Introduction
The study of epigenetic changes is experiencing a period of explosive growth, in part, due to the fact that epigenetic regulation has been linked to everything from cancer to obesity and from tissue differentiation to degeneration. An epigenetic change is an inheritable change in gene expression caused by mechanisms that do not include changes to the DNA sequence of the cell but rather changes to the chromatin structure and its interactions with various nuclear factors. Chromatin’s simplest unit is the nucleosome, which is composed of approximately 147 bp of DNA wrapped around a core of histone proteins. The histone core contains two each of histones H2A, H2B, H3, and H4. While the vast majority of the histone core has a globular, disc-like structure, the terminal tails of the histones are largely unstructured and extend out from the globular domains.
Epigenetic changes can be split into two main branches—posttranslational modifications (PTMs) of histones and DNA methylation. DNA methylation typically occurs in CpG islands resulting in 5′-methylcytosine. DNA methylation plays a role in stem cell differentiation and development and is largely associated with gene silencing [1, 2]. PTMs of histones are much more diverse chemically and include acetylation, phosphorylation, ubiquitination, sumoylation, ADP-ribosylation, and biotinylation [3]. PTMs of histones occur predominantly in the flexible N- and C-terminal tails of the nucleosomal core histone proteins, but they have also been found within the globular domain, as well as in the H1 linker histone [4, 5]. Histone modifications can have both cis and trans effects on nucleosomal arrangement. Changes in histones that directly alter their interaction with DNA or modify higher order chromatin structure are defined as cis effects [6]. The best characterized example of a cis effect is the change in charge on lysine residues following acetylation. The change from positive to neutral is theorized to weaken the association between the histone core and DNA, thereby making the DNA more accessible to transcription factors [7]. Conversely, trans effects are those that alter the association between the chromatin and any of a variety of nuclear complexes, often through the use of special protein domains that recognize various histone PTMs, such as the bromodomain of PCAF, a histone acetyltransferase, that specifically recognizes and binds acetylated lysine residues [8]. These specific changes are thought to create a specific “histone language” that provides both positive and negative signals that govern the binding of specific transcriptional molecules to different cis-regulatory modules on gene promoters [9]. Since modules can independently alter the temporal and tissue-specific expression of select genes [10], PTMs of histones are becoming recognized as a critical part of cell specification and differentiation during development.
This review will largely focus on the effect of histone modifications, particularly changes in acetylation, on ocular diseases, many of which involve the apoptotic loss of neurons. Because one of the hallmarks of apoptotic cell death is a widespread change in gene expression, epigenetic modifications in dying cells provide a feasible explanation for how the expression of a large number of genes with diverse regulatory elements may be rapidly adjusted.
Control of histone acetylation
Histone acetylation levels are controlled by two families of proteins with opposing functions—histone acetyltransferases (HATs) and histone deacetylases (HDACs). There are multiple members in each of these groups, which vary in their cellular localization and preferred substrate, including nonhistone substrates. There are three main subfamilies of HATs, GNAT, MYST, and p300/CBP [11]. The dysregulation of a large number of HATs has been implicated in diseases ranging from cancer to asthma [12]. Likewise, there are three subclasses of HDACs, classes I, II, and III. Class I and II HDACs are Zn+ dependent, whereas class III HDACs (sirtuins) are NAD dependent [13, 14]. Both HATs and HDACs function as members of large, multiprotein complexes, and complex formation plays a primary role in controlling HAT/HDAC activity [14].
The regulation of the balance between HAT and HDAC activity in a cell is tightly controlled, as tipping the balance in favor of one over the other can lead to drastic changes in cellular behavior, due to alterations in protein activity and gene expression. A disruption of the HAT:HDAC balance has been implicated in a number of neurodegenerative conditions, including Huntington’s disease, Alzheimer’s disease, and neuronal ischemia [15]. While most of these neurodegenerative diseases have been linked to a decrease in HAT activity, our work examining retinal ganglion cell (RGC) loss in an acute injury model has implicated an increase in HDAC activity as the problem. Regardless, the real issue is an upset in the balance between two opposing activities, which leads to an abnormal decrease in histone acetylation.
Histone acetylation and gene expression
Most histone acetylation occurs in the N-terminal tails of histones H3 and H4, and acetylation in these regions is associated with transcriptionally active genes [16]. While histone acetylation appears to be necessary for gene expression, it is not sufficient, as the various transcription factors and polymerase complex must still be recruited to the gene. However, histone acetylation may play a role in this recruitment as several transcription factors, including the SWI/SNF complex and TAFII250, contain bromodomains, which preferentially bind acetylated histones [17].
HATs and HDACs can act in sequence-specific and global manners. Global histone acetylation changes appear to be due to the ability of acetylated histones to recruit more HATs and the ability of unmodified histones to recruit complexes containing HDACs. In this manner, the existing modifications are propagated to the surrounding nucleosomes [18]. Due to the variety of genes affected in apoptotic cells, it is likely that any related histone acetylation changes are global rather than sequence specific; however, this has not been definitively shown.
