Abstract
Sialic-acid-binding immunoglobulin-like lectins (Siglecs) are a family of transmembrane receptors that are well documented to play roles in regulation of innate and adaptive immune responses. To see whether the features that define the molecular recognition of sialic acid were found in other sialic-acid-binding proteins, we analyzed 127 structures with bound sialic acids found in the Protein Data Bank database. Of these, the canine adenovirus 2-fiber knob protein showed close local structural relationship to Siglecs despite low sequence similarity. The fiber knob harbors a noncanonical sialic-acid recognition site, which was then explored for detailed specificity using a custom glycan microarray comprising 58 diverse sialosides. It was found that the adenoviral protein preferentially recognizes the epitope Neu5Acα2-3[6S]Galβ1-4GlcNAc, a structure previously identified as the preferred ligand for Siglec-8 in humans and Siglec-F in mice. Comparison of the Siglec and fiber knob sialic-acid-binding sites reveal conserved structural elements that are not clearly identifiable from the primary amino acid sequence, suggesting a Siglec-like sialic-acid-binding motif that comprises the consensus features of these proteins in complex with sialic acid.
Keywords: adenovirus, sialic acid, Siglecs, structural glycobiology
Introduction
The interaction of receptors with glycans underlies many aspects of biology such as cell adhesion, cell–cell communication and development (Varki 2009). The recognition event results from the interplay between the architecture of the glycan recognition site of the receptor and the conformational space adopted by the glycans (DeMarco and Woods 2008). Although important information can be obtained from cocrystal structures, only 8–10% of the structures in the Protein Data Bank (PDB) database contain carbohydrates (Lutteke and von der Lieth 2009), and most of these are covalently bound N- and O-linked glycosylations. In some cases, important information can be obtained from structurally related proteins. Although homologous receptors may be identified using algorithms to compare sequence analysis or overall fold, this alone might miss important examples where the similarity exists in the local area of the recognition site. Thus, algorithms that can better define the features of glycan recognition may be useful.
Sialic-acid-binding immunoglobulin-like lectins (Siglecs) are one family of glycan-binding proteins in need of a better description of the structural features required for their glycan recognition. These members of the I-type lectin family are predominantly found in immune cells (Crocker et al. 2007). Their involvement in many facets of innate and adaptive immune cell biology makes them attractive pharmacological targets and it is therefore of great interest to study their glycan specificity (O'Reilly and Paulson 2009). In recent years, X-ray crystallography provided structures of three Siglecs, namely Siglec-1, -5 and -7 (Attrill, Imamura et al. 2006; Attrill, Takazawa et al. 2006; Zaccai et al. 2007; Zhuravleva et al. 2008). Structural analysis of the complexes reveals common recognition features. One remarkable aspect of this interaction was distilled by the work of Zhuravleva et al. (2008) on Siglec-5. The authors highlight the sialic-acid extension of an antiparallel beta-sheet in all Siglecs.
Siglecs constitute only a fraction of glycan-binding proteins (lectins) that recognize sialosides. In addition to other endogenous vertebrate lectins, many pathogens also interact with terminal sialic-acid residues, thereby being key players in the life cycle of many bacteria and viruses (Angata and Varki 2002; Kumlin et al. 2008). Influenza viruses, polyomaviruses and adenoviruses are only a few examples of viruses for which structures in the presence of glycan ligands have been solved by X-ray crystallography. Therefore, a source of sialic-acid-recognizing proteins is available to expand our knowledge by homology searches. Here we present a strategy to identify structural homologs of Siglecs to gain insights into sialic-acid recognition, starting from the glycan structure rather than the protein. This allowed us to identify a glycan-binding site in the CAV-2 fiber head protein, which has high local similarity to Siglecs.
