Abstract.
We describe a nonscanning, fiber bundle endomicroscope that performs optically sectioned fluorescence imaging with fast frame rates and real-time processing. Our sectioning technique is based on HiLo imaging, wherein two widefield images are acquired under uniform and structured illumination and numerically processed to reject out-of-focus background. This work is an improvement upon an earlier demonstration of widefield optical sectioning through a flexible fiber bundle. The improved device features lateral and axial resolutions of 2.6 and 17 μm, respectively, a net frame rate of 9.5 Hz obtained by real-time image processing with a graphics processing unit (GPU) and significantly reduced motion artifacts obtained by the use of a double-shutter camera. We demonstrate the performance of our system with optically sectioned images and videos of a fluorescently labeled chorioallantoic membrane (CAM) in the developing . gallus embryo. HiLo endomicroscopy is a candidate technique for low-cost, high-speed clinical optical biopsies.
Keywords: endomicroscopy, fluorescence, optical sectioning, fiber-bundle imaging, double-shutter camera, graphics processing unit, chorioallantoic membrane
1. Introduction
Optical sectioning endomicroscopy has been gaining interest in the medical imaging community. Current techniques to obtain optical sectioning can be broadly separated into scanning and nonscanning. Scanning techniques can be implemented by raster scanning a laser beam across the proximal face of a flexible fiber bundle,1,2 by distal scanning obtained by vibrating the end of a single optical fiber,3,4 or by distal scanning with a microelectromechanical (MEMS) device.5,6 These techniques are limited by the requirement of fast, precise scanning mechanisms. The serial acquisition of pixels makes these techniques susceptible to streaking artifacts in the presence of sample or probe movement.
Nonscanning widefield techniques have been developed that do not rely on fast scanning systems. For example, a miniaturized plane illumination endomicroscope was demonstrated that limits excitation to a thin sheet in the objective focal plane.7 While this technique provides optical sectioning, it requires the insertion of a microprism into the tissue to generate the light sheet. Another widefield optical sectioning technique is structured illumination microscopy,8 where an illumination grid pattern is projected into the sample, and at least three images are acquired while laterally phase-stepping the grid. An image-processing routine then synthesizes an optically sectioned result. This technique has been implemented with a rigid Hopkins-type endoscope,9 as well as through a flexible fiber bundle.10 However, structured illumination microscopy is susceptible to artifacts due to imprecise grid translation or sample movement. Recently, we introduced a technique called HiLo imaging, which uses alternating uniform and structured illumination to synthesize an optically sectioned image.11 HiLo has reduced mechanical complexity compared with scanning techniques and is insensitive to imperfections in the illumination structure. We previously demonstrated an endomicroscope system that performed HiLo imaging through a flexible fiber bundle,12 but was limited by a low frame rate, fiber core artifacts, and the need for extensive post-processing. Here we present an improved HiLo endomicroscope system with reduced core and motion artifacts and with near video-rate acquisition capability with real-time image processing.
2. Methods
2.1. Setup
Our HiLo endomicroscope setup is illustrated in Fig. 1. A single-axis galvanometer mirror (Thorlabs GVS001) directs a laser beam (Omicron PhoxX 488, 80 mW) between two illumination paths. In the uniform path the beam travels through a expanding telescope and polarizing beam splitter and is focused onto the back aperture of a objective (Olympus Plan Achromat, 0.4 NA). This uniformly illuminates the proximal face of a flexible imaging fiber bundle (600 μm active dia; 30,000 cores). The distal face of the fiber bundle is equipped with a water-immersion micro-objective (Mauna Kea Technologies; 0.8 NA) to give additional magnification and working distance of 60 μm. The fiber bundle is 3 meters long with the distal tip 2.7 mm in diameter and 14 mm in length. The generated fluorescence from the sample is relayed through the fiber bundle, spectrally separated with a dichromatic mirror (Semrock FF506-Di03-25x36) and emission filter (Semrock FF01-536/40-25) and recorded with a CCD camera (PCO Pixelfly USB; binning ; double-shutter mode). The structured path is identical to the first with the addition of a transmission Ronchi ruling (Edmund Optics; ) in a plane conjugate to the objective and micro-objective focal planes. The image of the Ronchi ruling is projected into the sample, modulating the sample fluorescence with a high-contrast grid pattern.