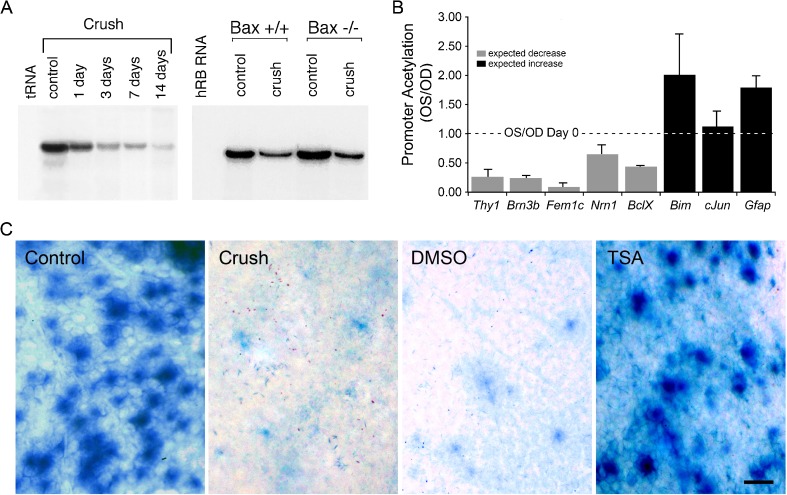
A widespread change in gene expression has been demonstrated in several models of neurodegeneration, including degenerative retinal conditions [19–24]. Several studies examining this phenomenon have been done in models of RGC degeneration, both acute and chronic models of damage. Using various detection methods in both models of RGC damage, researchers have found almost 500 genes that are downregulated prior to the loss of cells [25–28]. While most studies of gene expression changes in the eye used whole retinas as the tissue sample, they examined a number of RGC-specific genes, such as Brn3b, Thy1, Fem1c, Sncg, and Nrn1. This provides a much better snapshot of what is occurring in the dying RGCs since only 1–2 % of the retinal population in comprised of RGCs. Gene expression changes in damaged RGCs occur as early as 1 day after damage [21, 27] and even occur in Bax−/− mice, which do not complete the apoptotic process, after optic nerve injury [29] (Fig. 1). Associated with this decrease, the histones in the promoters of these genes exhibit dramatic deacetylation [21], while genes that are associated with apoptosis either remain unaffected or even increase in their acetylation levels (Fig. 1). The deacetylation process in dying RGCs may not be restricted to individual genes, however. Both wild-type and Bax−/− mice that undergo optic nerve crush (ONC) exhibit a global decrease in histone acetylation in the RGC layer as early as 1 day postcrush (Fig. 2), which is concurrent with the detected decrease in the expression of a number of genes [21, 30]. This has significant effects on nuclear architecture, which exhibit an increase in the formation of heterochromatin and change from round or oval in appearance to a highly convoluted appearance (Fig. 2). In another study of histone deacetylation in RGCs following ONC, valproic acid-mediated HDAC inhibition led to increases in DNA binding by CREB, which is known to mediate the expression of neuronal survival genes [31]. In a recent study examining the effect of histone acetylation in the DBA/2 J mouse model of glaucoma, Pelzel et al. found that histones in the RGCs of damaged eyes undergo deacetylation and that the timing of the histone deacetylation correlates with a decrease in expression of an RGC-marker gene [32].
Fig. 1.