Results
As a first step towards searching for Siglec-like sialic-acid-binding proteins, a comprehensive list of crystal structures of proteins in complex with sialic acids was compiled from the PDB database. A direct search of the PDB database using the nonchiral SMILES code representation of sialic acid identified 55 structures. A second search using the http://www.glycosciences.de web service identified 106 cocrystal structures, resulting in a combined list of 127 unique structures (see Supplementary data, Table S1). The coordinates of the ring atoms of the sialic acids were used for structural alignment of all complexes, and careful inspection of the alignment allowed for the comparison of the carbohydrate respective recognition features. We then conducted an in silico search focused on a structural feature of sialic-acid binding to Siglecs that has heretofore not been described in a non-Siglec protein, namely the glycerol side chain and the N-acetyl group of the sialic acid that mimics an extended antiparallel beta-sheet (Zhuravleva et al. 2008).
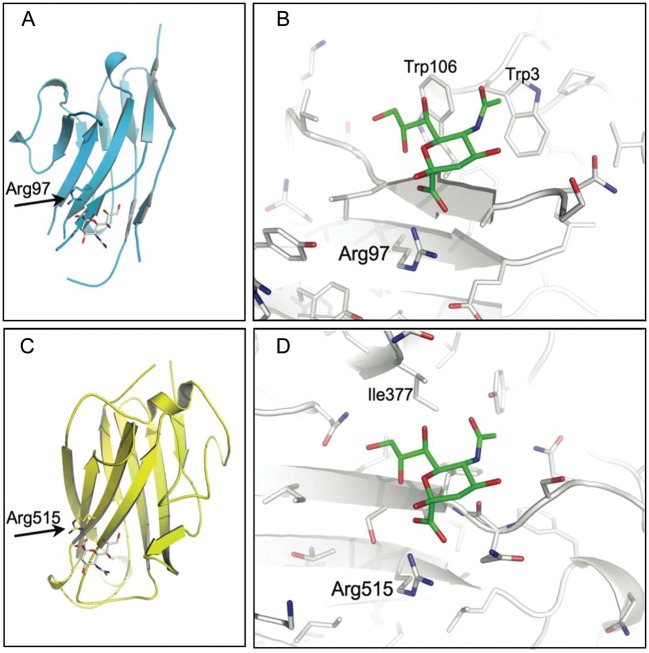
Not surprisingly, the database screen identified the complexes of Siglec-1, -5 and -7. However, out of the 127 queried structures, one additional non-Siglec protein was found to share a similar recognition site. This protein is the fiber knob protein of the canine adenovirus 2 (CAV-2) in complex with 3′-sialyllactose. Moreover, a ternary complex of CAV-2 with 3′-sialyllactose and the coxsackievirus and adenovirus receptor (CAR) reported by the same authors was also identified (Seiradake et al. 2009). In both structures, the Neu5Ac residue of the 3′-sialyllactose adopted a similar pose with respect to the protein backbone as found for the Siglecs (Figure 1). There was an arginine making the essential salt bridge to the Neu5Ac and, importantly, the pseudo-extension of the antiparallel beta-sheet was present. In contrast to the Siglec structures, there were no aromatic side chains interacting with the glycerol side chain and the N-acteyl group of the sialic acid.
Fig. 1.
Structural alignment of Siglec-1 and CAV-2 fiber knob. The structural alignment based on the ring atoms of the sialic acid is shown for the entire glycan binding site of the V-set domain of Siglec-1 (A, C) and CAV-2 fiber knob protein (B, D). The essential arginine is highlighted, and the sandwich fold of two antiparallel beta-sheets is shown for both structures. The extension of the beta-sheet by the sialic acid is also apparent for both structures. In contrast, the van der Waals interaction of the C9 methylene of the glycerol side chain to Trp-106 in Siglec-1 is not observed for the CAV-2 structure. Moreover, while the N-acteyl group recognition site in Siglec-1 is mediated by hydrophobic contacts originating from Trp-3, other hydrophobic side chains in CAV-2 replace this feature. Hydrogens were omitted for simplification (Siglec-1, pdb id: 1QFO; CAV-2, pdb id: 2WBV).
