Abstract
Purpose.
To describe an algorithm and software application (APP) for 100% optic nerve axon counting and to compare its performance with a semi-automated manual (SAM) method in optic nerve cross-section images (images) from normal and experimental glaucoma (EG) nonhuman primate (NHP) eyes.
Methods.
ON cross sections from eight EG eyes from eight NHPs, five EG and five normal eyes from five NHPs, and 12 normal eyes from 12 NHPs were imaged at 100×. Calibration (n = 500) and validation (n = 50) image sets ranging from normal to end-stage damage were assembled. Correlation between APP and SAM axon counts was assessed by Deming regression within the calibration set and a compensation formula was generated to account for the subtle, systematic differences. Then, compensated APP counts for each validation image were compared with the mean and 95% confidence interval of five SAM counts of the validation set performed by a single observer.
Results.
Calibration set APP counts linearly correlated to SAM counts (APP = 10.77 + 1.03 [SAM]; R2 = 0.94, P < 0.0001) in normal to end-stage damage images. In the validation set, compensated APP counts fell within the 95% confidence interval of the SAM counts in 42 of the 50 images and were within 12 axons of the confidence intervals in six of the eight remaining images. Uncompensated axon density maps for the normal and EG eyes of a representative NHP were generated.
Conclusions.
An APP for 100% ON axon counts has been calibrated and validated relative to SAM counts in normal and EG NHP eyes.
An algorithm and software application for counting 100% of the axons in nonhuman primate optic nerve cross sections (normal through end-stage experimental glaucoma) is presented and validated against a semi-automated manual method.
Introduction
Glaucoma and other optic neuropathies collectively represent a leading cause of blindness around the world. Thus, a great deal of laboratory research aims to determine their key pathophysiological mechanisms and to develop new treatment strategies. For many of these studies, the most direct and important outcome measure will be the number of axons remaining in the optic nerve. However, counting ON axons can be time consuming and error-prone, the latter especially if sampling techniques are employed.1–12 Study authors published a series of papers3,4 in which orbital ON axon counts were performed in nonhuman primate eyes using a semi-automated manual count method. In this technique, a randomized sample of the total ON area (8% for normal, 20% for experimental glaucoma nerves) was imaged and counted.3,4 This is extremely labor intensive and prone to sampling error. The purpose of the present study is to describe an automated algorithm for counting axons in 100% of the orbital ON cross-sectional area and to validate its results against SAM counts.
Achieving 100% ON axon counts is important for glaucoma research to improve the accuracy of axon loss estimates at any stage of glaucomatous disease and, more importantly, to allow for accurate estimates of regional loss within the orbital ON, thus enhancing the ability of all experimental glaucoma models to test core hypotheses regarding the mechanisms of glaucomatous damage. Furthermore, having a complete axonal map of the orbital ON is essential to fully understand complex structure-function relationships.13–16
Potts et al. reported the first automated axon counts covering 100% of orbital ON cross sections in two normal rhesus monkey and two normal human eyes.17,18 However, the method employed in that report was extremely labor intensive, in part due to the imaging and computer technology available at the time. Since those initial reports, a variety of manual or semi-automated techniques for estimating the total number of orbital ON axons have been employed to characterize the degree of orbital ON axon loss within experimental glaucoma models in a variety of animal species.1,2,7,11,12,19–24 Similar methods have also been used to estimate total axon number and size distributions within normal and glaucomatous human cadaver eyes.9,10,18,25,26 All such methodologies require the acquisition of high magnification images by light or electron microscopy to identify and count normal axons. The cross-sectional area sampled in these methods typically ranges from 3% to 20%. To achieve an estimate for the total orbital ON axon count, the axon densities from the small subset of sampled areas are averaged and extrapolated to the total cross-sectional area of the ON.
The nature and magnitude of the inaccuracies that can result from strategies that sample less than 100% of the orbital ON cross section to estimate the total ON axon count have been evaluated and discussed.4 Nevertheless, sampling methodologies have been employed because, up to now, it has been impractical to count 100% of the ON cross-sectional area. At 100X magnification, the typical number of images required to cover an average NHP or human ON ranges from 3000 to 7000; and a trained human observer is required to manually adjust intensity thresholds and other parameters for every image in order for existing image analysis software to effectively detect and count normal axons. Due to improvements in technology such as automated microscope stages and image acquisition software with auto-exposure and autofocus capabilities, the acquisition of a large number of high-quality images across an entire ON cross section is achievable in a relatively short period of time. The following sections describe the methodological approach and demonstrate the results of the study's automated strategy for counting ON axons.
