | 90.1 | 55 | 317.7 | 53 | 3.6 | 93 | 69 | 91 | 77.2 | 63 | 65 |
| CHB test1 Case11 | 1120.7 | 0 | 14 | 71 | 56.8 | 84 | 96 | 51 | 294.7 | 57 | 7.6 | 84 | 51.7 | 81 | 92 | 54 | 60 |
| CHB test1 Case12 | 32.4 | 95 | 2.9 | 94 | 27.7 | 67 | 60.7 | 73 | 32.7 | 95 | 3 | 94 | 28.2 | 68 | 69.2 | 68 | 82 |
| CHB test1 Case13 | 12.8 | 98 | 5.8 | 88 | 40 | 74 | 59.3 | 74 | 46.6 | 93 | 3.3 | 93 | 28.6 | 68 | 7.4 | 100 | 86 |
| CHB test1 Case15 | 337.4 | 51 | 4.5 | 91 | 83.6 | 99 | 90.3 | 55 | 476.8 | 30 | 5.6 | 89 | 93.6 | 100 | 95.1 | 52 | 71 |
| CHB test1 Case16 | 62.8 | 91 | 6 | 88 | 22.5 | 64 | 50 | 79 | 68 | 90 | 4.1 | 92 | 40 | 74 | 47.2 | 81 | 82 |
| CHB test1 Case17 | 123 | 82 | 7.3 | 85 | 32.1 | 70 | 58.9 | 74 | 9.4 | 99 | 3.4 | 93 | 22 | 64 | 43.8 | 83 | 81 |
| CHB test1 Case18 | 71.5 | 90 | 62.5 | 0 | 0 | 51 | 100 | 49 | 7.4 | 99 | 58.8 | 0 | 0 | 51 | 100 | 49 | 49 |
| All Average | 557.3 | 69 | 27.5 | 59 | 30.3 | 67 | 63.1 | 67 | 203.3 | 74 | 25.7 | 63 | 32.9 | 68 | 57.5 | 70 | 67 |
| All UNC | 96.3 | 79 | 45.3 | 39 | 8.8 | 53 | 41.6 | 74 | 102.1 | 78 | 43.7 | 43 | 14.4 | 56 | 36.6 | 77 | 62 |
| All CHB | 936.9 | 60 | 12.9 | 75 | 48.1 | 79 | 80.7 | 60 | 286.6 | 70 | 10.9 | 79 | 48.1 | 79 | 74.8 | 64 | 71 |
DISCUSSION
A novel method for MS lesions segmentation in multi-channels brain MR images has been developed. The segmentation process is based on three sectional views of the MRI data that are processed through three identical segmentation engines, one engine for each of the axial, sagittal and coronal views. Through each engine, the multi-channels MRI slices of the brain are preprocessed for intensity correction to remove the effect of noise and variations in brightness and contrast among corresponding sequences of different subjects. The next step in the single sectional view engine is the processing module which is used for the detection of initial MS lesions regions based mainly on textural features. The output segmentations of the axial, sagittal and coronal sectional views’ segmentation engines are aggregated to generate the final accurate segmentation. This work is an extension to our segmentation framework proposed for single view-single MRI channel [45].
The categories of features used within the MS blocks detection are selected to have similarity with the features used non-intentionally by the expert in the task of manual labeling of MS areas. When an expert labels MS lesions in the FLAIR slice, the hyper intense areas are the potential areas to have the lesion. These candidate areas are filtered by the expert based on the positions of these areas, the neighboring areas intensities, the other channels intensities of the same areas with aid of the experience about the brain tissues for the highlighted areas. This is emulated in our technique by using the textural features, position based features, neighboring features, the multi-channels grayscales and probabilistic tissues features.
Our first contribution in the method presented in this paper involves using textural features without manual selection of ROI, which was an area for future research and improvement [20, 21]. To the best of our knowledge, the common use of textural features in MS lesions detection was previously attempted with the aid of manual selection of regions of interests (ROIs). The second contribution of the presented method is introducing the concept of multi-sectional views segmentation to improve the quality of the overall segmentation by doing the segmentation from different perspectives that help, when aggregated, in the assessment of the MS lesion.
