Abstract
The link between experimental data and biophysically based mathematical models is key to computational simulation meeting its potential to provide physiological insight. However, despite the importance of this link, scrutiny and analysis of the processes by which models are parameterised from data are currently lacking. While this situation is common to many areas of physiological modelling, to provide a concrete context, we use examples drawn from detailed models of cardiac electro-mechanics. Using this biophysically detailed cohort of models we highlight the specific issues of model parameterization and propose this process can be separated into three stages: observation, fitting and validation. Finally, future research challenges and directions in this area are discussed.
Steven Niederer (left) received an Engineering Science Degree at the University of Auckland in 2003. He completed his DPhil on ‘Modelling the rat left ventricle’ in the Computing Laboratory at the University of Oxford in 2008. In 2009 he was awarded a UK Engineering and Physical Sciences Research Council Life Science Interface Fellowship to develop mathematical models of heart failure. This work led to a position at the Biomedical Engineering Department at King's College London where he works on the translation of cardiac modelling techniques into clinical applications. Nic Smith (right) has formally held academic positions at the Universities of Oxford and Auckland and is currently Professor and Head of Biomedical Engineering at King's College London. His research is characterised by the development of integrated multi-scale and multi-physics models of the heart, which provide the ability to link biophysically detailed experimental data to integrated function from sub-cellular to the whole organ level. He has particular interests in cardiac excitation–contraction coupling and fluid structure interaction in the coronary system.
 |
Introduction
Physiology is the study of both the constitutive parts and the integrated function of biological systems. However, the advent of ‘-omic’ science has increasingly resulted in the field being reposed within reductionist frameworks (Noble, 2008). Despite the aim of many of these emerging fields to consider all the parts of physiological systems collectively (Evans, 2000) research endeavours have consistently focused on quantitating the constituents. This has led to less attention being paid to understanding the functional mechanisms and consequences underpinning these measurements and observations. At the same time the cost of data storage is decreasing while tools and techniques for rapidly and robustly obtaining large volumes of biological data continue to develop. This improved ability to create and store data means that the underlying physiology of many systems risks becoming exceptionally well described but still relatively poorly understood.
Exploiting this growth in data to increase our understanding of physiological function is now necessitating the linking of this, often fractionated, information over multiple spatial and temporal scales, species and populations. Conventional intuition-driven analysis rapidly becomes limiting in these circumstances. Overcoming these limitations is increasingly motivating, and in some cases even requiring, the use of computational and mathematical models to integrate data within biophysically constrained physiological frameworks (Davidson et al. 1995; Hernandez & Kambhampati, 2004). The potential application of this type of computational approach is widespread and has been previously recognised in multiple reviews. A central theme in the exemplar cases provided in much of this literature is the study of complex systems where experimental data alone have failed to provide functional insight. In these systems, where sufficient experimental data are available to characterise the underlying complexity, computational techniques have been used to formalise and represent the underlying system biophysics. Using these quantitative frameworks multiple, often disparate, data sets have been linked together to provide insight not available through observation alone.
Specific examples of the successful application of this approach span multiple spatial scales. In genetics where, despite comprehensive genome sequences now being available for multiple species, many aspects of gene function and regulation remain elusive (Benfey & Mitchell-Olds, 2008). Directly addressing this issue, computational models have begun to expose and unravel some of the intrinsic complexity of gene regulation and the link between genotypes and phenotypes (Collins et al. 2003). The elements of non-linear feedback mechanisms, multiple agents and significant quantities of data, common in genetics, are also present in the simulation and computational analysis of cellular physiology and function in systems biology (Kitano, 2002; Butcher et al. 2004). These models at the cellular scale provide the additional ability to simulate interactions between proteins and molecules, offering a link between cellular patho-physiology with pharmacology and drug treatments (van de Waterbeemd & Gifford, 2003; Deisboeck et al. 2009). Cellular scale models also enable the integration of information at the increasing spatial scales of the whole organ and body. These measurements, when at the scale of the organ, often represent emergent behaviour resulting from the integrated behaviour of the systems constitutive components. Applying models to these data provides an essential mechanism to link between organ scale and clinically relevant measurements with underlying cellular and subcellular function (Lee et al. 2009).
Despite the potential of this approach to derive function from observations, linking models to data remains a significant challenge. While this issue is generic, to provide a specific context with a set of concrete, and we hope relevant, examples from our own research community, we will focus in the remainder of the article below on the challenges faced in linking coupled electrophysiological and mechanical cardiac models (Fig. 1) to experimental and clinical data. Although the examples provided below are taken from this specific research area, the breadth of spatial and temporal scales and applications means these examples are generally transferable to other computational physiology modelling systems.