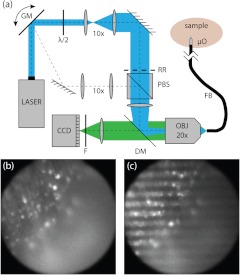
Fig. 1.
(a) HiLo endomicroscope setup. A galvanometer mirror (GM) directs a laser beam between two paths, one delivering uniform illumination and the other projecting the image of a Ronchi ruling (RR) into the sample. The paths are combined with a polarizing beam splitter (PBS). A flexible imaging fiber bundle (FB) is equipped with a micro-objective () for additional magnification and working distance. Generated fluorescence is spectrally separated by a dichromatic mirror (DM) and emission filter (), and images are recorded with a digital camera (CCD). (b) Representative uniform and (c) structured images.
2.2. Image Preprocessing
Several difficulties are associated with the use of a fiber bundle for fluorescence imaging. The core fill factor of our fiber bundle is only about 30%, meaning our raw images are sparsely sampled. This sampling is irregular and imparts a quasi-hexagonal honeycomb pattern to the raw images with contrast typically much larger than the sample contrast of interest. Moreover, the fiber cores are heterogeneous in size and thus transmission efficiency. Finally, the fiber core material is autofluorescent, which introduces an extraneous background in our raw images. These problems were found to significantly undermine the quality of our final HiLo images and needed to be addressed.
Several groups have suppressed the appearance of fiber cores by applying Gaussian blurring filters,2 disk filters,13 or spatial frequency domain filters.14–17 Glass fiber bundles have perfectly periodic hexagonal sampling that concentrates the core pattern in a few localized regions in spatial frequency space, making frequency domain filtering an attractive option. Quartz fiber bundles like those used in this work are distinctly aperiodic and are less amenable to spectral domain techniques. Histogram equalization can be used in conjunction with simple spatial filtering to enhance image contrast for visualization purposes,18 but the resulting images are no longer proportional to fluorescence concentration (i.e. are no longer quantitative). The main drawback of simple blurring is loss of resolution and image contrast since relatively wide kernels are required to sufficiently attenuate the core pattern. Also, due to core sampling sparsity, many of the camera pixels do not measure intensity from the sample, but rather from autofluorescence or extraneous reflections. A method proposed by Perchant and Le Goualher involves finding all 30,000 core centroids with a segmentation process, measuring the maximum intensity from each core, then interpolating the scattered data on a uniformly sampled rectilinear grid.1,19,20 This process is most comprehensive, but is computationally intensive and difficult to achieve in real time.
We introduce here a simple preprocessing routine that corrects for sparse sampling, core heterogeneity, and autofluorescence. This routine is based on an iterative segmentation-interpolation algorithm that removes the appearance of the cores without sacrificing spatial resolution or sample contrast. We begin our iterative procedure with the raw camera image itself, , which serves as the initial estimate of a core-suppressed image . For each iteration, a binary core mask, , is generated to distinguish core pixels from cladding pixels. This is done by setting a local, spatially varying threshold level determined by low-pass filtering the current estimate. Camera pixels above this threshold level are identified as core pixels because the cores are almost universally brighter than the cladding. Similarly, the cladding pixels are identified by the complementary mask . To obtain an improved estimate, , the cladding pixels are then assigned the low-pass filtered values while the core pixels retain their original values. The full width at half maximum (FWHM) of the Gaussian low-pass filter kernel is approximately one inter-core distance. This simple segmentation-interpolation procedure is iterated until a desired smoothness is achieved; typically two to five iterations suffice. We emphasize here that the low-pass filter used in this procedure is highly local and averages only over nearest neighbor cores. The core pixels thus retain their original intensity throughout the process, while the cladding pixels are filled in to approach the average of their nearest neighbor cores. The algorithm features the low complexity and fast execution time of Gaussian blurring, and minimal loss of sample resolution and contrast. However, it should not be regarded as simple filtering since the procedure is neither linear nor spatially invariant. The algorithm is summarized below.