Regarding the segmentation results presented in the previous section, for tests done on the synthetic data, two engines were prepared for testing the effect of the training set on the performance of the segmentation. It is clear from Fig. (8) that using the second engine (trained with the severe level of MS lesions and varying noise levels) provides better segmentation performance for the mild, moderate and severe cases for different noise levels compared to the first engine (trained with the moderate MS case at 0% noise level) in terms of and detected lesion load (DLL). Segmentation with the engine trained with the moderate case at 0% noise level fails to maintain DLL close to 1 for high noise levels in which noise results in high false positive rates that results in huge detected lesion load which represent failure in clinical practice. This makes us recommend that training set should involves datasets with several acquisition conditions which leads to robustness in segmentation as obtained in case of using the engine trained with the severe level of MS lesions and varying noise levels.
Comparison with Gracia-Lorenzo et al., [41] in Fig. (9) shows stability of the performance using the second engine against different noise levels even for the mild and moderate cases which are not included in the training set, which supports using different noise levels in the training set. Comparison with Leemput et al., [16] and Freifeld et al., [42] shows competitive performance while considering that we use an engine trained with a different brain template compared to the segmented brain data while both of techniques used the same brain template for training and segmentation with different noise levels.
For the evaluation done using the MS subjects datasets acquired at the University of Miami, the results shown in Table 3 demonstrate the success of the proposed technique to deal with the case when training is done using one source of data and conditions of acquisition and segmentation is performed using a different source of data and different conditions. Results for these datasets show weak performance when dealing with MRI studies that have very low total lesion load (TLL). Low performance in case of segmentation of studies having low TLL of MS lesion is mainly because of failure of texture properties to discriminate the very small structures. Besides, false positive MS regions obtained in case of low TLL will be much more than true positive, hence the dice similarity will dramatically falls to low value. This leads to the conclusion that extra effort is still needed to specifically handle the mild MS lesions cases.
For the test done on the datasets obtained from MS Lesion Challenge, two engines were trained namely Engine_CHB and Engine_UNC for the purpose of comparison and competition with other competitors have done the same. However, having a unified engine in clinical practice will provide better performance when dealing with other subjects.
According to the obtained results, we concluded set of findings. First, the proposed technique provides good segmentation when tested with datasets from a source which is different in acquisition conditions from the source of datasets used in training. Also, the textural based SVM provides good coarse segmentation that can be used as an initial step in any MS segmentation framework. Slice division into blocks enables the technique to benefit from the textural features in detection of the candidate lesion region without manually labeled ROIs. Slice division into blocks also makes the technique avoid voxel based learning which results in high rate of false positives and scattered automatic MS lesions. Besides, basing the classifier on SVM as a machine learning technique provides a robust classifier. Feature selection based on explicit use of human visible features and trying to emulate expert non-intended features aid the technique to find lesion areas that has high similarity (Dice similarity / Similarity Index) with the manually labeled areas. Training the blocks classifier with marking all blocks that contains at least one MS voxel as MS-blocks aids the classifier to find that blocks that are completely or partially are MS blocks. Using more sequences in MRI improves the performance of MS Segmentation. Using Tissues Probabilities based on the MNI atlas improves the performance of MS Segmentation. Performance evaluation that adds metrics such as Dice Similarity for regions (DSR) and Detected Lesion Load (DLL) ensures the clinically relevant performance. The multi-view segmentation pipeline is suitable in cases of high resolution 3D Images that provides Lesion Visibility from all sectional views and it adds to the accuracy of the final segmentation.
More efforts are needed to have overcome limitations of the proposed segmentation framework. First, the technique shows low performance when tested on datasets with low TLL. Also, the technique does not take into consideration the anatomic properties of the brain area that can be determined from the MNI atlas and thus using the probability based on statistics of having MS lesion in a specific area. Lesion regions shape improvement in post-processing does not take the contouring properties into consideration. The textural features are based on FLAIR sequence and does not benefit from the lesion pattern in the other sequences in multi-channels MRI. Performance of using SVM to learn lesion properties is affected by imbalanced training set due to the relative size of the lesion with respect to the normal brain tissues. Finally, Using SVM to learn lesion blocks properties within a slice does not benefit from the 3D information of the MRI in learning but left to the post-processing in case of single view pipelines or the aggregation step in multi-view pipeline.