Methods
Overview of Study Design
First, a calibration study was performed to compare total axon counts by each method within an initial group of 500 images. These images were qualitatively classified by a single experienced observer as normal, mild damage, moderate damage, severe damage, and end-stage damage in order to balance the total sample with 100 images from each damage level. Next, a validation study was performed to compare APP counts with the SAM counts by assessing whether the APP counts fell within the range of repeated SAM counts. For this experiment, SAM counts were repeated five individual times on a separate group of 50 images representing a similar, broad range of damage (10 images per damage group). The APP counts were compared with the mean SAM counts and with the range of SAM recount variability (95% confidence interval). Finally, to demonstrate the overall strategy for 100% ON axon counting, APP axon counts, image composites, and density maps were generated for the total ON area of the normal and experimental glaucoma eye of a single representative NHP.
Optic Nerve Tissues
ON tissues from 17 normal and 13 experimental glaucoma eyes from a total of 25 previously studied NHPs (19 rhesus, six cynomolgus, ranging in age from 4 to 19 years at the time of sacrifice) were included in this study. All animals were treated in accordance with the ARVO Statement for the Use of Animals in Ophthalmic and Vision Research. Before induction of experimental glaucoma in any NHP, its eyes were deemed normal if they passed a complete ocular examination by an experienced observer, which included slit lamp examination, dilated fundoscopy, ON stereo-photographic evaluation, and tonometry. Unilateral chronic IOP elevation was induced following laser damage to the trabecular meshwork leading to glaucomatous damage to the ON head as detected by confocal scanning laser tomography as previously described.27 ONs were either perfusion- or immersion-fixed with either 4% paraformaldehyde or 6% glutaraldehyde (depending upon individual study protocols) at the time of sacrifice.
After enucleation, three longitudinal cuts (one superior and two nasal) were made in the orbital ON, which was then transversely cut 3–6 mm posterior to the sclera (measured along the superior surface of the nerve and cut perpendicular to its longitudinal axis) and stored in either 4% paraformaldehyde or 6% glutaraldehyde for further processing. A 500-μm piece was removed from the cut end using a vibratome (Leica VT 100S; Leica Microsystems Inc., Deerfield, IL) and each ON piece was post-fixed in 4% osmium tetroxide and embedded in epoxy resin. ON cross sections (1-μm thick) were cut using an ultramicrotome (RMC Ultramicrotome MT6000; Boeckeler Instruments Inc., Tucson, AZ), stained with p-phenylenediamine, and mounted on glass slides.4
Image Acquisition of 100% of Each ON Cross Section
Images covering 100% of the ON cross section were automatically captured under oil immersion with a 100× objective (Leica PL Fluotar NA = 1.3) using an inverted light microscope (Leica DM IRB; Leica Microsystems Inc.), imaging software (Bioquant imaging system, NOVA, R&M Biometrics, Inc., Nashville, TN) and a computer-controlled (X-Y-Z) stage (Applied Scientific Instrumentation, Inc., Eugene, OR). Each acquired image was 640 × 480 pixels (W x H) covering an area of 1412 μm2. To ensure full ON cross-sectional coverage, images were captured with a 15% overlap in both x and y directions. This overlap was necessary to overcome the small misalignment between the main axis of the camera and the stage (1–5 μm) over large distance spans.
While the acquisition software has auto-exposure functionality, the upper exposure limit is controlled by a user-selected value that must be optimized for the entire ON so as to minimize the number of under- or overexposed images. Special consideration was also given to finding auto-exposure and illumination settings that enhanced the myelin's contrast with the surrounding neural area, allowing the autofocus mechanism to converge on the myelin's plane rather than on debris or other background noise that might be present within the field of a given image. Each image was saved in Bioquant's BIF format, which contains the image's x, y coordinates relative to a user-selected center.