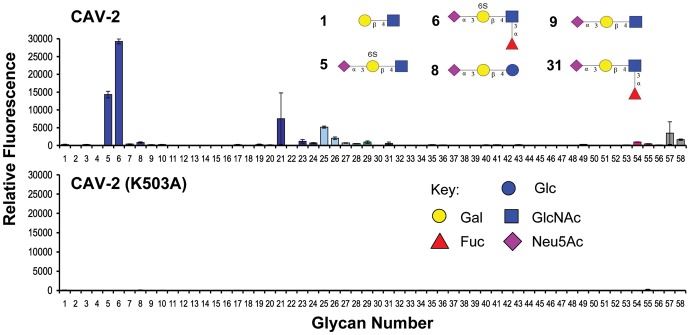
The reported structure of CAV-2 was solved with 3′-sialyllactose in the binding pocket and, to our knowledge, no additional information on the glycan specificity is available. Accordingly, a sialoside glycan microarray (Nycholat et al. 2012; Xu et al. 2012) was employed to investigate the specificity of CAV-2. For this analysis, we used a recombinant CAV-2 with a His tag, which can be multimerized using anti-His tag antibodies (Seiradake et al. 2009). Using this construct, microarray data clearly reveal a preferred specificity for the Neu5Acα2-3[6S]Galβ1-4GlcNAc trisaccharide (5) and the related 6′sulfo-sialyl-LewisX tetrasaccharide (6) (Figure 2A). The CAV-2 protein did not bind to either the nonsulfated analogs 9 and 31 or to the disaccharide lacking the terminal Neu5Ac (1), a specificity previously identified for other Siglecs. At higher concentrations of the CAV-2 protein, additional lower avidity glycans were identified including extended N-glycans with terminal Neu5Acα2-3-Gal epitopes (24, 25, 57) and the GM2 tetrasaccharide (29) (see Supplementary data, Figure S1a).
Fig. 2.
Glycan array analysis of the CAV-2 fiber knob protein binding to a printed sialoside library. (Top) The adenoviral capsid protein (50 µg/mL) strongly recognizes the two 6′-sulfated α2,3-linked sialosides 5 and 6. (Bottom) Site-directed mutagenesis of lysine 503 to an alanine abrogated binding to the printed glycans (see Supplementary data for the complete list of glycans, Supplementary data, Table S2 and Figure S1 for glycan array analysis at higher protein concentrations).
To explain the affinity gain of the two sulfated α2-3-sialosides, we conducted molecular modeling based on the available cocrystal structure of the CAV-2 protein with Neu5Acα2-3-Galβ1-4-Glc (Figure 3). Strikingly, the recognition of the negatively charged sulfate at the 6-position of the galactose in 5 and 6 was readily accessible for the positively charged side chain of lysine 503. The substitution of the glucose residue of the lactose moiety by a glucosamine, as found in 5 and 6, did not alter the interaction of the glycans. Only minor side chain motions in the protein had to be applied during the modeling process to allow for optimal recognition of the sialosides, which strongly indicates a preformed binding site for α2-3-sialosides sulfated at the 6-position of galactose. In order to access the importance of this interaction experimentally, we conducted site-directed mutagenesis of lysine 503. No binding of the mutant CAV-2 to the printed sialoside array was detected, an observation extending the functional role of this residue also to the recognition of the nonsulfated glycans picked up at higher CAV-2 concentrations (Figure 2B and Supplementary data, Figure S1).
Fig. 3.
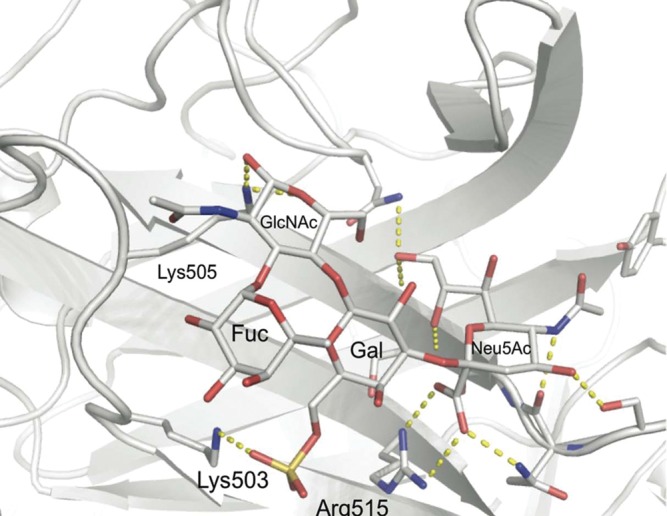
Model of 6′-sulfo-Lewisx (6) bound to CAV-2 fiber knob protein. The 3′-sialyllactose cocrystal structure of CAV-2 was used as a template to model the 6′sulfo-sialyl-LewisX antigen (6) into the binding site. The sulfate occupies a positively charged subsite with favorable interactions with Lys-503. An identical pose of the sulfated trisaccharide (5) was found following the same modeling protocol (data not shown).