CONCLUSION
We have developed an automated method for detection of MS lesions in brain MR images using aggregation of segmentations generated by applying a textural-based SVM to multiple sectional views of multi-channels MRI data. The main contributions of the presented method are using textural features without manual selection of ROIs and the concept of the multi-sectional views’ segmentations of volumes of MS lesions that can be generalized to segment any other MRI volume lesions segmentation techniques. The method has been tested using 51 real multi-channels MRI datasets. The performance evaluation and comparative results with other automated techniques demonstrate that our method provides competitive results for the detection of MS lesions.
ACKNOWLEDGMENT
This work was supported in part by National Institutes of Health (NIH), National Institute of Neurological Disease and Stroke (NINDS) through grant #R41NS060473.
The authors would like to thank the MS Lesion Segmentation Challenge 2008 organizers who kindly provided their pre-processed images o allow a fair comparison of results and for their excellent technical support and assistance in reading the data.
The authors would also like to thank Pradip M. Pattany and Efrat Saraf-Lavi for the acquisition and manual labeling of the datasets from the University of Miami Miller School of Medicine.
APPENDIX A TEXTURAL FEATURES EXTRACTION TECHNIQUES
Textural features can be categorized according to the matrix or vector used to calculate the feature. In this section, we are interested with histogram, gradient, run-length matrix and co-occurrence based features. These categories include features that are selected after being tested to be identifying for the texture of regions that suffer from the multiple sclerosis lesions. For all feature calculations, the image is represented by a function f(x,y) of two space variables x and y, x=0,1,…N-1 and y=0,1,…, M-1. The function f(x,y) can take any value i=0,1,…,G-1 where G is total number of intensity levels in the image.
A.1. Histogram Based Features
The intensity level histogram is a function h(i) providing, for each intensity level i, the number of pixels in the whole image having this intensity.
| (A-1) |
The histogram is a concise and simple summary of the statistical information contained in the image. Dividing the histogram h(i) by the total number of pixels in the image provides the approximate probability density of the occurrence of the intensity levels p(i), given by:
| (A-2) |
The following set of textural features is calculated from the normalized histogram:
| (A-3) |
| (A-4) |
| (A-5) |
| (A-6) |
A.2. Gradient Based Features
The gradient matrix element g(x,y) is defined for each pixel in the image based on the neighborhood size. For a 3x3 pixels neighborhood, g is defined as follows:
| (A-7) |
The following set of textural features is calculated from the gradient matrix:
| (A-8) |
| (A-9) |
Skewness and kurtosis of the absolute gradient can be calculated similar to those calculated for histogram.