Selection of Representative Images for the Calibration and Validation Studies
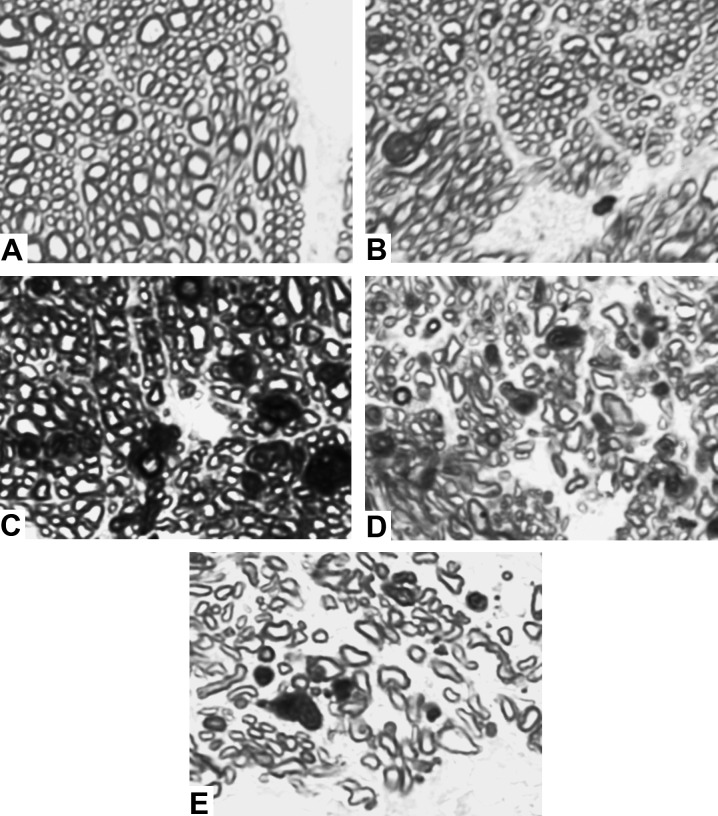
ON images from a total of 17 normal eyes and 13 eyes with experimental glaucoma were reviewed by a trained observer and assigned a damage grade (i.e., normal, mild, moderate, severe, end-stage) based on a qualitative estimate of the number and condition of the axons within each image. Representative images from each group are shown in Figure 1 below. All selected images for the calibration and validation studies were deemed to be of “countable” quality and no attempt was made to select only high-quality images. The images used in the calibration and validation studies were in some instances taken from the same ON cross sections; however, all images used in each study were unique.
Figure 1. .
Representative 100× images by damage grade used in the calibration and validation studies. (A) Normal. (B) Mild. (C) Moderate. (D) Severe. (E) End-stage. See Methods section for further description.
Automated Axon Counting Method and Software Application
The study's current approach to ON axon counting is the result of work that has evolved initially from a method that was optimized for normal ONs28,29 to one that was designed to count axons in damaged ONs.30,31 However, these previous algorithms proved to be too complex or often failed to perform satisfactorily on images with moderate levels of axonal damage. To address these two issues, an APP was developed that, given an image, provides an estimate of the number of “normal” axons (i.e., axons with normal morphological characteristics as outlined below) regardless of the overall axonal damage level present in the image. Other parameters such as axon size and shape were also obtained. The APP is written in Java and currently utilizes the ImageJ32 package for shape analysis and filtering. For details on the algorithm and other software utilized in developing the APP, please refer to Appendix 1.
Semi-Automated Manual Axon Counts
Within the subset of images selected for the calibration and validation studies (see Fig. 1), an experienced operator counted the normal axons present in the images using the Bioquant imaging system.3,4 The steps performed by the operator were similar to the ones done by the APP. First, a hard threshold was applied to the images to produce the binary maps. Depending on the quality of the stain, illumination, or level of damage in the images, the operator may have had to manually “clean” the resulting binary maps by removing areas that were clearly not axons and by closing gaps in valid myelin to recover axons that were otherwise lost.
Once images were cleaned, a shape filter was applied to remove shapes that were clearly not axons. This was, again, an iterative manual task that required direct user intervention in order to determine the proper shape filter parameters and to indicate when all non-axon shapes were successfully removed.