Gene expression changes in RGCs associated with deacetylation of histones. a RNase protection assay for transcripts of the RGC-specific gene Thy1. Shortly after optic nerve crush in mice, Thy1 levels are rapidly depleted (left panel). This precedes the loss of RGCs, which begin to drop out of the retina at 1 week after optic nerve damage. The loss of Thy1 also occurs in Bax-deficient mice, which have RGCs completely resistant to the optic nerve crush protocol (right panel, 1 week after ONC). b ChIP assays showing the relative acetylation of promoter histones in genes that decrease in expression after ONC (gray bars) and genes that exhibit an increase in expression (black bars) at 3 days postcrush. Promoter histone deacetylation is correlated with genes that are silenced. c Blocking the activity of class I and II HDACs with TSA prevents the silencing of the Fem1cR3 RGC reporter gene in mice after ONC. In this experiment, TSA was administered to mice 1 day prior to nerve damage, and expression of Fem1c (as a function of β-galactosidase activity) was examined 5 days after surgery. a was reprinted from [29] with permission. b and c were reprinted from [21], which is an open access journal
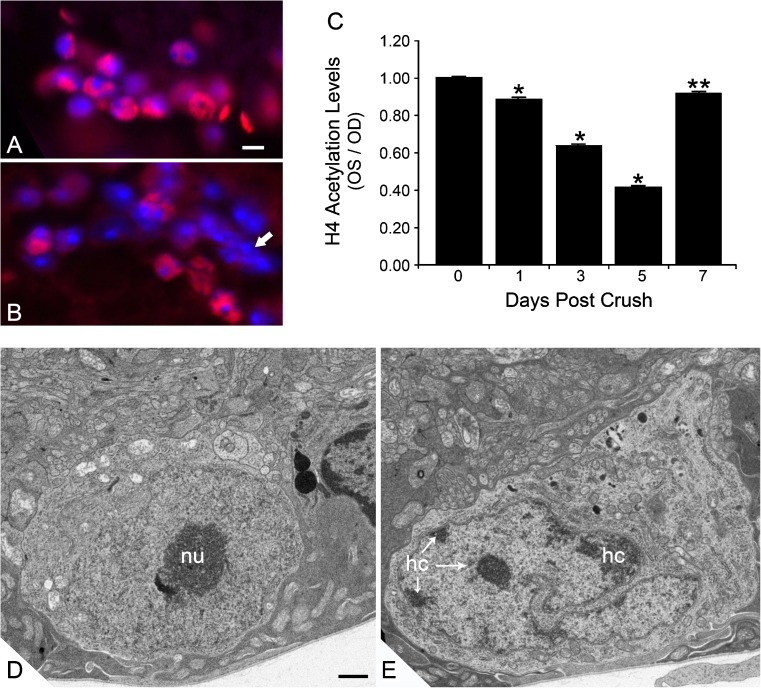
Fig. 2.
Histone deacetylation and nuclear changes in RGCs after ONC. Immunofluorescent labeling of acetylated histone H4 (AcH4) in the ganglion cell layer of a control mouse eye (a) and an eye 5 days after optic nerve crush (b). Nuclei have been counterstained with DAPI. Eyes with crush exhibit nuclei with reduced or absent staining for AcH4. Nuclear morphology often demonstrates highly condensed chromatin (arrow in b). Scale bar = 10 μm. c Quantification of histone H4 acetylation in the ganglion cell layer of mouse eyes after ONC. Data from this experiment were collected by measuring the pixel density of AcH4 label per total area of each nucleus examined and normalizing this value to the pixel density of unaffected nuclei in the inner nuclear layer. The data are depicted as the ratio of experimental (OS) and control (OD) retinas after ONC. *P ≤ 0.0001 and **P = 0.041 (OS vs. OD at given time point). Transmission electron micrographs of control (d) and experimental (e) mouse retinas. Images were taken in the ganglion cell layer of eyes 5 days after ONC. Control eyes d exhibit round or oval nuclei with lightly staining euchromatin and prominent nucleoli (nu). In crush eyes, nuclei predominantly appear highly convoluted and exhibit the formation of varying degrees of heterochromatin (hc), typically forming along the inner side of the nuclear envelope. Scale bar = 500 nm. c was reprinted from [21], which is an open access journal
In ischemic models of retinal degeneration, changes in histone modifications also appear to play a role in the altered expression of some genes. Work by Crosson et al. demonstrated that retinal TNF-α expression increases following retinal ischemia but that this increase can be attenuated following treatment with trichostatin A (TSA), a class I and II HDAC inhibitor, indicating that expression of this gene is controlled by histone acetylation levels [33]. Similarly, other inflammatory stimuli, such as high glucose or oxidized lipid concontrations, as would be found in diabetic individuals, are associated with hyperacetylation of the TNF-α and COX2 promoters, with corresponding increases in gene expression in cultured monocytes [34]. However, the epigenetic effects of diabetic retinopathy in the retina appear to be due to a decrease in histone acetylation rather than an increase, as the expression of HDACs 1, 2, and 8 increased in retinas from streptozotocin-treated rats [35].
Increases in clusterin, a secreted chaperone protein, occur during aging and particularly in those afflicted with age-related macular degeneration (AMD). Recent studies by Suuronen et al. have demonstrated that clusterin expression in RPE cells is effected by epigenetic changes, as seen after treatment with 5-aza-2′-deoxycytidine for DNA hypomethylation or treatment with TSA or valproic acid (VPA) to induce histone hyperacetylation [36]. While it is unclear what role clusterin plays in AMD pathology, this information relating expression to epigenetic modification provides a possible treatment strategy.
HDAC activity changes in retinal degeneration
There is a growing body of evidence of HAT:HDAC imbalance in retinal degenerative diseases. While most of this evidence implies that an imbalance favoring histone deacetylation leads to neurodegeneration, there are some contradictions, particularly in undeveloped retinas and undifferentiated cells.