Discussion
We followed a strategy for finding a sialic-acid-binding proteins by comparing local structural homology of the Siglec sialic-acid-binding site. A search for such features fails using global measures (such as sequence identity or structural superposition) as the local homology is not necessarily a property of the overall sequence or structure. For example, the sequence identity of Siglec-1 and Siglec-5 V-set domains is only 29%. Yet, both proteins share the same fold and recognize sialic acids in an identical fashion. However, the low sequence identity does not permit finding one Siglec simply using the sequence of the other, since the statistical significance of the aligned sequences is almost lost (Rost 1999). Building a homology model on the basis of this alignment would not be reliable for extracting relevant information on the ligand recognition (Evers et al. 2003). Moreover, the overall fold does not provide sufficient information. Too many potential structures are identified conducting the search for proteins with a similar recognition motif based on a global 3D structural alignment. For instance, the root mean square deviation (RMSD) of the alpha carbons (Cα) of the two Siglecs is 2.2 Å and that between Siglec-1 and CAV-2 protein is 12.6 Å. Using the jFATCAT-rigid algorithm, already more than 180 structures with an RMSD below 2.2 Å to Siglec-1 were found in the PDB database (Ye and Godzik 2003). A comprehensive analysis of proteins with an RMSD below 12.6 Å would not be straightforward.
Therefore, our database query was initially based on carbohydrate structures rather than protein homologs. Sialic acids are unique glycans mimicking an extended antiparallel beta-sheet of the V-set domain in Siglecs (Zhuravleva et al. 2008). The glycerol side chain of sialic acids is known to adopt a stretched structure in solution (Siebert et al. 1992; Sawada et al. 2006). The presentation of the hydrogen bond donors and acceptors by this particular conformation allows the Neu5Ac residue to interact with protein backbone. This rather unusual motif led us to the analysis of all sialosides in complex with proteins as found in the PDB depository. Unfortunately, these complexes are not readily accessible by the search engine provided. However, combining a direct search of the PDB database with a query using http://www.glycosciences.de tools (Lutteke and von der Lieth 2009) led to a more comprehensive set of 127 cocrystal structures. This search identified the fiber knob domain of the CAV-2 to bind the sialic acid in an identical pose combined with a very similar local geometry of the binding site. To the best of our knowledge, this resembles a novel approach to find cases of convergently evolved glycan-binding proteins.
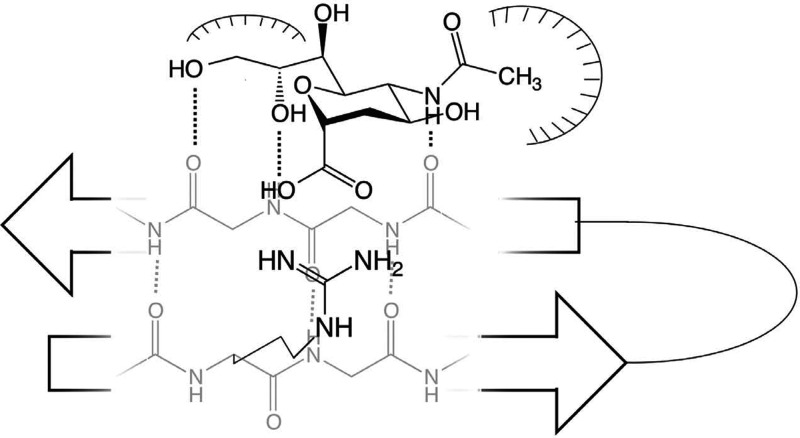
The Siglec-like sialic-acid recognition motif found in CAV-2 covers essential recognition features of the interaction found in Siglecs. However, several features believed to be important for the coordination of the Neu5Ac were not found. For example, the van der Waals interaction of a tryptophan or a tyrosine with the C9 methylene of the stretched glycerol side chain in Siglecs lacks an equivalent in the fiber knob. Also absent in CAV-2 is the hydrophobic interactions between the methyl group and an aromatic amino acid (see Figure 4).