Calibration Study—Determining the Relationship of the APP to SAM Axon Counts within a Representative Group of 500 Images
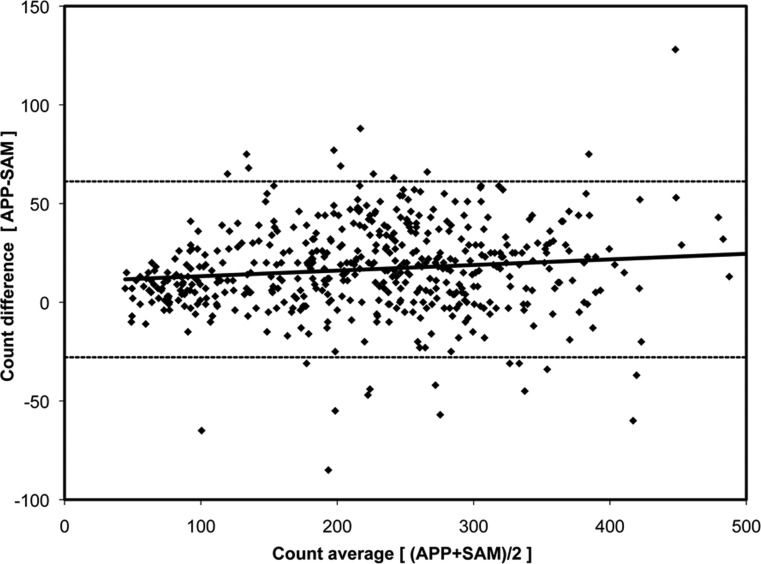
The purpose of this study was to directly compare total axon counts by each method within a group of images that were considered to be normal or that qualitatively demonstrated mild, moderate, severe, or end-stage levels of axonal damage (see Fig. 1). Because of the qualitative nature of the classification, some overlap between damage levels will undoubtedly exist. To account for this overlap and to still provide a large enough sample, 500 images (n = 100 for each damage level) were selected. Output data for each image was the total number of normal axons counted by the APP and SAM. The relation between APP and SAM counts was assessed by Deming regression,33 which assumes equal variances for both methods under the null hypothesis that the two methods are equivalent. A Bland-Altman plot was generated to assess agreement between both methods. A small bias was found (as described in the Results section) and accounted for in the validation study as described in the section that follows.
Validation Study—Comparison of Compensated APP to SAM Counts within a Second Representative Group of n = 50 Images with Multiple (5) SAM Counts
In order to validate the algorithm, compensated APP axon counts were compared with the mean and range of repeatability (95% confidence interval) derived from five separate repeat SAM counts. For this validation study, a separate group of 50 representative images was used. The images in this group were considered to be normal (n = 10) or that qualitatively demonstrated mild (n = 10); moderate (n = 10); severe (n = 10); or end-stage (n = 10) levels of axonal damage. SAM counts were performed on all 50 images by the same operator (GC) on five separate occasions at least 1 week apart. Correlation between the APP and SAM counts was assessed by Deming regression as outlined above. APP counts for each image were then compensated using the equation resulting from the linear regression of the difference versus average axon counts from the calibration set (see Fig. 3). The number of images for which the compensated APP counts fell outside of the 95% confidence interval of the SAM counts was recorded and the potential reasons for disagreement were assessed.
Figure 3. .
Bland-Altman plot for the 500 calibration images. The dashed lines represent the 95% limits of agreement between the APP and SAM (−28 and 61). The solid black line represents the linear regression defined by the equation y = 10.36 + 0.03x; R2 = 0.01.
Automated Whole ON (100%) Normal Axon Counts in Both Eyes of a Single Representative NHP with Unilateral Early Experimental Glaucoma
To demonstrate the utility of the study's automated 100% optic nerve axon counting strategy, ON cross sections from both the normal eye and the experimental glaucoma eye of a single NHP were imaged (as described in the image acquisition section above) yielding a set of 5000+ images per ON. All images were then counted by the APP at an average rate of 4 images/minute. For every counted image, an individual output file was generated as well as an output image highlighting the centroid of all recognized axons. No compensation of the automated counts was performed in these images (see the Discussion section).