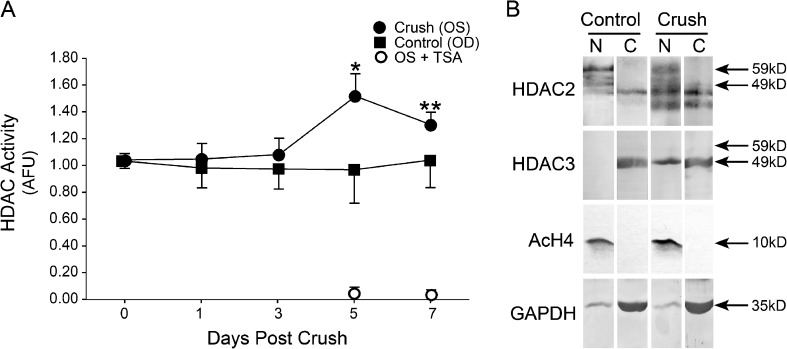
Investigations into HDAC activity following ONC indicate that there is an increase in nuclear HDAC activity in whole retina lysates that begins at 1 day postcrush and reaches significant levels by 5 days postcrush [21]. In addition, this increase in HDAC activity in the whole retina occurs concurrently with an increased nuclear presence of HDAC3 in RGCs (Fig. 3), the cells affected by ONC, indicating that the increase in HDAC activity in whole retinas may be due to increases in apoptotic cells [21]. Increased retinal HDAC activity has also been detected in streptozotocin-treated rats with poor glycemic control [35]. Interestingly, rats that experienced 6 months of poor glycemic control followed by 6 months of good glycemic control exhibited even higher levels of HDAC activity, providing evidence for the cause of the metabolic memory phenomenon observed in diabetic patients [35].
Fig. 3.
Changes in HDAC activity in mouse nuclei after ONC. a Total nuclear HDAC activity in control (OD) and experimental (OS) retinas after ONC. Crush retinas exhibit a slow progressive increase in nuclear HDAC activity relative to unaffected eyes. All activity can be inhibited by the broad002Dspectrum inhibitor TSA. b HDAC2 and HDAC3 localization after ONC. HDAC2 is predominantly present in the nuclear fraction of both control and crush retinas. HDAC3, however, redistributes to the nuclear fraction in crush eyes, consistent with the increase in nuclear HDAC activity after crush. Reprinted from [21], which is an open access journal
In addition to increased HDAC activity, streptozotocin-treated rats also exhibited a decrease in HAT activity responsible for H3 acetylation, but no decrease in H4 acetylation [35]. This indicates possible inhibition of the SAGA or SLIK Gcn5-containing complexes, which preferentially acetylate H3 over H4 [37, 38]. Since an imbalance favoring deacetylation may also be due to a decrease in HAT activity, these rats have compounded issues with histone acetylation. In a spinocerebellar ataxia type 7 (SCA7) cell culture model of neurodegeneration, decreases in another Gcn5-containing HAT complex, STAGA, have also been demonstrated [39]. In this model, the molecular mechanism of HAT inhibition has been identified as sequestration via the polyglutamine-expanded ataxin-7 protein.
In AMD, it appears that a decrease in HDAC activity may actually play a role in the disease pathology. There is an age-related decrease in expression of SIRT1, a class III HDAC, which is significantly worse in age-matched eyes of patients with AMD [40]. While this appears to contradict the HAT:HDAC imbalance indicated in other retinal degeneration models, it should be noted that the role of SIRT1 appears to be acetylation of a nonhistone substrate, FOXO3, which acts as a transcriptional regulatory protein [41].
HDAC3 involvement in neuronal degeneration
Studies of RGC death have implicated an important role for HDAC3 in the process of global histone deacetylation and gene silencing, early in the apoptotic pathway. HDAC3 resides in both cytosolic and nuclear compartments in many cell types, but in normal retina, it is predominantly detected in the cytosol. Shortly after acute optic nerve damage, HDAC3 redistributes in affected cells, becoming nuclear [21] (Fig. 3). This correlative evidence supports recently reported studies that HDAC3 expression increased neuronal susceptibility to apoptotic stimuli. Bardai and D’Mello [42] reported that exogenous overexpression of HDAC3 in neuronal cell lines promoted their death, while nonneuronal cells are unaffected under the same conditions. Conversely, silencing of HDAC3 expression in neuronal cells increases their resistance to apoptotic stimuli. Supporting these studies, treatment of mice with a form of Friedrich’s ataxia with selective HDAC3 inhibitors provides a neuroprotective effect [43].
The role of HDAC3 in neuronal death is not well characterized. Pelzel and colleagues [21] documented an association between HDAC3 and gene silencing, and HDAC3 inhibitors may affect neuronal death in the models of Friedrich’s axtaxia by preventing downregulation of frataxin (Fxn) gene expression [44]. Silencing of transcription may be a consequence of a more systemic function of HDAC3, however, to precipitate the global formation of heterochromatin and nuclear condensation associated with apoptosis.