Algorithm
for to do
end for
The next steps in our preprocessing routine involve subtracting the contributions from autofluorescence and correcting for heterogeneities in fiber core efficiency. We follow a modified procedure laid out by Perchant and Zhong.20,21 Three calibration images are acquired before imaging begins. The first, , is acquired with the laser turned off while imaging a nonfluorescent blank solution, such as water, in a dark compartment. This provides a measurement of camera bias and diffuse room light that passes the emission filter. The second, , is acquired under the same conditions but with the laser turned on, providing an estimate of the autofluorescence of the fiber core and cladding material. Since the autofluorescence is a function of laser power while the bias and room light are not, the two images need to be acquired separately. The third image, , is acquired while imaging a homogeneously labeled fluorescent fluid. When camera bias, room light, and autofluorescence signals are subtracted out, this third image provides a mapping of the core transmission and collection efficiencies. In practice, each calibration image is derived from averaging many () raw images to improve signal-to-noise ratio. Calibration images are reacquired each time the fiber bundle probe is connected and aligned to the system.
Image preprocessing can be summarized as a two-step process. First, raw images are subject to iterative segmentation-interpolation to fill in the cladding pixels, yielding . Background subtraction and normalization then yield the preprocessed image
| (1) |
where and are the illumination powers used during the acquisition of and , respectively. Note that because the reference images containing laser-induced and laser-independent backgrounds are acquired separately, the former can be scaled to the laser power in use. Our preprocessing is thus robust to changes in illumination power. In particular, this allows us to dynamically adjust the laser power to properly fill the camera dynamic range. After preprocessing the raw structured- and uniform-illumination images, these are then ready for HiLo processing.
2.3. HiLo Principle
The benefit of HiLo over standard widefield imaging is that it provides out-of-focus background rejection. As described in previous work, the uniform and structured images can be roughly decomposed into in-focus and out-of-focus components.
| (2) |
| (3) |
where is the lateral position vector, is the grid spatial frequency and is the modulation depth (note: for ease of presentation, we have used a sine function to model the grid function). The imaged in-focus signal appears modulated by the grid pattern, while the out-of-focus signal does not. It follows that a local evaluation of the grid contrast scales with the in-focus signal. Here we use a rectified subtraction of normalized images to compute the local contrast,
| (4) |
where denotes a local average evaluated by Gaussian low-pass filtering with frequency cutoff much lower than . This not only ensures the modulated component is locally centered about zero, but also makes the evaluation insensitive to differences in global illumination profile. Since the contrast is normalized and thus scales as the proportion of in-focus signal, it is used as a weighting function to select the in-focus portion of . This demodulation technique is reliable for low spatial frequencies below . An accurate measure of in-focus high spatial frequencies above is found by applying a complementary high-pass filter to directly. The weighting function is not needed for these high spatial frequencies because the frequencies are already inherently axially resolved.22 The final HiLo image is the addition of the high and low spatial frequency images. A scaling factor, , is used to level the intensities and ensure a seamless fusion. More detailed descriptions of the algorithm, including how we calculate and deal with noise-induced bias in the contrast measurement, are found in Refs. 11, 23, 24.
| (5) |
| (6) |
| (7) |
The final HiLo image is axially resolved across the entire spatial frequency spectrum, from dc to the bandwidth of our imaging optics. The lateral resolution, , is limited by the sampling of the fiber cores and is given by , where is the average fiber inter-core separation, and is the micro-objective magnification. The axial resolution of the low-frequency component, , can be approximated by considering a thin fluorescent plane and calculating the decay of the grid contrast as a function of defocus. The contrast of the grid pattern is attenuated by both the illumination and detection optical transfer functions (OTFs), and the axial resolution is defined as the FWHM of this function. Using the Stokseth approximation25 for the 3D OTF, assuming similar excitation and detection wavelengths, and assuming is much smaller than the imaging bandwidth, the axial resolution is given by . The transmission of low frequencies is found to decay as for large defocus . In comparison, the axial resolution of the high-frequency component at the filter cutoff frequency is given by . This is slightly broader due to the action of only the detection OTF providing axial confinement and is found to decay with defocus as for large . A matching of the axial resolution at the transition between high- and low-frequency components (i.e. at ) involves selecting the cutoff frequency appropriately. We have found that choosing to be slightly lower than provides a reasonably accurate match while still maintaining a reduced artifact in the low channel found at twice the grid frequency. The lateral and axial resolutions of the current system are approximately (governed by the Nyquist sampling criterion associated with the fiber cores) and (governed by the structured illumination grid period).