Figure 1.
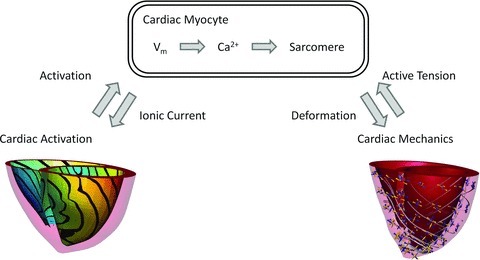
Schematic showing the multi-scale relationship between cellular electrophysiology and contraction and whole organ activation and deformation
Models of cardiac electromechanics have been applied at the genetic (Campbell & McCulloch, 2011), cellular (Niederer & Smith, 2007; Campbell et al. 2008), tissue (Niederer & Smith, 2008; Land et al. 2011; Nordsletten et al. 2011) and organ scales (Nickerson et al. 2005; Campbell et al. 2009; Niederer & Smith, 2009; Gurev et al. 2010, 2011; Kerckhoffs et al. 2010). Most recently, significant technical advances have led to a renewed focus on translation of this approach to the clinic (Rudy et al. 2008) with computational models being used to simulate human cardiac function (Keldermann et al. 2010) and specific patient cases (Aguado-Sierra et al. 2011; Niederer et al. 2011a,b). A selection of these models is presented in Fig. 2. This transition towards clinical applications in electromechanics models parallels advances in defibrillation (Luther et al. 2011; Tandri et al. 2011) and drug interaction (Mirams et al. 2011) modelling studies and places renewed emphasis on the issues discussed above. Specifically, while it has significant potential to positively impact human health, the use of simulation results to inform treatment decisions only increases the importance of rigorously describing the link between data to model parameters and simulation results. The links between models and data are formed during three distinct phases in the model development process: observation, fitting and validation.
Figure 2. Example multi-scale excitation contraction models.
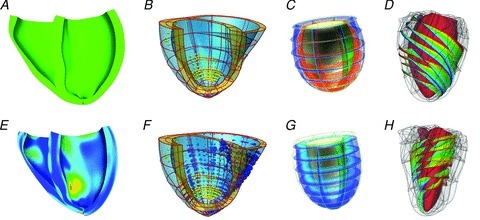
Top and bottom panels show end diastolic and end systolic cardiac models, respectively. A, fibre strain patterns in human CRT patient heart, –0.15 blue to +0.15 red (Aguado-Sierra et al. 2011), B, regional work patterns in CRT patient case; yellow spheres correspond to scar, blue and red spheres correspond to shortening and elongating myocardium, and the size of the spheres correspond to the magnitude of the work rate (Niederer et al. 2011a). C, generic heart model, showing membrane potential, blue –80mV to red +20 mV (Nickerson et al. 2005). D, fibre orientation in the canine myocardium, where colours trace individual fibres (Gurev et al. 2011).
Observation
Consistent with the scientific method, the motivation for developing a computational physiology model begins with an observation, question or hypothesis. Intrinsically mathematical models are approximations providing a finite caricature of the unbounded real system. The initial observation is used to define the limits and specifics of the system under study, including the species, temperature, age and spatial and temporal scales of the model (Smith et al. 2007; Niederer et al. 2009). In clinical and experimental studies the impact of this information is generally recognized and these values are routinely reported. Unlike wet lab studies, where new and intrinsically relevant data from the system of interest are typically clearly presented as results and set apart from contextual and potentially less relevant external data introduced in the discussion, computational models are agnostic to the source or type of data. Unless otherwise specified, the relevance of data to the system understudy is not represented in the model. As a result the conclusions of computational studies are often derived from a mix of data with varying degrees of relevance to the original observation.
Defining the limits of the system of interest, explicitly stating approximations and clearly defining the envelope within which the model can be applied, provides the necessary context for interpreting modelling results. In some cases the system under study is general; for example, a ventricular, atrial or Purkinje cell, with specific details such as the temperature, species or specific cell location not explicitly stated. These models often retain the potential to provide important generic insights but are clearly more limited when aiming to represent specific experimental or clinical systems. While this issue remains in computational physiology studies, recognition of the importance of these factors is the trend in the number of recent computational physiology models now focused on addressing these issues (Fink et al. 2011). This work has resulted in second generation models, which integrate additional data to enhance the temperature and species consistency of a model parameterisation. This progression is demonstrated in Fig. 3, which shows the change in the experimental data dependence between an initial foundation mouse model (Bondarenko et al. 2004) and a more recent refinement (Li et al. 2010). Most recently, this evolution of model specificity has been further focused in our group to account for specific genetic strains within a species (Li et al. 2010, 2011).