Fig. 4.
Schematic representation of the Siglec-like sialic-acid recognition motif highlighting the extension of the antiparallel beta-sheet, the salt bridge between essential arginine and the carboxylic acid of the Neu5Ac, the hydrophobic interaction coordinating the glycerol side chain and the hydrophobic pocket harboring the N-acteyl group of the sugar.
Glycan array analysis showed that the CAV-2 protein exhibited a preference for two sialylated and sulfated structures, the 6′sulfo-sialyl LewisX tetrasaccharide (6) and the corresponding nonfucosylated trisaccharide (5). No binding to 3′-sialyllactose (8), the ligand that the structure of CAV-2 was solved with, was detected, suggesting that sulfate in 5 and 6 provides significant affinity increase. Our modeling studies readily explained the affinity gain to result from a salt bridge with lysine at 503, which was confirmed with glycan array analysis of the K503A mutant (Figures 2 and 3). The modeled structure of 6 bound to CAV-2 is in accordance with other reported complexes of the nonsulfated glycans.
It is also noticeable that the sulfated glycans (5 and 6) are also ligands of Siglec-8 and -F, two Siglecs expressed in murine and human eosinophils, respectively (Bochner et al. 2005; Tateno et al. 2005). Although the structures of these Siglecs are not yet available, structural alignment of CAV-2 with Siglec-5 suggests good candidates for amino acids making favorable contacts to the sulfate group, such as Arg72 and Asn61 in Siglec-8 and -F, respectively (data not shown).
The expression profile of the keratan sulfate Gal-6-sulfotransferase (KS6ST) in human tissue has been reported recently, an enzyme potentially involved in the biosynthesis of the sulfated epitope (Fukuta et al. 1997; Tateno et al. 2010). In particular, high mRNA levels in lung and brain samples are reported, corresponding well with the tropism of the virus. Moreover, analyzing the fiber head protein family, six sequences bearing the lysine and the essential arginine have been revealed, indicating that more members of this family may evolved this noncanonical binding site recognizing sulfated sialosides (Supplementary data, Figure S2).
Materials and methods
Reagents and proteins
Mutated and wild-type forms of CAV-2 fiber head cloned in pPROEX HTb were expressed in Escherichia coli strain BL21 Star (DE3) (Invitrogen, Carlsbad, CA) at 37°C. Cells were resuspended in Lysis-Equilibration-Wash (LEW) buffer (50 mM NaH2PO4, 300 mM NaCl, pH 8) complemented with 1 mg/mL lysozyme and protease inhibitor cocktail (Roche, Mannheim, Germany) and lysed by sonication. The cell lysate was centrifuged for 30 min at 10,000 × g and the supernatant was loaded onto Protino Ni-TED columns (Macherey-Nagel, Düren, Germany). Columns were washed with LEW buffer and eluted with elution buffer (50 mM NaH2PO4, 300 mM NaCl, 250 mM imidazole, pH 8). The point mutant CAV-2 K503A was generated using the following mutagenesis PCR primer and the corresponding complement, 5′-GCATGCTAATTATTAACGCTCCGAAAGGCGT TGCCACTTACACC-3′, with the mutated codon being underscored. Before use, proteins were stored at 1.4 mg/mL at 4°C.
Database analysis
A list of protein complexes with sialic acids was established by using a direct search of the PDB database (25 March 2010) employing nonchiral SMILES code filtering and http:// www.glycosciences.de (Berman et al. 2000; Lutteke et al. 2006). A script written in BioPython (version 1.53) was employed to align the complexes on the basis of the ring atoms of the sialic acids (Cock et al. 2009). Inspection of the sialic-acid-binding poses was performed in PyMOL (DeLano, W.L. The PyMOL Molecular Graphics System. (2008) DeLano Scientific LLC, Palo Alto, CA, USA., version 1.2r0).