A.3. Run Length Matrix Based Features
The run length matrix is defined for a specific direction. Usually a matrix is calculated for the horizontal, vertical, 45° and 135° directions. The matrix element r(i,j) is defined as the number of times there is a run of length j having gray level i. Let G be the number of gray levels and Nr be the number of runs. The following set of textural features is calculated from the run length matrix:
| (A-10) |
| (A-11) |
| (A-12) |
| (A-13) |
where the normalization coefficient C is defined as follows:
| (A-14) |
A.4. Co-occurrence Matrix Based Features
The co-occurrence matrix is a form of second order histogram that is defined for certain angle θ and certain distance d. The matrix element hdθ(i,j) is the number of times f(x1,y1) = i and f(x2,y2) = j where (x2,y2)=(x1,y1) + (d cos θ, d sin θ). Usually the co-occurrence matrix is calculated for d = 1 and 2 with angles θ = 0°, 45°, 90° and 135°. When the matrix element hdθ (i,j) is divided by the total number of neighboring pixels, the matrix becomes the estimate of the joint probability codθ (i,j) of two pixels, a distance d apart along a given direction θ having co-occurring values i and j. Let µx, µy, σx and σy denote the mean and standard deviation of the row and column sums of the matrix co, respectively. The following set of textural features is calculated from the co-occurrence matrix:
| (A-15) |
| (A-16) |
| (A-17) |
| (A-18) |
| (A-19) |
REFERENCES
- 1.C Zhu, T Jiang. Knowledge guided information fusion for segmentation of multiple sclerosis lesions in MRI images. SPIE03. 2003;5032 [Google Scholar]
- 2.C Confavreux, H Vukusic, S J Grimaud. Clinical progression and decision making process in multiple sclerosis. Multiple Sclerosis. 1999;5:212–5. doi: 10.1177/135245859900500403. [DOI] [PubMed] [Google Scholar]
- 3.KV Leemput. PhD Thesis. Leuven, Belgium: Katholieke Universiteit Leuven; 2001. Quantitative analysis of signal abnormalities in MR imaging for multiple sclerosis and creutzfeldtjakob disease. [Google Scholar]
- 4.LJ Rosner, S Ross. Multiple Sclerosis. New York: Simon and Schuster; 1992. [Google Scholar]
- 5.S Bricq, C Collet, J-P Armspach. Lesions detection on 3D brain MRI using trimmmed likelihood estimator and probabilistic atlas. Biomed Imag: From Nano to Macro. ISBI. 5th IEEE Int Sym, Paris. 2008. pp. 93–6.
- 6.CJ Wallace, TP Seland, and TC Fong. Multiple sclerosis: the impact of MR imaging. Am. J. Roentg. 1992;158:849–57. doi: 10.2214/ajr.158.4.1546605. [DOI] [PubMed] [Google Scholar]
- 7.S Wiebe, DH Lee, SJ Karlik, M Hopkins, MK Vandervoort, CJ Wong, L Hewitt, GP Rice, GC Ebers, JH Noseworthy. "Serial cranial and spinal cord magnetic resonance imaging in multiple sclerosis". Ann. Neurol. 1992;32:643–50. doi: 10.1002/ana.410320507. [DOI] [PubMed] [Google Scholar]
- 8.L Truyen. Magnetic resonance imaging in multiple sclerosis: a review. Acta Neurol. Belg. 1994;94:98–102. [PubMed] [Google Scholar]
- 9.F Fazekas, et al. The contribution of magnetic resonance imaging to the diagnosis of multiple sclerosis. Neurology. 1999;53:448–56. doi: 10.1212/wnl.53.3.448. [DOI] [PubMed] [Google Scholar]
- 10.D Yamamoto, et al. Computer-aided detection of multiple sclerosis lesions in brain magnetic resonance images: False positive reduction scheme consisted of rule-based, level set method, and support vector machine. Comput. Med. Imaging Graph. 2010 Jul;34(5):404–13. doi: 10.1016/j.compmedimag.2010.02.001. [DOI] [PubMed] [Google Scholar]
- 11.Ezequiel Geremia, et al. Spatial Decision Forests for MS Lesion Segmentation in Multi-Channel MR Images. Med. Image Comput. Comput. Assist. Interv. 2010;6361:111–118. doi: 10.1007/978-3-642-15705-9_14. [DOI] [PubMed] [Google Scholar]
- 12.P Anbeek, KL Vincken, MA Viergever. Automated MSLesion Segmentation by K-Nearest Neighbor Classification. MIDAS J . 2008. (workshop) (workshop)
- 13.F Rousseau, F Blanc, J de Seze, L Rumbach, J Armspach. An a contrario approach for outliers segmentation: Application to Multiple Sclerosis in MRI. "Biomedical Imaging: From Nano to. Macro. ISBI. 5th IEEE International Symposium, Paris. 2008. pp. 9–12.