2.4. Fast Image Processing with a GPU
Our image processing routines were developed and tested in MATLAB (MathWorks). The processing speed was found to be unacceptably slow for real-time processing (). To enable real-time processing and display, a library of image management and manipulation functions (memory allocation and deallocation, arithmetic operations, filtering in Fourier space) were written in CUDA-C and compiled to a graphics processing unit (GPU) target (NVIDIA GeForce GTX 280). The library was compiled as a dynamic linked library for maximum utility. A LabVIEW interface (National Instruments) was created to coordinate communication among the camera, CPU memory, and GPU memory. The multicore processor architecture (Intel i7 quad core) and inherent parallelism of LabVIEW greatly improved the speed of the endomicroscope. The entire processing pipeline—preprocessing, HiLo processing, and memory transfers between the CPU and GPU—occurs in 30 ms for an image resolution . The endomicroscope is thus capable of real-time image processing at speeds up to .
3. Results
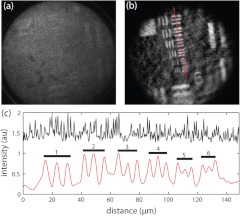
Figure 2 shows a 1951 USAF resolution target (Edmund Optics) before and after preprocessing. The weak contrast of the resolution elements is overwhelmed by autofluorescence background and the high contrast of the cores. After preprocessing, the resolution elements become clearly visible. A line profile through the preprocessed image shows clear contrast of elements in group 7, including the smallest features (element 6, ) spanning approximately two fiber cores. This result demonstrates that image resolution and contrast are not significantly degraded by our iterative segmentation-interpolation algorithm.
Fig. 2.
USAF 1951 resolution target. (a) Raw and (b) after preprocessing. The weak sample contrast is initially overwhelmed by fiber core contrast, efficiency heterogeneity, and autofluorescence background. Preprocessing improves sample contrast. (c) Line profile through raw (top trace) and preprocessed (bottom trace) images. The preprocessed image reveals the resolution elements in group 7 with discernible contrast (elements 1–6 identified with bars). The fundamental spatial frequency of the smallest feature (element 6, ) is 55% of the Nyquist limit imposed by the average fiber core separation.
To demonstrate the suitability of our HiLo endomicroscope for in vivo imaging we utilized a chick embryo, which is commonly studied in developmental biology research.26 In particular, the chorioallantoic membrane (CAM) was examined as it is a model for angiogenesis and lymphangiogenesis27–29 and is readily accessed by producing a small hole in the egg shell. Several videos were acquired to showcase the image resolution, background-rejection capacity and speed of HiLo imaging (9.5 Hz).
3.1. Chick Embryo Preparation
Fertilized eggs of the Silkie chick (Gallus gallus) were obtained from a local farm (Golden Egg Farm; Hardwick, MA) and incubated at 38°C and 80% humidity until approximately embryonated day E5. Imaging of the embryo was performed through a small hole in the egg shell. A fluorescent solution (50 mM fluorescein in ) was injected into the amnion and allowed to diffuse for approximately 1 hr before imaging. For some samples, the solution was injected intravenously whereupon it bound to serum albumin and diffused through the embryo. After imaging the embryos were euthanized by hypothermia by storing the eggs at .
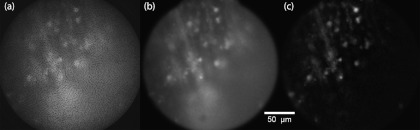
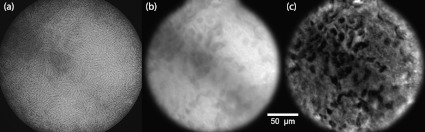
A video of a thin fluorescently-labeled membrane floating through focus provides a demonstration of our sectioning capacity. The raw uniform image directly acquired by the camera [Fig. 3(a)] is included to illustrate the deleterious effects of the fiber cores. The preprocessed uniform image [Fig. 3(b)] shows a marked improvement in sample contrast and features small in-focus structures with full resolution. However, this image also features considerable out-of-focus background haze. The final HiLo image [Fig. 3(c)] preserves the in-focus structures while rejecting this out-of-focus background haze.