Figure 3. Computational model phylogenic tree showing the experimental data dependencies of the Bondorenko et al. (2004) (A) and Li et al. (2010) (B) models.
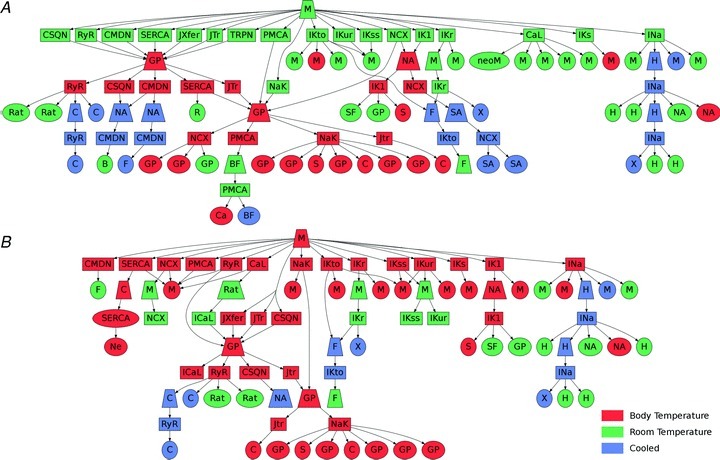
Trapeziums correspond to modelling studies. Squares correspond to model components (ion channels, buffers, transporters, etc.). Ellipses correspond to experimental studies. The colour scheme corresponds to the temperature of the data and the letters correspond to the species where M is mouse, GP is guinea pig, H is human, Rat is rat, S is sheep, SF is starfish, X is Xenopus, C is canine, F is ferret, Ca is calf, Ne is neuron, BF is bullfrog, SA is squid axon, B is bovine, R is rabbit and NA is not defined.
Fitting
The second link between experimental data and models in the development process is fitting parameter values. This fitting process is underpinned by three elements: labelling data and dependencies, processing data and determining parameters. To fully describe the fitting of model parameters to data requires a transparent description of the link between the value used in the final simulations and the actual recording made. This description is provided by accurately labelling the source of experimental data used to fit model parameters and labelling the dependency of model parameters on specific data. Despite the relatively low cost and high value of comprehensive labelling it is often absent in modelling studies.
Determining model parameters from experimental data often requires a degree of data processing. When measurements are made from the system under study, data processing can involve multiple steps with the raw measurements being transformed, calibrated, filtered and smoothed to remove noise and infer more physiologically meaningful measurements. For example, tension development in single cardiac myocytes can be calculated by measuring the bending of carbon tweezers with known stiffness (Iribe et al. 2007), the cytosolic Ca2+ concentration or the cell membrane potential can be calculated from fluorescence measurements (Bishop et al. 2007; Li et al. 2010) and endocardial electrical activation times can be extracted from non-contract catheter mapping systems (Niederer et al. 2011a). The process of transforming these indirect measurements into physiological values that feed into the model development process is thus clearly important for model fidelity and reproducibility. Once the data are processed the physiological values can then be used to determine model parameters.
In some cases it is not possible to make direct measurements in situ or the desired measurement is confounded by other signals. In cardiac electromechanics ion channels currents can be isolated in expression systems, sarcomere kinetics can be measured in skinned preparations and SERCA-ATPase can be characterized in sarcoplasmic reticulum vesicles. Although all of these approaches allow the properties of specific proteins to be characterised, the isolation process can often have an unknown effect on function. Expression systems may lack small but important channel subunits or post-translational regulation (Tseng-Crank et al. 1990; Blair et al. 1991; Paulmichl et al. 1991), altering the channel kinetics. Skinning alters the chemical environment of the sarcomere and may compromise sarcomere proteins causing the observed decrease in active tension Ca2+ sensitivity in these preparations (Niederer et al. 2006). Creating sarcoplasmic reticulum vesicles compromises the cellular environment, removing potentially important regulatory mechanisms of SERCA function. Mapping parameters from these reduced or altered systems to cardiac myocytes and whole organ systems poses significant challenges. Previously these differences have been assumed nominal or parameters from the altered system have been scaled to match a limited set of in situ observations (Niederer et al. 2006). However, in the case of expression systems, recent models that link channel structure at the protein scale to function (Silva et al. 2009) may provide a novel method for quantitatively linking expression system measurements to in situ channel kinetics.