- 14.B Johnston, MS Atkins, B Mackiewich, M Anderson. "Segmentation of multiple sclerosis lesions in intensity corrected multispectral MRI. IEEE Trans. Med. Imaging, 1996;15(2):154–169. doi: 10.1109/42.491417. [DOI] [PubMed] [Google Scholar]
- 15.AO Boudraa, SM Dehak, YM Zhu, C Pachai, YG Bao, J Grimaud. Automated segmentation of multiple sclerosis lesions in multispectral MR imaging using fuzzy clustering. Comput. Biol. Med. 2000;30(1 ):23–40. doi: 10.1016/s0010-4825(99)00019-0. [DOI] [PubMed] [Google Scholar]
- 16.KV Leemput, F Maes, D Vandermeulen, A Colchester, P Suetens. Automated segmentation of multiple sclerosis lesions by model outlier detection. IEEE Trans. Med. Imaging. 2001;20(8 ):677–688. doi: 10.1109/42.938237. [DOI] [PubMed] [Google Scholar]
- 17.AP Zijdenbos, R Forghani, AC Evans. Automatic “pipeline” analysis of 3-D MRI data for clinical trials: application to multiple sclerosis. IEEE. Trans. Med. Imaging. 2002;21(10):1280–1291. doi: 10.1109/TMI.2002.806283. [DOI] [PubMed] [Google Scholar]
- 18.F Kruggel, SP Joseph, G Hermann-Josef. Texturebased segmentation of diffuse lesions of the brain's white matter. NeuroImage. 2008;39(3 ):987–996. doi: 10.1016/j.neuroimage.2007.09.058. [DOI] [PubMed] [Google Scholar]
- 19.W Liu, Z Xiaoxia, G Jiang, L Tong. Texture analysis of MRI in patients with multiple sclerosis based on the gray-level difference statistics, ". Educ. Tech. Comp. Sci., ETCS. 2009;3:771–4. [Google Scholar]
- 20.J Zhang, L Tong, L Wang, N Li. Texture analysis of multiple sclerosis: a comparative study. Magn. Reson. Imaging. 2008;26(8):1160–6. doi: 10.1016/j.mri.2008.01.016. [DOI] [PubMed] [Google Scholar]
- 21.M Ghazel, A Traboulsee, RK Ward. Optimal Filter Design for Multiple Sclerosis Lesions Segmentation from Regions of Interest in Brain MRI. IEEE Int. Symp. Signal Proc. Inf. Tech. 2006. pp. 1–5.
- 22. A Pozdnukhov, M Kanevski. Monitoring network optimisation for spatial data classification using support vector machines. Int. J. Environ. Pollut. 2006;28:20. [Google Scholar]
- 23.M Kanevski, M Maignan, A Pozdnukhov. Active Learning of Environmental Data Using Support Vector Machines. Conference of the International Association for Mathematical Geology; 2005; Toronto. [Google Scholar]
- 24.RR Edelman, JR Hesselink, MB Zlatkin, JV Crues. Clinical Magnetic Resonance Imaging. 3rd. Vol. 2. New York: Elsevier; 2006. [Google Scholar]
- 25. R Khayati, M Vafadust, F Towhidkhah, SM Nabavi. Fully automatic segmentation of multiple sclerosis lesions in brain MR FLAIR images using adaptive mixtures method and Markov random field model. Comput. Biol. Med. 2008;38:379–90. doi: 10.1016/j.compbiomed.2007.12.005. [DOI] [PubMed] [Google Scholar]
- 26.BrainWeb: Simulated Brain Database. [Online] Avalible from http://www.bic.mni.mcgill.ca/brainweb/
- 27.CA Cocosco, V Kollokian, RKS Kwan, AC Eva. "BrainWeb: Online Interface to a 3D MRI Simulated Brain Database," in NeuroImage; Proceedings of 3rd International Conference on Functional Mapping of the Human Brain; 1997; Copenhagen. [Google Scholar]
- 28.RKS Kwan, AC Evans, GB Pike. MRI simulationbased evaluation of image-processing and classification methods. IEEE Trans. Med. Imaging. 1999 Nov;18(11 ):1085–97. doi: 10.1109/42.816072. [DOI] [PubMed] [Google Scholar]
- 29.RKS Kwan, AC Evans, GB Pike. An Extensible MRI Simulator for Post-Processing Evaluation. Visualization in Biomedical Computing (VBC'96). Lecture Notes in Computer Science. 1996;1131:135–140. [Google Scholar]
- 30.DL Collins, AP Zijdenbos, V Kollokian, JG Sled, NJ Kabani, CJ Holmes, AC Evans. Design and construction of a realistic digital brain phantom. IEEE Trans. Med. Imaging . 1998;17(3 ):463–8. doi: 10.1109/42.712135. [DOI] [PubMed] [Google Scholar]
- 31.M Styner, et al. 3D segmentation in the clinic: A grand challenge II: MS lesion segmentation. MIDAS J. 2008. pp. 1–5.