A global summary file containing the image coordinates, axon count, average size, and density for all counted images was created for each nerve. The global summary file was used by two final applications. The first application generated a composite of the optic nerve cross sections by placing scaled (1/10) thumbnails of every image in a large canvas at the appropriate x, y coordinates. The second application generated a composite in which each scaled image was colorized according to its axon density value defined to be the axon count divided by the total image area (axons that resided in the overlap areas were included in the density calculation for each image but not in the final counts). The assignment of superior, inferior, nasal, and temporal to each optic nerve cross section was accomplished by matching the superior and nasal cuts within that image to the same three cuts in the optic nerve stump of the digital 3D optic nerve head reconstruction of that eye (as previously described).4
Results
The Table reports the mean ± SD and range of axon counts by each method for the normal, mild, moderate, severe, and end-stage images used in the calibration and validation studies, respectively. These data confirm that the five damage groups represent a broad range of axon numbers and that the range is similar in both studies, though the mild, moderate, and severe damage groups appear to have a slightly lower axon count (i.e., to be more severely damaged), on average, in the validation study set.
Table. .
Normal Axon Counts within the Normal, Mild, Moderate, Severe, and End-Stage Images Used in the Calibration and Validation Studies by Counting Method
|
Calibration Study |
Validation Study |
t-Test |
|||||||
|
SAM |
APP |
SAM* |
APP† |
||||||
|
Mean ± SD |
Range |
Mean ± SD |
Range |
Mean ± SD |
Range |
Mean ± SD |
Range |
||
| Normal | 308 ± 75 | 129–495 | 330 ± 75 | 144–512 | 314 ± 69 | 222–453 | 311 ± 71 | 202−432 | P = 0.63 |
| Mild | 291 ± 41 | 232–433 | 303 ± 43 | 199–418 | 256 ± 66 | 132–387 | 252 ± 70 | 126–363 | P = 0.61 |
| Moderate | 225 ± 33 | 157–309 | 243 ± 35 | 154–350 | 182 ± 30 | 124–218 | 185 ± 34 | 134–226 | P = 0.46 |
| Severe | 162 ± 25 | 90–236 | 182 ± 33 | 68–261 | 106 ± 41 | 50–196 | 121 ± 51 | 52–236 | P = 0.01 |
| End-stage | 82 ± 20 | 38–118 | 94 ± 27 | 44–171 | 92 ± 41 | 23–160 | 87 ± 40 | 28–147 | P = 0.16 |
*SAM counts in the validation study are the mean of five separate counting sessions.
†APP counts in the validation study are compensated using the equation for the regression line generated in the calibration study.
Calibration of APP to SAM Counts within an Initial Group of 500 images
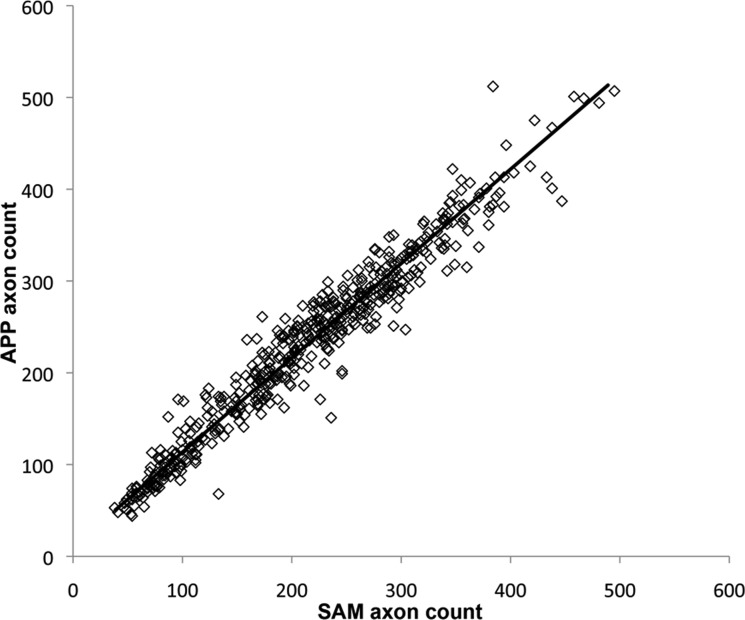
The APP counts were plotted versus SAM counts for all 500 images in Figure 2. The relationship between APP (ordinate) and SAM counts (abscissa) for these 500 images was APP = 10.77 + 1.03 (SAM) with an R2 of 0.94 (P < 0.0001, Deming regression). These data indicated that there was close correlation between APP and SAM counts in images from normal NHP eyes as well as through the complete range of mild to end-stage glaucomatous damage.