Parameters are typically determined by minimizing a cost function describing the difference between a model result and the corresponding experimental or clinical measurement. In some cases measurements relate directly to model parameters; for example, the length–tension relationship in a cardiac myocyte can be measured directly and can be represented explicitly in the model (Niederer et al. 2006; Rice et al. 2008). Alternately, model parameters can be fitted to integrated emergent phenomena, such as fitting the sodium channel density to achieve a desired action potential upstroke (Li et al. 2010). In these, often numerous, situations it is only possible to fit parameters to these system scale observations. However, the difficulty in fitting parameters to systemic phenomena is that any errors from fitting other model components accumulate in the parameters that are fitted to system scale observations. This process, thus, can significantly decrease confidence in the value of parameters estimated using system responses (Niederer et al. 2009). Once the cost function is defined it can then be minimized manually or using one of many minimization algorithms. However, it is important to note that these approaches are not always successful or viable due to (1) disparities in model complexity and information contained in experimental data and/or (2) non-linearities in the model leading to fitting algorithms identifying local minima. For cardiac cell models it has been shown that in some cases it is impossible to fit a unique set of parameters to the model equations (Sarkar & Sobie, 2010). This difficulty in fitting parameters has been addressed by either manually fitting parameters or by developing the models in a form that enables parameters to be directly identifiable (Fink & Noble, 2009). The inclusion of sensitivity studies has also been adopted to provide additional information regarding the dependence of model outcomes on specific model parameters (Li et al. 2010; O’Hara et al. 2011). The goal of these sensitivity analyses is to separate model parameters into those that have a significant impact on model outcomes and those that have only a nominal effect. The parameters that have a significant impact must all be well constrained by experimental data; if an important parameter is poorly constrained this significantly reduces the confidence in any conclusions drawn from the model results. Conversely the data requirements underpinning parameters whose variation is identified as not producing large changes in key predictions are lower.
Validation
The final crucial stage in model development is validation. Rather than the validation of a model being considered binary i.e. a model is validated or it is not, we argue the degree of validation should be thought of as a spectrum. At one end there is no validation where the model can only replicate the data it was fitted to. On the other end of the spectrum you can show that a unique set of parameters has been determined from the available training data and that the model predicts measurements from an identified set of system-level data not included in the fitting process (Niederer et al. 2011a; Provost et al. 2011). The multi-physics nature of cardiac electromechanics models and an insufficient quantity and quality of data has limited validation in the past. Examples of previous studies include the combination of models of rat contraction at room temperature with guinea pig electrophysiology models at body temperature (Nickerson et al. 2001), using a guinea pig electrophysiology model within a rabbit geometry (Jie et al. 2010), the development of whole organ models, where the cellular models were based on available data at room temperature, which could not be compared directly against in vivo measurements (Niederer & Smith, 2009).
As is often the case in computational modeling, comprehensive validation is not achievable. However, we assert that in these cases, models compared against limited data demonstrate that the proposed set of parameters and equations provide only a plausible representation of the system under study. Moving to the concept of a plausibility spectrum would lead to models demonstrating that they are highly plausible through multiple independent comparisons of model predictions and experimental data. This would, in turn, provide a more transparent description of the model validation process.
Future perspectives
Improving the link between models and experimental data is essential if models are to fulfill their role in linking multiple physiological observations across disparate spatial and temporal scales with underlying function. Cardiac electromechanical models have begun to consider the impact of temperature and species on model outcomes. However, at present the vast majority of these models still ignore the dominant predictor of disease: age (Houle et al. 2010). Models have begun to account for the effects of age by simulating neonatal cells (Wang & Sobie, 2008) and the progression of heart failure following genetic knockout of SERCA over periods of weeks (Li et al. 2011). Accounting for age in computational physiology models will need to increase as models are used in clinical applications. This is particularly important in the use of computational models in drug screening (Mirams et al. 2011; Moreno et al. 2011), where the failure to identify drugs that adversely affect older unhealthy patients is because of efficacy being based on experimental studies using young healthy animals (Lin, 1995).
In addition to age, models will also need to start to account for biological variations associated with inter-subject differences in experimental data. Current cardiac electromechanics models often implicitly aim to represent population averages or a single individual. Studies have begun to include the impact of spatial variability in models with cell types fitted to data from cells in specific regions of the heart (Campbell et al. 2008; Fink et al. 2011; O’Hara et al. 2011). However, these models have yet to account for the intrinsic variation between individual cells (Bahar et al. 2006) or between individuals (Lerner & Kannel, 1986). Accounting for this variation motivates the need for computational frameworks to move away from a single model methodology to studying populations of models that can represent the underlying variability present in an individual's cells or across a population of individuals.