Glycan array data acquisition and analysis
Printing of glycan arrays was carried out as previously described (Blixt et al. 2004). Glycan samples were printed on to NHS-ester-coated glass slides (SlideH, Schott/Nexterion, Jena, Germany), in six replicate spots, using a MicroGrid II (Digilab, Ann Arbor, MI) printing robot equipped with Stealth SMP4B microarray pins, followed by quenching of the unreacted NHS-esters prior to probing with the protein sample.
To probe the array, 100 µL of CAV-2 recombinant protein at 500 µg/mL was mixed in PBS + 0.05% Tween-20 (Fisher, St. Louis, MO) with mouse anti-HIS-AlexaFluor532 antibody (Qiagen, Hilden, Germany) and goat-anti-mouse-IgG-R-PE (Invitrogen, Carlsbad, CA) in a 4:2:1 molar ratio and allowed to mix for 15 min. A 1:5 dilution was made to achieve two 100 µL samples at 500 and 50 µg/mL. Samples were allowed to incubate in a humidified chamber at room temperature for 2 h, followed by a wash by dipping successively in PBS-Tween, PBS and water. The washed slide was then scanned using a confocal slide scanner (ProScan Array, Perkin Elmer, Waltham, MA). Images were analyzed using Imagene (v6.0, Biodiscovery), and the averaged mean signal minus background values for four of the six replicate spots were calculated.
Molecular modeling
Models of the Neu5Acα2-3-Galβ1-4-GlcNAc trisaccharide and the LewisX antigen were built using the Glycam server (http://glycam.ccrc.uga.edu). All modeling were performed in Molecular Operating Environment (MOE; MOE 2009.10, Chemical Computing Group). Sulfates were added at the 6-position of the galactose yielding glycans 5 and 6, respectively. The docking was achieved by rigid body alignment of these models with the cocrystalized α2-3-sialyl-lactose using 200 iterations under standard conditions (Labute et al. 2001). To probe the protein for favorable contact with the modeled glycan ligands, the MOE internal side chain rotamer library was employed. This was followed by a short energy minimization using the AMBER99 force field using 100 iterations, keeping heavy atoms of the protein and the carbohydrate ring atoms under restrained movement.
Supplementary data
Supplementary data for this article is available online at http:// glycob.oxfordjournals.org/.
Funding
This work was supported by the National Institutes of Health (P01AI058113) and CDC 200-2009-32562.
Supplementary Material
Acknowledgements
C.R. thanks EMBO for a long-term fellowship. We thank Ian Wilson for letting us use of the MOE software; the Consortium for Functional Glycomics for providing several of the glycans used for the glycan microarray (U54GM62116).
Abbreviations
CAR, coxsackievirus and adenovirus receptor; CAV-2, canine adenovirus 2; Fuc, fucose; Gal, galactose; GlcNAc, N-acetyl glucosamine; LEW, Lysis-Equilibration-Wash; MOE, Molecular Operating Environment; Neu5Ac, 5-N-acetyl neuraminic acid; PDB, Protein Data Bank; RMSD, root mean square deviation; Siglec, Sialic-acid-binding immunoglobulin-like lectin.