HDAC inhibitors as treatment for ocular diseases
Because the HAT:HDAC balance in retinal degeneration appears to favor deacetylation, several groups have studied the effects of HDAC inhibitor (HDACi) treatment as a mode of restoring balance. In all cases where treatment is applied to damaged retinas or differentiated neurons on culture, HDACi appears to have a neuroprotective effect.
In an ischemic model of damage, treatment with TSA prevented retinal thinning at 7 days post ischemia/reperfusion, in addition to having the functional benefit of attenuating the loss of a- and b-wave amplitude detected by ERG that is normally seen following ischemia [33]. A separate study examining the effect of VPA on protein expression in ischemic retinas found that VPA stimulated increased H3 acetylation that was associated with a decrease in the stress response proteins, GRP78/BiP and C/EBP homologous protein, as well as a decrease in caspase-12 activation [45].
In ONC models of RGC degeneration, TSA has been shown to attenuate cell loss as long as 2 weeks postcrush, a time when the vast majority of RGCs has disappeared in untreated eyes [21]. While treatment with VPA appears to have a slightly less robust effect, it also attenuated RGC loss following ONC for up to 8 days postcrush [31]. Purified cultures of RGCs also benefit from HDACi treatment, as VPA, TSA, and sodium butyrate (SB) all increase histone acetylation and have neuroprotective effects [46]. In addition to increasing cell survival, HDACi has been shown to block damage-related gene silencing and decrease caspase activation in retina following ONC [21, 31]. In retinal explants, VPA was found to stimulate neurite outgrowth, indicating that HDACi may not only be neuroprotective but may also increase the regenerative potential of damaged neurons [31]. More recently, HDACi treatment has demonstrated effectiveness at attenuating the decrease in expression of an RGC-specific gene in the DBA/2 J model of chronic age-related glaucoma [32].
In the SCA7 model of retinal degeneration, which is characterized by an inhibition of HAT activity, treatment of the cultures with SB and suberoylanilide hydroxamic acid, both HDACi, reversed the inhibition of CRX/NRL-dependent transactivation of the rhodopsin promoter [39].
DNA methylation and ocular disease susceptibility
Many ocular diseases are considered complex genetic disorders, since they do not present with classical Mendelian inheritance patterns, but family history still clearly plays a role as a major risk factor [47, 48]. There is still more evidence that environmental factors could also influence the prevalence of some ocular diseases, and epigenetic changes to the DNA, which influence gene expression patterns, are likely an intersection of these two variables (for example, see [49]). In this scenario, genetic susceptibility is enhanced or augmented by environmental factors that alter an otherwise “healthy” gene expression pattern. The epigenetic influence of environment is most likely mediated by the methylation of CpG islands in the genome. Studies of the methylome (the overall pattern of DNA methylation in the genome) in monozygotic twins show that cells in young twins have nearly identical patterns of methylation, while cells in older twins exhibit marked differences [50]. Since the genetic information in these individuals is identical, epigenetic changes to their genomes have been proposed as the principal mechanism that leads to disconcordant diseases they may acquire [51]. Environmental cues that can affect methylomes include diet, smoking, and pollution [52–56].
Although still a fledgling area of study, AMD presents the most compelling and likely candidate for a disease influenced by both genetics and environment. Several large population-based studies have definitively assigned smoking history and dietary intake with an increased risk of developing AMD [57–59]. The importance of these environmental factors in a controlled genetic background has been further examined in monozygotic twins disconcordant for developing AMD revealed that both cigarette smoking and dietary habits were associated with the disease [60–62]. Although these associations are compelling, the methylomes of individuals affected with AMD have not been examined. This likely underscores that relative naiveté of the ophthalmic community of the influence of epigenetics in the pathology of ocular disease.
Acknowledgments
The authors wish to thank Dr. Cassandra Schlamp for assistance with the figures and critically reading of the manuscript. This work was supported by grants from the National Eye Institute (R01 EY012223 to RWN, and a CORE grant P30 EY016665 to the Department of Ophthalmology and Visual Sciences) and from Research to Prevent Blindness, Inc.