- 32.MS Lesion. Segmentation Challenge 2008 [Online] http://www.ia.unc.edu/MSseg/
- 33.AC Evans, et al. 3D statistical neuroanatomical models from 305 MRI volumes. IEEE-Nuclear Science Symposium and Medical Imaging Conference; 1993. pp. 1813–7. [Google Scholar]
- 34.S Prima, S Ourselin, N Ayache. Computation of the midsagittal plane in 3d brain images. IEEE. Trans. Med. Imaging. 2002;21(2 ):122–38. doi: 10.1109/42.993131. [DOI] [PubMed] [Google Scholar]
- 35.RP Woods, ST Grafton, CJ Holmes, SR Cherry, JC Mazziotta. Automated image registration: General methods and intrasubject, intramodality validation. J. Comput. Assist. Tomogr. 1998;22:39–152. doi: 10.1097/00004728-199801000-00027. [DOI] [PubMed] [Google Scholar]
- 36.A Younis, M Ibrahim, Mansur Kabuka, Nigel John. An Artificial Immune-Activated Neural Network Applied to Brain 3D MRI Segmentation. J. Digit. Imaging. 2008;21: 569–88. doi: 10.1007/s10278-007-9081-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.J Rexilius, HK Hahn, H Bourquain, H-O Peitgen. Ground Truth in MS Lesion Volumetry –A Phantom Study," Medical Image Computing and Computer-Assisted Intervention - MICCAI03 Lect. Notes Comput. Sci. 2003;2879:546–53. [Google Scholar]
- 38.CC Chang, CJ Lin. LIBSVM: A library for support vector machines. ACM Transactions on Intelligent Systems and Technology. 2011;2(3 ):1–27. [Google Scholar]
- 39.J Lecoeur, et al. Multiple Sclerosis Lesions Segmentation using Spectral Gradient and Graph Cuts. Proceedings of MICCAI workshop on Medical Image Analysis on Multiple Sclerosis (validation and methodological issues) .2008. [Google Scholar]
- 40.AP Zijdenbos, BM Dawant, RA Margolin, AC Palmer. "Morphometric analysis of white matter lesions in mr images: method and validation. IEEE. Trans. Med. Imaging . 1994;13(4 ):716–724. doi: 10.1109/42.363096. [DOI] [PubMed] [Google Scholar]
- 41.D Garcia-Lorenzo, L Lecoeur, D Arnold, DL Collins. "Multiple Sclerosis lesion segmentation using an automatic multimodal graph cuts. MICCAI09. 2009. pp. 584–91. [DOI] [PubMed]
- 42.O Freifeld, H Greenspan, J Goldberger. Multiple sclerosis lesion detection using constrained gmm and curve evolution. Int. J. Biomed. Imaging. 2009. pp. 124–715. [DOI] [PMC free article] [PubMed]
- 43.JC Souplet, C Lebrun, N Ayache, G Malandain. An automatic segmentation of T2-FLAIR multiple sclerosis lesions. The MIDAS Journal - - MS Lesion Segmentation (MICCAI Workshop) 2008.
- 44.Results - MS Lesion Segmentation Challenge 2008. [Online] http://www.ia.unc.edu/MSseg/results_table php .
- 45.BA Abdullah, AA Younis, PM Pattany, E Saraf-Lavi. Textural based SVM for MS Lesion Segmentation in FLAIR MRIs. Open J. Med. Imaging. 2011;1(2 ):15–52. [Google Scholar]