References
- Angata T, Varki A. Chemical diversity in the sialic acids and related alpha-keto acids: An evolutionary perspective. Chem Rev. 2002;102:439–469. doi: 10.1021/cr000407m. doi:10.1021/cr000407m. [DOI] [PubMed] [Google Scholar]
- Attrill H, Imamura A, Sharma RS, Kiso M, Crocker PR, van Aalten DM. Siglec-7 undergoes a major conformational change when complexed with the alpha(2,8)-disialylganglioside GT1b. J Biol Chem. 2006;281:32774–32783. doi: 10.1074/jbc.M601714200. doi:10.1074/jbc.M601714200. [DOI] [PubMed] [Google Scholar]
- Attrill H, Takazawa H, Witt S, Kelm S, Isecke R, Brossmer R, Ando T, Ishida H, Kiso M, Crocker PR, et al. The structure of siglec-7 in complex with sialosides: Leads for rational structure-based inhibitor design. Biochem J. 2006;397:271–278. doi: 10.1042/BJ20060103. doi:10.1042/BJ20060103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. doi:10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blixt O, Head S, Mondala T, Scanlan C, Huflejt ME, Alvarez R, Bryan MC, Fazio F, Calarese D, Stevens J, et al. Printed covalent glycan array for ligand profiling of diverse glycan binding proteins. Proc Natl Acad Sci USA. 2004;101:17033–17038. doi: 10.1073/pnas.0407902101. doi:10.1073/pnas.0407902101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochner BS, Alvarez RA, Mehta P, Bovin NV, Blixt O, White JR, Schnaar RL. Glycan array screening reveals a candidate ligand for Siglec-8. J Biol Chem. 2005;280:4307–4312. doi: 10.1074/jbc.M412378200. doi:10.1074/jbc.M412378200. [DOI] [PubMed] [Google Scholar]
- Cock PJ, Antao T, Chang JT, Chapman BA, Cox CJ, Dalke A, Friedberg I, Hamelryck T, Kauff F, Wilczynski B, et al. Biopython: Freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics. 2009;25:1422–1423. doi: 10.1093/bioinformatics/btp163. doi:10.1093/bioinformatics/btp163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crocker PR, Paulson JC, Varki A. Siglecs and their roles in the immune system. Nat Rev Immunol. 2007;7:255–266. doi: 10.1038/nri2056. doi:10.1038/nri2056. [DOI] [PubMed] [Google Scholar]
- DeMarco ML, Woods RJ. Structural glycobiology: A game of snakes and ladders. Glycobiology. 2008;18:426–440. doi: 10.1093/glycob/cwn026. doi:10.1093/glycob/cwn026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evers A, Gohlke H, Klebe G. Ligand-supported homology modelling of protein binding-sites using knowledge-based potentials. J Mol Biol. 2003;334:327–345. doi: 10.1016/j.jmb.2003.09.032. doi:10.1016/j.jmb.2003.09.032. [DOI] [PubMed] [Google Scholar]
- Fukuta M, Inazawa J, Torii T, Tsuzuki K, Shimada E, Habuchi O. Molecular cloning and characterization of human keratan sulfate Gal-6-sulfotransferase. J Biol Chem. 1997;272:32321–32328. doi: 10.1074/jbc.272.51.32321. doi:10.1074/jbc.272.51.32321. [DOI] [PubMed] [Google Scholar]
- Kumlin U, Olofsson S, Dimock K, Arnberg N. Sialic acid tissue distribution and influenza virus tropism. Influenza Other Respi Viruses. 2008;2:147–154. doi: 10.1111/j.1750-2659.2008.00051.x. doi:10.1111/j.1750-2659.2008.00051.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labute P, Williams C, Feher M, Sourial E, Schmidt JM. Flexible alignment of small molecules. J Med Chem. 2001;44:1483–1490. doi: 10.1021/jm0002634. doi:10.1021/jm0002634. [DOI] [PubMed] [Google Scholar]
- Lutteke T, Bohne-Lang A, Loss A, Goetz T, Frank M, von der Lieth CW. GLYCOSCIENCES.de: An Internet portal to support glycomics and glycobiology research. Glycobiology. 2006;16:71R–81R. doi: 10.1093/glycob/cwj049. doi:10.1093/glycob/cwj049. [DOI] [PubMed] [Google Scholar]
- Lutteke T, von der Lieth CW. Data mining the PDB for glyco-related data. Methods Mol Biol. 2009;534:293–310. doi: 10.1007/978-1-59745-022-5_21. doi:10.1007/978-1-59745-022-5_21. [DOI] [PubMed] [Google Scholar]