References
- 1.Yeo S, et al. Characterization of DNA methylation change in stem cell marker genes during differentiation of human embryonic stem cells. Biochem Biophys Res Commun. 2007;359(3):536–42. doi: 10.1016/j.bbrc.2007.05.120. [DOI] [PubMed] [Google Scholar]
- 2.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16(1):6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 3.Vaquero A, Loyola A, Reinberg D. The constantly changing face of chromatin. Sci Aging Knowledge Environ. 2003;2003(14):RE4. doi: 10.1126/sageke.2003.14.re4. [DOI] [PubMed] [Google Scholar]
- 4.Happel N, Doenecke D. Histone H1 and its isoforms: contribution to chromatin structure and function. Gene. 2009;431(1–2):1–12. doi: 10.1016/j.gene.2008.11.003. [DOI] [PubMed] [Google Scholar]
- 5.Cosgrove MS, Boeke JD, Wolberger C. Regulated nucleosome mobility and the histone code. Nat Struct Mol Biol. 2004;11(11):1037–43. doi: 10.1038/nsmb851. [DOI] [PubMed] [Google Scholar]
- 6.Allis CD, Jenuwein T, Reinberg D. In: Epigenetics. 1. Allis CD, Jenuwein T, Reinberg D, editors. New York: Cold Spring Harbor Laboratory Press; 2007. p. 502. [Google Scholar]
- 7.Wolffe AP, Hayes JJ. Chromatin disruption and modification. Nuc Acids Res. 1999;27:711–20. doi: 10.1093/nar/27.3.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Taverna SD, et al. How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers. Nat Struct Mol Biol. 2007;14(11):1025–40. doi: 10.1038/nsmb1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oliver SS, Denu JM. Dynamic interplay between histone H3 modifications and protein interpreters: emerging evidence for a “histone language”. ChemBioChem. 2011;12(2):299–307. doi: 10.1002/cbic.201000474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peter IS, Davidson EH. Evolution of gene regulatory networks controlling body plan development. Cell. 2011;144(6):970–85. doi: 10.1016/j.cell.2011.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roth SY, Denu JM, Allis CD. Histone acetyltransferases. Annu Rev Biochem. 2001;70:81–120. doi: 10.1146/annurev.biochem.70.1.81. [DOI] [PubMed] [Google Scholar]
- 12.Dekker FJ, Haisma HJ. Histone acetyl transferases as emerging drug targets. Drug Discov Today. 2009;14(19–20):942–8. doi: 10.1016/j.drudis.2009.06.008. [DOI] [PubMed] [Google Scholar]
- 13.Ruijter AJ, et al. Histone deacetylases (HDACs): characterization of the classical HDAC family. Biochem J. 2003;370(Pt 3):737–49. doi: 10.1042/BJ20021321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grozinger CM, Schreiber SL. Deacetylase enzymes: biological functions and the use of small-molecule inhibitors. Chem Biol. 2002;9(1):3–16. doi: 10.1016/S1074-5521(02)00092-3. [DOI] [PubMed] [Google Scholar]
- 15.Saha RN, Pahan K. HATs and HDACs in neurodegeneration: a tale of disconcerted acetylation homeostasis. Cell Death Differ. 2006;13:539–50. doi: 10.1038/sj.cdd.4401769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shahbazian MD, Grunstein M. Functions of site-specific histone acetylation and deacetylation. Ann Rev Biochem. 2007;76:75–100. doi: 10.1146/annurev.biochem.76.052705.162114. [DOI] [PubMed] [Google Scholar]
- 17.Agalioti T, Chen G, Thanos D. Deciphering the transcriptional histone acetylation code for a human gene. Cell. 2002;111(3):381–92. doi: 10.1016/S0092-8674(02)01077-2. [DOI] [PubMed] [Google Scholar]
- 18.Vogelauer M, et al. Global histone acetylation and deacetylation in yeast. Nature. 2000;408(6811):495–8. doi: 10.1038/35044127. [DOI] [PubMed] [Google Scholar]
- 19.Guo Y, et al. Early gene expression changes in the retinal ganglion cell layer of a rat glaucoma model. Invest Ophthalmol Vis Sci. 2011;52(3):1460–73. doi: 10.1167/iovs.10-5930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Panagis L, et al. Retinal gene expression changes related to IOP exposure and axonal loss in DBA/2J mice. Invest Ophthalmol Vis Sci. 2011;52(11):7807–16. doi: 10.1167/iovs.10-7063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pelzel HR, Schlamp CL, Nickells RW. Histone H4 deacetylation plays a critical role in early gene silencing during neuronal apoptosis. BMC Neurosci. 2010;11:62. doi: 10.1186/1471-2202-11-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rattner A, et al. The genomic response of the retinal pigment epithelium to light damage and retinal detachment. J Neurosci. 2008;28(39):9880–9. doi: 10.1523/JNEUROSCI.2401-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Swiderski RE, et al. Gene expression analysis of photoreceptor cell loss in bbs4-knockout mice reveals an early stress gene response and photoreceptor cell damage. Invest Ophthalmol Vis Sci. 2007;48(7):3329–40. doi: 10.1167/iovs.06-1477. [DOI] [PubMed] [Google Scholar]