In addition to intrinsic variability within experimental data, as already discussed, measurements also inevitably suffer from uncertainty due to noise, under-sampling or bias. Regardless of the cause, models need to take into account the impact of uncertainty on predictions. This has motivated our own work in cardiac electromechanics models to provide confidence intervals or ranges of potential outcomes (Niederer & Smith, 2009) as opposed to single deterministic predictions.
Current community efforts have led to the use of standardized languages for publishing models (Hucka et al. 2003; Garny et al. 2008), open source and/or freely available simulation software (Garny et al. 2003; Pitt-Francis et al. 2009; Bradley et al. 2011), and the availability of models from individual groups and community repositories (Fink et al. 2011) provides a transparent representation of the model equations, parameters and solution methods. However, these efforts have yet to provide a comprehensive format to describe experimental measurements, data processing, mapping and fitting methods and a link between data to model parameters.
As demonstrated by cardiac electromechanics models, computational physiology has a significant role to play in rationalising data, linking observations to function and translating laboratory results into clinical contexts. Embedded within this role there are significant challenges and opportunities, a number of which have been outlined above. Ensuring that models transparently define the system under study, link parameters with data and model results are compared with data to demonstrate increased plausibility is a major challenge for the modelling community. Towards this goal, extensive supplements describing the model development methodology have been provided alongside recent modelling studies. If this trend continues, models will reach their potential of providing a succinct, complete and transparent representation of physiological data and function and, in turn, provide a valuable approach for revealing physiological insight.
References
- Aguado-Sierra J, Krishnamurthy A, Villongco C, Chuang J, Howard E, Gonzales MJ, et al. Patient-specific modeling of dyssynchronous heart failure: a case study. Prog Biophys Mol Biol. 2011;107:147–155. doi: 10.1016/j.pbiomolbio.2011.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahar R, Hartmann CH, Rodriguez KA, Denny AD, Busuttil RA, Dolle MET, et al. Increased cell-to-cell variation in gene expression in ageing mouse heart. Nature. 2006;441:1011–1014. doi: 10.1038/nature04844. [DOI] [PubMed] [Google Scholar]
- Benfey PN, Mitchell-Olds T. From genotype to phenotype: systems biology meets natural variation. Science. 2008;320:495–497. doi: 10.1126/science.1153716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop MJ, Rodriguez B, Qu F, Efimov IR, Gavaghan DJ, Trayanova NA. The role of photon scattering in optical signal distortion during arrhythmia and defibrillation. Biophys J. 2007;93:3714–3726. doi: 10.1529/biophysj.107.110981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair TA, Roberds SL, Tamkun MM, Hartshorne RP. Functional characterization of RK5, a voltage-gated K+ channel cloned from the rat cardiovascular system. FEBS Lett. 1991;295:211–213. doi: 10.1016/0014-5793(91)81420-d. [DOI] [PubMed] [Google Scholar]
- Bondarenko VE, Szigeti GP, Bett GCL, Kim S-J, Rasmusson RL. Computer model of action potential of mouse ventricular myocytes. Am J Physiol Heart Circ Physiol. 2004;287:H1378–H1403. doi: 10.1152/ajpheart.00185.2003. [DOI] [PubMed] [Google Scholar]
- Bradley C, Bowery A, Britten R, Budelmann V, Camara O, Christie R, et al. OpenCMISS: a multi-physics & multi-scale computational infrastructure for the VPH/Physiome project. Prog Biophys Mol Biol. 2011;107:32–47. doi: 10.1016/j.pbiomolbio.2011.06.015. [DOI] [PubMed] [Google Scholar]
- Butcher EC, Berg EL, Kunkel EJ. Systems biology in drug discovery. Nat Biotech. 2004;22:1253–1259. doi: 10.1038/nbt1017. [DOI] [PubMed] [Google Scholar]