Figure 2. .
APP versus SAM counts in 500 images. The correlation produced an R2 = 0.94 (P < 0.0001). The relation of APP to SAM counts established by Deming regression is defined by the formula APP = 10.77 + 1.03 (SAM) and shown by the solid line.
A Bland-Altman plot (Fig. 3) demonstrated an average difference of 16.7 axons (APP > SAM) that decreased slightly as the number of axons per image decreased. The limits of agreement extended from −28 to +61 axons.
Validation of the APP Calibration to SAM Counts within a Second Group of 50 Images with Multiple Repeats (5) of SAM Counts
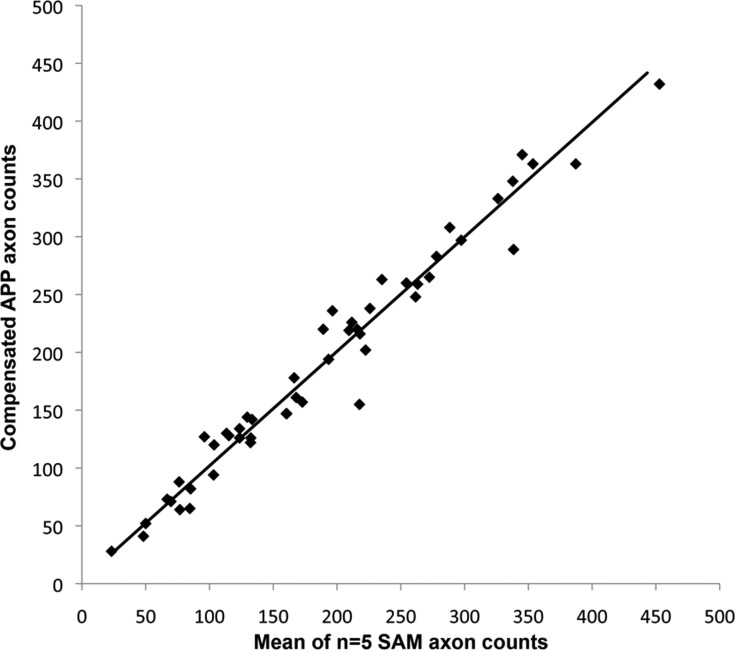
The relationship between uncompensated APP counts and the mean of five repeated SAM counts for these 50 images was APP = 12.03 + 1.03 (SAM) with an R2 of 0.96 (P < 0.0001, Deming regression). These data indicated that in a separate group of 50 images with glaucomatous axonal damage (normal to end-stage), there was again close correlation between the APP and SAM counts. Similar to the first group of 500 images, APP counted on average 12 more axons per image than the SAM with this difference declining for images with fewer axons. After APP counts were compensated (by the regression from data in Fig. 3), the mean ± SD of the SAM counts (190 ± 100) was not significantly different from the mean ± SD of the APP counts (191 ± 99) for the group of 50 images used in this validation set (paired, two-sided t-test [P = 0.71]). Compensated APP counts are plotted versus the mean of five SAM counts for each of the 50 images in Figure 4.
Figure 4. .
Validation of the calibration equation between APP counts and the mean of five SAM counts within a second group of 50 images. APP counts were compensated using the regression line equation established by the initial calibration study (Fig. 3) and compared to the mean of five repeated SAM counts of each image. The solid line represents the result of the Deming regression; y = 2.87 + 0.99x, R2 = 0.97.