- 24.Kirk CA, et al. Age-related alterations in retinal neurovascular and inflammatory transcripts. Mol Vis. 2011;17:1261–74. [PMC free article] [PubMed] [Google Scholar]
- 25.Ahmed F, et al. Microarray analysis of changes in mRNA levels in the rat retina after experimental elevation of intraocular pressure. Invest Ophthalmol Vis Sci. 2004;45:1247–58. doi: 10.1167/iovs.03-1123. [DOI] [PubMed] [Google Scholar]
- 26.Piri N, et al. Gene expression changes in the retina following optic nerve transection. Mol Vis. 2006;12:1660–73. [PubMed] [Google Scholar]
- 27.Yang Z, et al. Changes in gene expression in experimental glaucoma and optic nerve transection: the equilibrium between protective and detrimental mechanisms. Invest Ophthalmol Vis Sci. 2007;48:5539–48. doi: 10.1167/iovs.07-0542. [DOI] [PubMed] [Google Scholar]
- 28.Soto I, et al. Retinal ganglion cells downregulate gene expression and lose their axons within the optic nerve head in a mouse glaucoma model. J Neurosci. 2008;28(2):548–61. doi: 10.1523/JNEUROSCI.3714-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schlamp CL, et al. Changes in Thy1 gene expression associated with damaged retinal ganglion cells. Mol Vis. 2001;7:192–201. [PubMed] [Google Scholar]
- 30.Janssen KT et al. Nuclear atrophy of retinal ganglion cells precedes the Bax-dependent stage of apoptosis., 2012; in press. [DOI] [PMC free article] [PubMed]
- 31.Biermann J, et al. Valproic acid-mediated neuroprotection and regeneration in injured retinal ganglion cells. Invest Ophthalmol Vis Sci. 2010;51(1):526–34. doi: 10.1167/iovs.09-3903. [DOI] [PubMed] [Google Scholar]
- 32.Pelzel HR et al. Silencing of Fem1cR3 gene expression in the DBA/2J mouse precedes retinal ganglion cell death and is associated with histone deacetylase activity. submitted, 2012. [DOI] [PMC free article] [PubMed]
- 33.Crosson CE, et al. Inhibition of histone deacetylase protects the retina from ischemic injury. Invest Ophthalmol Vis Sci. 2010;51(7):3639–45. doi: 10.1167/iovs.09-4538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miao F, et al. In vivo chromatin remodeling events leading to inflammatory gene transcription under diabetic conditions. J Biol Chem. 2004;279(17):18091–7. doi: 10.1074/jbc.M311786200. [DOI] [PubMed] [Google Scholar]
- 35.Zhong Q, Kowluru RA. Role of histone acetylation in the development of diabetic retinopathy and the metabolic memory phenomenon. J Cell Biochem. 2010;110(6):1306–13. doi: 10.1002/jcb.22644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Suuronen T, et al. Epigenetic regulation of clusterin/apolipoprotein J expression in retinal pigment epithelial cells. Biochem Biophys Res Commun. 2007;357(2):397–401. doi: 10.1016/j.bbrc.2007.03.135. [DOI] [PubMed] [Google Scholar]
- 37.Kuo MH, et al. Gcn4 activator targets Gcn5 histone acetyltransferase to specific promoters independently of transcription. Mol Cell. 2000;6(6):1309–20. doi: 10.1016/S1097-2765(00)00129-5. [DOI] [PubMed] [Google Scholar]
- 38.Suka N, et al. Highly specific antibodies determine histone acetylation site usage in yeast heterochromatin and euchromatin. Mol Cell. 2001;8(2):473–9. doi: 10.1016/S1097-2765(01)00301-X. [DOI] [PubMed] [Google Scholar]
- 39.Palhan VB, et al. Polyglutamine-expanded ataxin-7 inhibits STAGA histone acetyltransferase activity to produce retinal degeneration. Proc Natl Acad Sci U S A. 2005;102(24):8472–7. doi: 10.1073/pnas.0503505102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peng CH, et al. Delivery of Oct4 and SirT1 with cationic polyurethanes-short branch PEI to aged retinal pigment epithelium. Biomaterials. 2011;32(34):9077–88. doi: 10.1016/j.biomaterials.2011.08.008. [DOI] [PubMed] [Google Scholar]
- 41.Wu Z, et al. Oxidative stress modulates complement factor H expression in retinal pigmented epithelial cells by acetylation of FOXO3. J Biol Chem. 2007;282(31):22414–25. doi: 10.1074/jbc.M702321200. [DOI] [PubMed] [Google Scholar]