Fig. 3.
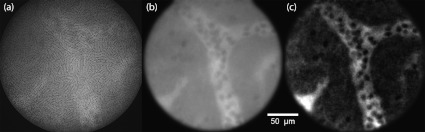
Figure 4 shows blood vessels and other features of the CAM membrane after intravenous injection of fluorescein. Figure 5 shows a torrent of red blood cells (RBCs) passing through a CAM sinus and later collecting into a venule. The RBCs do not take up the fluorescein stain and appear as shadows on a bright background. These videos illustrate the capacity of our HiLo endomicroscope to capture fast dynamics.
Fig. 4.
Fig. 5.
3.2. Imaging Speed and Motion Artifacts
As observed in Fig. 5, our HiLo endomicroscope is largely immune to artifacts arising from sample motion (e.g. respiration, blood flow) or probe motion. Part of the reason for this immunity comes from the fact that all the high-frequency information is derived from a single image (). On the other hand, the low-frequency information is more susceptible to motion artifacts since it is derived from a comparison of two serially acquired images. While the consequence of intra-frame motion during the camera integration time is loss of spatial resolution in the raw images, a much more pronounced motion artifact arises from inter-frame motion when significant translation occurs between the two raw image exposures. The magnitude of this artifact can be roughly estimated by considering two serially acquired images of a sharp edge, both under uniform illumination. In the absence of sample motion, both images are the same, and we have [see Eq. (4)]. If instead the sample translates laterally between images, an artifactual signal occurs in the neighborhood of the edge that appears to be in focus. The trapezoidal profile of this signal can be approximated as a box-top function of unity height and width , where is the translation velocity (assumed constant), is the exposure time and is the inter-frame delay. The low-pass filtering that occurs to generate serves to spatially distribute this artifact, reducing the maximum error at a given pixel. For small , the error bound is , where is the low-pass filter cutoff frequency. Under typical imaging conditions and the error is governed by the camera readout time.
To minimize , we made use of a double-shutter camera (PCO Pixelfly USB). Such a camera acquires images pairwise with an inter-frame delay of only , followed by a paired readout. While the net frame rate for image pairs (i.e. the HiLo frame rate) is the same with a double-shutter camera as with a traditional camera, the key benefit is the reduction in , which greatly suppresses . We note that in our case was limited to a minimum by the need to switch illumination paths with the galvanometer mirror. In principle, two dedicated, electronically shuttered illumination sources could be used to overcome this limitation, though with diminishing returns when . In practice, motion artifacts are minimal when displacements are kept below .
To illustrate the improved immunity to motion artifacts obtained with a double-shutter camera, we compare HiLo videos acquired with double-shutter versus traditional timing (see Fig. 6). The videos share the same raw data actually acquired in double-shutter mode, and traditional camera timing was simulated here by staggering the raw data (i.e. using uniform and structured raw images from neighboring frame pairs instead of the same frame pair). The effective inter-frame delay for the traditional camera [Fig. 6(a)] was 106 ms, while the double-shutter camera [Fig. 6(b)] was 1 ms. Motion artifacts arising from the misregistration of raw images are easily identified as bright low-frequency blobs in the neighborhood of moving RBCs. The reduced inter-frame delay from the double-shutter timing manifestly attenuates these artifacts and provides crisper imaging. This demonstration is somewhat exaggerated as scientific camera readout times are typically faster than 100 ms. Nevertheless, it serves as an illustration of motion artifact reduction.
Fig. 6.
4. Summary
We have demonstrated some of the performance properties of our improved HiLo endomicroscope. Our key improvements are (1) the introduction of image preprocessing to mitigate the effects of fiber core sparsity, heterogeneity and autofluorescence background; (2) faster speed achieved by the operation of our preprocessing and HiLo algorithms on a GPU, enabling us to attain a net frame rate of 9.5 Hz in real time; and (3) significantly enhanced immunity to motion artifacts with the use of a double-shutter camera. Our goal with these improvements is to further lay the groundwork in establishing HiLo endomicroscopy as a viable technique for clinical applications.