- Campbell SG, Flaim SN, Leem CH, McCulloch AD. Mechanisms of transmurally varying myocyte electromechanics in an integrated computational model. Philos Trans A Math Phys Eng Sci. 2008;366:3361–3380. doi: 10.1098/rsta.2008.0088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell SG, Howard E, Aguado-Sierra J, Coppola BA, Omens JH, Mulligan LJ, et al. Effect of transmurally heterogeneous myocyte excitation;contraction coupling on canine left ventricular electromechanics. Exp Physiol. 2009;94:541–552. doi: 10.1113/expphysiol.2008.044057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell SG, McCulloch AD. Multi-scale computational models of familial hypertrophic cardiomyopathy: genotype to phenotype. J R Soc Interface. 2011;8:1550–1561. doi: 10.1098/rsif.2011.0184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins FS, Green ED, Guttmacher AE, Guyer MS. A vision for the future of genomics research. Nature. 2003;422:835–847. doi: 10.1038/nature01626. [DOI] [PubMed] [Google Scholar]
- Davidson SB, Overton C, Buneman P. Challenges in integrating biological data sources. J Comput Biol. 1995;2:557–572. doi: 10.1089/cmb.1995.2.557. [DOI] [PubMed] [Google Scholar]
- Deisboeck TS, Zhang L, Yoon J, Costa J. In silico cancer modeling: is it ready for prime time? Nat Clin Prac Oncol. 2009;6:34–42. doi: 10.1038/ncponc1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans GA. Designer science and the “omic” revolution. Nat Biotech. 2000;18:127–127. doi: 10.1038/72480. [DOI] [PubMed] [Google Scholar]
- Fink M, Niederer SA, Cherry EM, Fenton FH, Koivumäki JT, Seemann G, et al. Cardiac cell modelling: observations from the heart of the cardiac physiome project. Prog Biophys Mol Biol. 2011;104:2–21. doi: 10.1016/j.pbiomolbio.2010.03.002. [DOI] [PubMed] [Google Scholar]
- Fink M, Noble D. Markov models for ion channels: versatility versus identifiability and speed. Philos Transact A Math Phys Eng Sci. 2009;367:2161–2179. doi: 10.1098/rsta.2008.0301. [DOI] [PubMed] [Google Scholar]
- Garny A, Kohl P, Noble D. Cellular Open Resource (COR): a public CellML based environment for modeling biological function. Int J Bifurcat Chaos. 2003;12:3579–3590. [Google Scholar]
- Garny A, Nickerson DP, Cooper J, Weber dos Santos R, Miller AK, McKeever S, et al. CellML and associated tools and techniques. Philos Transact A Math Phys Eng Sci. 2008;366:3017–3043. doi: 10.1098/rsta.2008.0094. [DOI] [PubMed] [Google Scholar]
- Gurev V, Constantino J, Rice JJ, Trayanova NA. Distribution of electromechanical delay in the heart: insights from a three-dimensional electromechanical model. Biophys J. 2010;99:745–754. doi: 10.1016/j.bpj.2010.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurev V, Lee T, Constantino J, Arevalo H, Trayanova N. Models of cardiac electromechanics based on individual hearts imaging data. Biomech Model Mechanobiol. 2011;10:295–306. doi: 10.1007/s10237-010-0235-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez T, Kambhampati S. Integration of biological sources: current systems and challenges ahead. SIGMOD Rec. 2004;33:51–60. [Google Scholar]
- Houle D, Govindaraju DR, Omholt S. Phenomics: the next challenge. Nat Rev Genet. 2010;11:855–866. doi: 10.1038/nrg2897. [DOI] [PubMed] [Google Scholar]
- Hucka M, Finney A, Sauro HM, Bolouri H, Doyle JC, Kitano H, et al. The systems biology markup language (SBML): a medium for representation and exchange of biochemical network models. Bioinformatics. 2003;19:524–531. doi: 10.1093/bioinformatics/btg015. [DOI] [PubMed] [Google Scholar]
- Iribe G, Helmes M, Kohl P. Force-length relations in isolated intact cardiomyocytes subjected to dynamic changes in mechanical load. Am J Physiol Heart Circ Physiol. 2007;292:H1487–H1497. doi: 10.1152/ajpheart.00909.2006. [DOI] [PubMed] [Google Scholar]
- Jie X, Gurev V, Trayanova N. Mechanisms of mechanically induced spontaneous arrhythmias in acute regional ischemia. Circ Res. 2010;106:185–192. doi: 10.1161/CIRCRESAHA.109.210864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keldermann RH, Nash MP, Gelderblom H, Wang VY, Panfilov AV. Electromechanical wavebreak in a model of the human left ventricle. Am J Physiol Heart Circ Physiol. 2010;299:H134–H143. doi: 10.1152/ajpheart.00862.2009. [DOI] [PubMed] [Google Scholar]