- 42.Bardai FH, D’Mello SR. Selective toxicity by HDAC3 in neurons: regulation by Akt and GSK3beta. J Neurosci. 2011;31(5):1746–51. doi: 10.1523/JNEUROSCI.5704-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sandi C, et al. Prolonged treatment with pimelic o-aminobenzamide HDAC inhibitors ameliorates the disease phenotype of a Friedreich ataxia mouse model. Neurobiol Dis. 2011;42(3):496–505. doi: 10.1016/j.nbd.2011.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rai M, et al. Two new pimelic diphenylamide HDAC inhibitors induce sustained frataxin upregulation in cells from Friedreich’s ataxia patients and in a mouse model. PLoS One. 2010;5(1):e8825. doi: 10.1371/journal.pone.0008825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang Z, et al. Valproate protects the retina from endoplasmic reticulum stress-induced apoptosis after ischemia–reperfusion injury. Neurosci Lett. 2011;504(2):88–92. doi: 10.1016/j.neulet.2011.09.003. [DOI] [PubMed] [Google Scholar]
- 46.Biermann J, et al. Histone deacetylase inhibitors sodium butyrate and valproic acid delay spontaneous cell death in purified rat retinal ganglion cells. Mol Vis. 2011;17:395–403. [PMC free article] [PubMed] [Google Scholar]
- 47.Shahid H et al. Age-related macular degeneration: the importance of family history as a risk factor. Br J Ophthalmol, 2011. [DOI] [PubMed]
- 48.Wiggs JL. Genetic etiologies of glaucoma. Arch Ophthalmol. 2007;125:30–7. doi: 10.1001/archopht.125.1.30. [DOI] [PubMed] [Google Scholar]
- 49.Zanke B, et al. A genetic approach to stratification of risk for age-related macular degeneration. Can J Ophthalmol. 2010;45(1):22–7. doi: 10.3129/i09-209. [DOI] [PubMed] [Google Scholar]
- 50.Fraga MF, et al. Epigenetic differences arise during the lifetime of monozygotic twins. Proc Natl Acad Sci U S A. 2005;102(30):10604–9. doi: 10.1073/pnas.0500398102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Poulsen P, et al. The epigenetic basis of twin discordance in age-related diseases. Pediatr Res. 2007;61(5 Pt 2):38R–42. doi: 10.1203/pdr.0b013e31803c7b98. [DOI] [PubMed] [Google Scholar]
- 52.Bollati V, et al. Changes in DNA methylation patterns in subjects exposed to low-dose benzene. Cancer Res. 2007;67(3):876–80. doi: 10.1158/0008-5472.CAN-06-2995. [DOI] [PubMed] [Google Scholar]
- 53.Franco R, et al. Oxidative stress, DNA methylation and carcinogenesis. Cancer Lett. 2008;266(1):6–11. doi: 10.1016/j.canlet.2008.02.026. [DOI] [PubMed] [Google Scholar]
- 54.Liu H, et al. Cigarette smoke induces demethylation of prometastatic oncogene synuclein-gamma in lung cancer cells by downregulation of DNMT3B. Oncogene. 2007;26(40):5900–10. doi: 10.1038/sj.onc.1210400. [DOI] [PubMed] [Google Scholar]
- 55.Milagro FI, et al. A dual epigenomic approach for the search of obesity biomarkers: DNA methylation in relation to diet-induced weight loss. FASEB J. 2011;25(4):1378–89. doi: 10.1096/fj.10-170365. [DOI] [PubMed] [Google Scholar]
- 56.Rampersaud GC, et al. Genomic DNA methylation decreases in response to moderate folate depletion in elderly women. Am J Clin Nutr. 2000;72(4):998–1003. doi: 10.1093/ajcn/72.4.998. [DOI] [PubMed] [Google Scholar]
- 57.Clemons TE, et al. Risk factors for the incidence of advanced age-related macular degeneration in the age-related eye disease study (AREDS) AREDS report no. 19. Ophthalmology. 2005;112(4):533–9. doi: 10.1016/j.ophtha.2004.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jia L, et al. Risk factors for age-related macular degeneration in elderly Chinese opulation in Shenyang of China. Biomed Environ Sci. 2011;24(5):506–11. doi: 10.3967/0895-3988.2011.05.008. [DOI] [PubMed] [Google Scholar]
- 59.Chiu CJ, Taylor A. Dietary hyperglycemia, glycemic index and metabolic retinal diseases. Prog Retin Eye Res. 2011;30(1):18–53. doi: 10.1016/j.preteyeres.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Seddon JM, et al. The US twin study of age-related macular degeneration: relative roles of genetic and environmental influences. Arch Ophthalmol. 2005;123(3):321–7. doi: 10.1001/archopht.123.3.321. [DOI] [PubMed] [Google Scholar]
- 61.Seddon JM, George S, Rosner B. Cigarette smoking, fish consumption, omega-3 fatty acid intake, and associations with age-related macular degeneration: the US twin study of age-related macular degeneration. Arch Ophthalmol. 2006;124(7):995–1001. doi: 10.1001/archopht.124.7.995. [DOI] [PubMed] [Google Scholar]
- 62.Seddon JM, et al. Smoking, dietary betaine, methionine, and vitamin D in monozygotic twins with discordant macular degeneration: epigenetic implications. Ophthalmology. 2011;118(7):1386–94. doi: 10.1016/j.ophtha.2010.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]