HiLo endomicroscopy is not without its limitations. The main limitation comes from the fact that we use structured illumination to identify out-of-focus background. The contrast imparted by this structured illumination must be large enough to be visible. That is, it must be greater than noise-induced contrast—in particular shot-noise-induced contrast introduced by the background itself. When the background is large, this can impose limitations on the maximum grid frequency, which, in turn, limits background rejection capacity and axial resolution. Nevertheless, as we have shown here, HiLo endomicroscopy provides significant background rejection even when imaging thick in vivo samples. This, along with its remarkable simplicity, speed, and robustness, make HiLo endomicroscopy an attractive imaging technique.
Finally, to further advertise HiLo imaging in general, we have made available our HiLo algorithm as an ImageJ plug-in. This may be found on our website http://biomicroscopy.bu.edu.
Acknowledgments
We thank Kengyeh Chu and Nenad Bozinovic for help in the initial development of HiLo imaging. This work was supported by the NIH (R01-EB010059).
References
- 1.Le Goualher G., et al. , “Toward optical biopsies with an integrated fibered confocal fluorescence microscope,” Medical Image Computing and Computer-Assisted Intervention, Saint-Malo, France: (2004). [Google Scholar]
- 2.Göbel W., et al. , “Miniaturized two-photon microscope based on a flexible coherent fiber bundle and a gradient-index lens objective,” Opt. Lett. 29(21), 2521–3 (2004). 10.1364/OL.29.002521 [DOI] [PubMed] [Google Scholar]
- 3.Polglase A. L., et al. , “A fluorescence confocal endomicroscope for in vivo microscopy of the upper- and the lower-gi tract,” Gastrointest. Endosc. 62(5), 686–695 (2005). 10.1016/j.gie.2005.05.021 [DOI] [PubMed] [Google Scholar]
- 4.Engelbrecht C. J., et al. , “Ultra-compact fiber-optic two-photon microscope for functional fluorescence imaging in vivo,” Opt. Exp. 16(8), 5556–5564 (2008). 10.1364/OE.16.005556 [DOI] [PubMed] [Google Scholar]
- 5.Ra H., et al. , “Two-dimensional MEMS scanner for dual-axes confocal microscopy,” J. Microelectromech. Syst. 16(4), 969–976 (2007). 10.1109/JMEMS.2007.892900 [DOI] [Google Scholar]
- 6.Fu L., et al. , “Nonlinear optical endoscopy based on a double-clad photonic crystal fiber and a MEMS mirror,” Opt. Exp. 14(3), 1027–32 (2006). 10.1364/OE.14.001027 [DOI] [PubMed] [Google Scholar]
- 7.Engelbrecht C. J., Voigt F., Helmchen F., “Miniaturized selective plane illumination microscopy for high-contrast in vivo fluorescence imaging,” Opt. Lett. 35(9), 1413–5 (2010). 10.1364/OL.35.001413 [DOI] [PubMed] [Google Scholar]
- 8.Neil M. A. A., Juškaitis R., Wilson T., “Method of obtaining optical sectioning by using structured light in a conventional microscope,” Opt. Lett. 22(24), 1905–1907 (1997). 10.1364/OL.22.001905 [DOI] [PubMed] [Google Scholar]
- 9.Karadaglić D., Juškaitis R., Wilson T., “Confocal endoscopy via structured illumination,” Scanning 24(6), 301–304 (2002). 10.1002/sca.4950240604 [DOI] [PubMed] [Google Scholar]
- 10.Bozinovic N., et al. , “Fluorescence endomicroscopy with structured illumination,” Opt. Exp. 16(11), 8016–8025 (2008). 10.1364/OE.16.008016 [DOI] [PubMed] [Google Scholar]
- 11.Lim D., et al. , “Optically sectioned in vivo imaging with speckle illumination hilo microscopy,” J. Biomed. Opt. 16(1), 016014 (2011). 10.1117/1.3528656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Santos S., et al. , “Optically sectioned fluorescence endomicroscopy with hybrid-illumination imaging through a flexible fiber bundle,” J. Biomed. Opt. 14(3), 030502 (2009). 10.1117/1.3130266 [DOI] [PubMed] [Google Scholar]