- Kerckhoffs RCP, Omens JH, McCulloch AD, Mulligan LJ. Ventricular dilation and electrical dyssynchrony synergistically increase regional mechanical nonuniformity but not mechanical dyssynchrony: a computational model. Circ Heart Fail. 2010;3:528–536. doi: 10.1161/CIRCHEARTFAILURE.109.862144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitano H. Systems biology: a brief overview. Science. 2002;295:1662–1664. doi: 10.1126/science.1069492. [DOI] [PubMed] [Google Scholar]
- Land S, Niederer SA, Smith NP. Efficient computational methods for strongly coupled cardiac electromechanics. IEEE Trans Biomed Eng. 2011 doi: 10.1109/TBME.2011.2112359. (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- Lee J, Niederer S, Nordsletten D, Le Grice I, Smail B, Kay D, Smith N. Coupling contraction, excitation, ventricular and coronary blood flow across scale and physics in the heart. Philos Transact A Math Phys Eng Sci. 2009;367:2311–2331. doi: 10.1098/rsta.2008.0311. [DOI] [PubMed] [Google Scholar]
- Lerner DJ, Kannel WB. Patterns of coronary heart disease morbidity and mortality in the sexes: a 26-year follow-up of the Framingham population. Am Heart J. 1986;111:383–390. doi: 10.1016/0002-8703(86)90155-9. [DOI] [PubMed] [Google Scholar]
- Li L, Louch WE, Niederer SA, Andersson KB, Christensen G, Sejersted OM, Smith NP. Calcium dynamics in the ventricular myocytes of SERCA2 knockout mice: a modeling study. Biophys J. 2011;100:322–331. doi: 10.1016/j.bpj.2010.11.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Niederer SA, Idigo W, Zhang YH, Swietach P, Casadei B, Smith NP. A mathematical model of the murine ventricular myocyte: a data-driven biophysically based approach applied to mice overexpressing the canine NCX isoform. Am J Physiol Heart Circ Physiol. 2010;299:H1045–H1063. doi: 10.1152/ajpheart.00219.2010. [DOI] [PubMed] [Google Scholar]
- Lin JH. Species similarities and differences in pharmacokinetics. Drug Metab Dispos. 1995;23:1008–1021. [PubMed] [Google Scholar]
- Luther S, Fenton FH, Kornreich BG, Squires A, Bittihn P, Hornung D, et al. Low-energy control of electrical turbulence in the heart. Nature. 2011;475:235–239. doi: 10.1038/nature10216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirams GR, Cui Y, Sher A, Fink M, Cooper J, Heath BM, et al. Simulation of multiple ion channel block provides improved early prediction of compounds’ clinical torsadogenic risk. Cardiovasc Res. 2011;91:53–61. doi: 10.1093/cvr/cvr044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno JD, Zhu ZI, Yang P-C, Bankston JR, Jeng M-T, Kang C, et al. A computational model to predict the effects of class i anti-arrhythmic drugs on ventricular rhythms. Sci Transl Med. 2011;3:98ra83. doi: 10.1126/scitranslmed.3002588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickerson D, Smith N, Hunter P. New developments in a strongly coupled cardiac electromechanical model. Europace. 2005;7:S118–S127. doi: 10.1016/j.eupc.2005.04.009. [DOI] [PubMed] [Google Scholar]
- Nickerson DP, Smith NP, Hunter PJ. A model of cardiac cellular electromechanics. Philos Transact A Math Phys Eng Sci. 2001;359:1159–1172. [Google Scholar]
- Niederer SA, Fink M, Noble D, Smith NP. A meta-analysis of cardiac electrophysiology computational models. Exp Physiol. 2009;94:486–495. doi: 10.1113/expphysiol.2008.044610. [DOI] [PubMed] [Google Scholar]
- Niederer SA, Hunter PJ, Smith NP. A quantitative analysis of cardiac myocyte relaxation: a simulation study. Biophys J. 2006;90:1697–1722. doi: 10.1529/biophysj.105.069534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niederer SA, Plank G, Chinchapatnam P, Ginks M, Lamata P, Rhode KS, et al. Length-dependent tension in the failing heart and the efficacy of cardiac resynchronization therapy. Cardiovasc Res. 2011a;89:336–343. doi: 10.1093/cvr/cvq318. [DOI] [PubMed] [Google Scholar]
- Niederer SA, Shetty AK, Plank G, Bostock J, Razavi R, Smith NP, Rinaldi CA. Biophysical modeling to simulate the response to multisite left ventricular stimulation using a quadripolar pacing lead. Pacing Clin Electrophysiol. 2011b doi: 10.1111/j.1540-8159.2011.03243.x. (Epub ahead of print½. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niederer SA, Smith NP. A mathematical model of the slow force response to stretch in rat ventricular myocytes. Biophys J. 2007;92:4030–4044. doi: 10.1529/biophysj.106.095463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niederer SA, Smith NP. An improved numerical method for strong coupling of excitation and contraction models in the heart. Prog Biophys Mol Biol. 2008;96:90–111. doi: 10.1016/j.pbiomolbio.2007.08.001. [DOI] [PubMed] [Google Scholar]