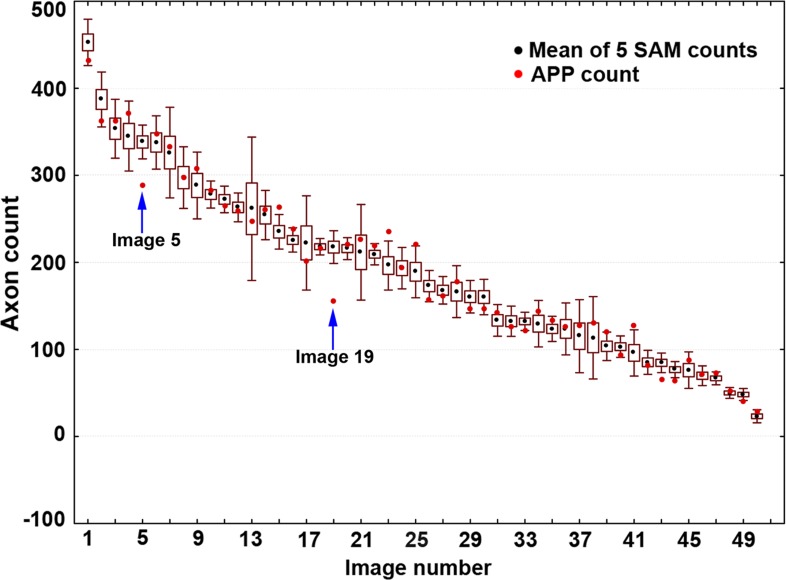
Figure 5 shows the compensated APP counts from the validation study plotted relative to the mean and 95% confidence interval of the five SAM counts sorted by rank. APP counts fell within the 95% confidence interval of SAM counts in 42 of the 50 images (84%). In the remaining eight images, six had APP counts that fell outside of the 95% confidence interval by a margin of 12 axons or less. The mean ± SD difference between the APP and SAM counts for the 50 validation study images was 1.15 ± 18.7 axons. The 95% confidence interval for this difference ranged from −35 to +38 axons. The two remaining images (#5 and #19) differed more substantially, which can be attributed to their relatively poor image quality (see Fig. 6).
Figure 5. .
Compensated APP counts plotted relative to the mean ±95% confidence interval (box = one standard deviation, whiskers = 95% CI) of five SAM counts for each of the 50 validation study images. The APP count for each image was compensated using the factor derived from the regression line of the calibration study (see Fig. 3). Compensated APP counts for 42 of the 50 images fell within the mean ±95% CI of five SAM repeated counts. Of the remaining eight images, six had compensated APP counts that fell outside of the 95% CI by a margin of 12 axons or less and two images (images #5 and #19, highlighted with arrows above) exhibited a difference greater than 30 axons from the SAM 95% CI, most likely due to their relatively poor quality.
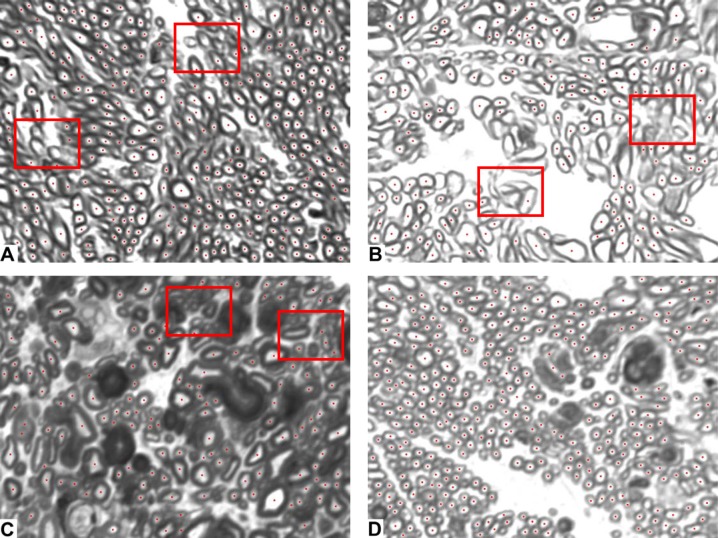
Figure 6. .
Representative images to evaluate the agreement between the APP (red dots) and SAM counts. The images in this figure show instances where the agreement between the APP and SAM counts varied. (A, B) Poor (images 5 and 19 from Fig. 5, respectively). (C) Fair. (D) Excellent. All results herein refer to compensated APP and the average of five repeat SAM counts. In these images, the SAM counts were 338, 218, 115, and 297 and the APP counts were 289, 151, 128, and 297 [(A)–(D), respectively]. (A) and (B) have multiple axons that are either very small or poorly defined, highlighted by the red boxes. Panel C shows an image from an area of severe damage in which the contrast between the myelin border and the inner axon area is reduced (red boxes). Finally, panel D shows an image with good illumination and high contrast.