- 13.Oh W. Y., et al. , “Spectrally modulated full-field optical coherence microscopy for ultrahigh-resolution endoscopic imaging,” Opt. Exp. 14(19), 8675–84 (2006). 10.1364/OE.14.008675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dickens M. M., et al. , “Method for depixelating micro-endoscopic images,” Opt. Eng. 38(11), 1836–1842 (1999). 10.1117/1.602235 [DOI] [Google Scholar]
- 15.Alaruri S., et al. , “An endoscopic imaging system for turbine engine pressure sensitive paint measurements,” Opt. Laser Eng. 36(3), 277–287 (2001). 10.1016/S0143-8166(01)00049-5 [DOI] [Google Scholar]
- 16.Suter M., et al. , “Bronchoscopic imaging of pulmonary mucosal vasculature responses to inflammatory mediators,” J. Biomed. Opt. 10(3), 034013 (2005). 10.1117/1.1924714 [DOI] [PubMed] [Google Scholar]
- 17.Winter C., et al. , “Automatic adaptive enhancement for images obtained with fiberscopic endoscopes,” IEEE Trans. Biomed. Eng. 53(10), 2035–2046 (2006). 10.1109/TBME.2006.877110 [DOI] [PubMed] [Google Scholar]
- 18.Han J.-H., Lee J., Kang J. U., “Pixelation effect removal from fiber bundle probe based optical coherence tomography imaging,” Opt. Exp. 18(7), 7427–7439 (2010). 10.1364/OE.18.007427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vercauteren T., “Image registration and mosaicing for dynamic in vivo fibered confocal microscopy,” PhD Thesis, Mines, Paris (2008). [Google Scholar]
- 20.Perchant A., Le Goualher G., Berier F., “Method for processing an image acquired through a guide consisting of a plurality of optical fibers,” US Patent US2005207668 (2004).
- 21.Zhong W., et al. , “In vivo high-resolution fluorescence microendoscopy for ovarian cancer detection and treatment monitoring,” Br. J. Canc. 101(12), 2015–2022 (2009). 10.1038/sj.bjc.6605436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mertz J., Introduction to Optical Microscopy, Roberts & Company; (2010). [Google Scholar]
- 23.Lim D., Chu K. K., Mertz J., “Wide-field fluorescence sectioning with hybrid speckle and uniform-illumination microscopy,” Opt. Lett. 33(16), 1819–1821 (2008). 10.1364/OL.33.001819 [DOI] [PubMed] [Google Scholar]
- 24.Mertz J., Kim J., “Scanning light-sheet microscopy in the whole mouse brain with hilo background rejection,” J. Biomed. Opt. 15(1), 016027 (2011). 10.1117/1.3324890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stokseth P. A., “Properties of a defocused optical system,” J. Opt. Soc. Am. 59(10), 1314–1321 (1969). 10.1364/JOSA.59.001314 [DOI] [Google Scholar]
- 26.Gilbert S. F., Developmental biology, 8th edition, Sinauer Associates Inc., (2006). [Google Scholar]
- 27.Chin W. W. L., et al. , “Chlorin e6-polyvinylpyrrolidone as a fluorescent marker for fluorescence diagnosis of human bladder cancer implanted on the chick chorioallantoic membrane model,” Canc. Lett. 245(1–2), 127–133 (2007). 10.1016/j.canlet.2005.12.041 [DOI] [PubMed] [Google Scholar]
- 28.Oh S.-J., et al. , “VEGF and VEGF-c: specific induction of angiogenesis and lymphangiogenesis in the differentiated avian chorioallantoic membrane,” Dev. Bio. 188(1), 96–109 (1997). 10.1006/dbio.1997.8639 [DOI] [PubMed] [Google Scholar]
- 29.Wilting J., Neeff H., Christ B., “Embryonic lymphangiogenesis,” Cell and Tissue Res. 297(1), 1–11 (1999). 10.1007/s004410051328 [DOI] [PubMed] [Google Scholar]