- Niederer SA, Smith NP. The role of the Frank–Starling law in the transduction of cellular work to whole organ pump function: a computational modeling analysis. PLoS Comput Biol. 2009;5:e1000371. doi: 10.1371/journal.pcbi.1000371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble D. Claude Bernard, the first systems biologist, and the future of physiology. Exp Physiol. 2008;93:16–26. doi: 10.1113/expphysiol.2007.038695. [DOI] [PubMed] [Google Scholar]
- Nordsletten DA, Niederer SA, Nash MP, Hunter PJ, Smith NP. Coupling multi-physics models to cardiac mechanics. Prog Biophys Mol Biol. 2011;104:77–88. doi: 10.1016/j.pbiomolbio.2009.11.001. [DOI] [PubMed] [Google Scholar]
- O’Hara T, Virág L, Varró A, Rudy Y. Simulation of the undiseased human cardiac ventricular action potential: model formulation and experimental validation. PLoS Comput Biol. 2011;7:e1002061. doi: 10.1371/journal.pcbi.1002061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulmichl M, Nasmith P, Hellmiss R, Reed K, Boyle WA, Nerbonne JM, et al. Cloning and expression of a rat cardiac delayed rectifier potassium channel. Proc Natl Acad Sci U S A. 1991;88:7892–7895. doi: 10.1073/pnas.88.17.7892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitt-Francis J, Pathmanathan P, Bernabeu MO, Bordas R, Cooper J, Fletcher AG, et al. Chaste: a test-driven approach to software development for biological modelling. Comput Phys Commun. 2009;180:2452–2471. [Google Scholar]
- Provost J, Gurev V, Trayanova N, Konofagou EE. Mapping of cardiac electrical activation with electromechanical wave imaging: an in silico–in vivo reciprocity study. Heart Rhythm. 2011;8:752–759. doi: 10.1016/j.hrthm.2010.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice JJ, Wang F, Bers DM, de Tombe PP. Approximate model of cooperative activation and crossbridge cycling in cardiac muscle using ordinary differential equations. Biophys J. 2008;95:2368–2390. doi: 10.1529/biophysj.107.119487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudy Y, Ackerman MJ, Bers DM, Clancy CE, Houser SR, London B, et al. Systems approach to understanding electromechanical activity in the human heart. Circulation. 2008;118:1202–1211. doi: 10.1161/CIRCULATIONAHA.108.772715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar AX, Sobie EA. Regression analysis for constraining free parameters in electrophysiological models of cardiac cells. PLoS Comput Biol. 2010;6:e1000914. doi: 10.1371/journal.pcbi.1000914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva JR, Pan H, Wu D, Nekouzadeh A, Decker KF, Cui J, et al. A multiscale model linking ion-channel molecular dynamics and electrostatics to the cardiac action potential. Proc Natl Acad Sci U S A. 2009;106:11102–11106. doi: 10.1073/pnas.0904505106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith NP, Crampin EJ, Niederer SA, Bassingthwaighte JB, Beard DA. Computational biology of cardiac myocytes: proposed standards for the physiome. J Exp Biol. 2007;210:1576–1583. doi: 10.1242/jeb.000133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tandri H, Weinberg SH, Chang KC, Zhu R, Trayanova NA, Tung L, Berger RD. Reversible cardiac conduction block and defibrillation with high-frequency electric field. Sci Transl Med. 2011;3:102ra196. doi: 10.1126/scitranslmed.3002445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng-Crank JCL, Tseng G-N, Schwartz A, Tanouye MA. Molecular cloning and functional expression of a potassium channel cDNA isolated from a rat cardiac library. FEBS Lett. 1990;268:63–68. doi: 10.1016/0014-5793(90)80973-m. [DOI] [PubMed] [Google Scholar]
- van de Waterbeemd H, Gifford E. ADMET in silico modelling: towards prediction paradise? Nat Rev Drug Discov. 2003;2:192–204. doi: 10.1038/nrd1032. [DOI] [PubMed] [Google Scholar]
- Wang LJ, Sobie EA. Mathematical model of the neonatal mouse ventricular action potential. Am J Physiol Heart Circ Physiol. 2008;294:H2565–H2575. doi: 10.1152/ajpheart.01376.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]