- Nycholat CM, McBride R, Ekiert DC, Xu R, Rangarajan J, Peng W, Razi N, Gilbert M, Wakarchuk WW, Wilson IA, et al. Recognition of sialylated poly-LacNAc on N- and O-linked glycans by human and avian influenza A virus hemagglutinins. Angew Chem Int Ed Engl. 2012;51:4860–4863. doi: 10.1002/anie.201200596. doi:10.1002/anie.201200596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Reilly MK, Paulson JC. Siglecs as targets for therapy in immune-cell-mediated disease. Trends Pharmacol Sci. 2009;30:240–248. doi: 10.1016/j.tips.2009.02.005. doi:10.1016/j.tips.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rost B. Twilight zone of protein sequence alignments. Protein Eng. 1999;12:85–94. doi: 10.1093/protein/12.2.85. doi:10.1093/protein/12.2.85. [DOI] [PubMed] [Google Scholar]
- Sawada T, Hashimoto T, Nakano H, Shigematsu M, Ishida H, Kiso M. Conformational study of Œ±-N-acetyl-D-neuraminic acid by density functional theory. J Carbohyd Chem. 2006;25:387–405. doi:10.1080/07328300600778801. [Google Scholar]
- Seiradake E, Henaff D, Wodrich H, Billet O, Perreau M, Hippert C, Mennechet F, Schoehn G, Lortat-Jacob H, Dreja H, et al. The cell adhesion molecule “CAR” and sialic acid on human erythrocytes influence adenovirus in vivo biodistribution. PLoS Pathog. 2009;5:e1000277. doi: 10.1371/journal.ppat.1000277. doi:10.1371/journal.ppat.1000277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebert HC, Reuter G, Schauer R, von der Lieth CW, Dabrowski J. Solution conformations of GM3 gangliosides containing different sialic acid residues as revealed by NOE-based distance mapping, molecular mechanics, and molecular dynamics calculations. Biochemistry. 1992;31:6962–6971. doi: 10.1021/bi00145a014. doi:10.1021/bi00145a014. [DOI] [PubMed] [Google Scholar]
- Tateno H, Crocker PR, Paulson JC. Mouse Siglec-F and human Siglec-8 are functionally convergent paralogs that are selectively expressed on eosinophils and recognize 6′-sulfo-sialyl Lewis X as a preferred glycan ligand. Glycobiology. 2005;15:1125–1135. doi: 10.1093/glycob/cwi097. doi:10.1093/glycob/cwi097. [DOI] [PubMed] [Google Scholar]
- Tateno H, Ohnishi K, Yabe R, Hayatsu N, Sato T, Takeya M, Narimatsu H, Hirabayashi J. Dual specificity of Langerin to sulfated and mannosylated glycans via a single C-type carbohydrate recognition domain. J Biol Chem. 2010;285:6390–6400. doi: 10.1074/jbc.M109.041863. doi:10.1074/jbc.M109.041863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varki A. Essentials of Glycobiology. 2nd ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2009. [PubMed] [Google Scholar]
- Xu R, McBride R, Nycholat CM, Paulson JC, Wilson IA. Structural characterization of the hemagglutinin receptor specificity from the 2009 H1N1 influenza pandemic. J Virol. 2012;86:982–990. doi: 10.1128/JVI.06322-11. doi:10.1128/JVI.06322-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y, Godzik A. Flexible structure alignment by chaining aligned fragment pairs allowing twists. Bioinformatics. 2003;19(Suppl 2):ii246–ii255. doi: 10.1093/bioinformatics/btg1086. doi:10.1093/bioinformatics/btg1086. [DOI] [PubMed] [Google Scholar]
- Zaccai NR, May AP, Robinson RC, Burtnick LD, Crocker PR, Brossmer R, Kelm S, Jones EY. Crystallographic and in silico analysis of the sialoside-binding characteristics of the Siglec sialoadhesin. J Mol Biol. 2007;365:1469–1479. doi: 10.1016/j.jmb.2006.10.084. doi:10.1016/j.jmb.2006.10.084. [DOI] [PubMed] [Google Scholar]
- Zhuravleva MA, Trandem K, Sun PD. Structural implications of Siglec-5-mediated sialoglycan recognition. J Mol Biol. 2008;375:437–447. doi: 10.1016/j.jmb.2007.10.009. doi:10.1016/j.jmb.2007.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.