Generation of APP Density Maps and Total ON Axon Counts for Both Eyes of a Representative Unilateral Early Experimental Glaucoma NHP
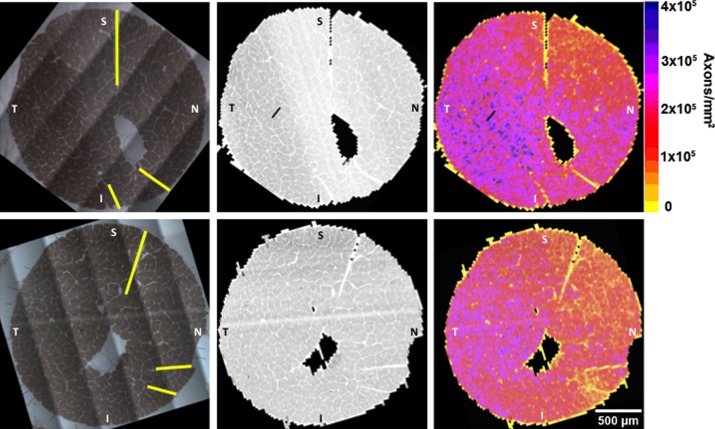
Orbital ON cross sections along with their image composites and APP density maps are displayed for both the normal and early experimental glaucoma eye of a representative NHP in Figure 7. A total of 5768 and 6000 images make up the image composites for the contralateral control and experimental glaucoma eyes, respectively. Every image in each of the two composites was counted by the APP. The total uncompensated ON axon counts for the normal (1,184,801 axons) and early EG (1,008,103 axons) eyes indicate a loss of 14.9% in the glaucomatous eye relative to the contra-lateral control eye.
Figure 7. .
Orbital ON cross-section image stained for myelin (left column), 100× composite image (middle column) and 100× density map (right column) for both the control (upper row) and early EG (bottom row) eyes of an NHP.34 All images are in right eye configuration and in approximate clinical orientation (superior [S], inferior [I], nasal [N], and temporal [T] based on the optic nerve cross-section cuts outlined in yellow in the upper and lower left panels). A total of 5768 and 6000 images at 100× magnification make up the image composites and were counted by APP for the control and early EG eyes, respectively. The colored density plots (right) are generated from the uncompensated (see Methods section) total axon count of every image divided by the image area (including the axons that lie in overlap areas). The total uncompensated ON axon counts (excluding the overlap areas) for the control (1,184,801 axons—upper row) and early EG (1,008,103 axons—lower row) eyes indicate a 14.9% between–eye difference in this animal.
Discussion
The purpose of this paper was to describe an automated algorithm and software application for counting axons with normal morphology across 100% of the cross-sectional area of the orbital ON, and to assess its performance relative to the previously published semi-automated method in a representative sample of images taken from control and glaucomatous eyes of NHPs with unilateral experimental glaucoma. The study method proved to be highly correlated to SAM counts in an initial calibration set of 500 images. It also showed that its performance was similar to that of an experienced operator in a validation set of 50 images counted five times each by demonstrating that its results fell within the operator's 95% CI in 42 out of 50 images. Since the images in both sets demonstrated axonal damage ranging from normal to end-stage, the results obtained showed that the study method achieved the requirements set forth at the start of the work: to develop a simple solution that is applicable to all levels of ON axonal damage and that is computationally robust and efficient. It is important to stress that—like any other image processing algorithm—the performance of the described method depends entirely on the quality of the input images that is affected not only by proper exposure and focus settings, but also (and perhaps more importantly) by proper tissue processing, sectioning, mounting, and staining.
In terms of performance, it took the system approximately 12 hours to acquire the 11,768 images required to cover 100% of the cross-sectional areas of one normal (5768 images) and one EG (6000 images) NHP ON. On the test system used for this study (MacBook Pro laptop, 2.2 GHz Core2 Duo, 3GB RAM; Apple, Cupertino, CA), the APP processed an average of 4 images/minute (49 hours for 11,768 images). However, due to the independent nature of the problem (i.e., each image is an independent sample), the APP can be easily parallelized, moving it from its single-threaded model to one that uses all CPU cores available in the computer, to reduce the processing time required by a factor equivalent to the number of cores available. The software can also be distributed to different computers on the network and executed simultaneously to further improve its performance. The application will be modified to implement these parallel features.
Combining 100% axon counts with precise alignment of the orbital ON cross section to the ON head and retina34 should allow a new level of hypothesis testing regarding structure-function relationships and the cellular mechanisms underlying RGC axonal damage. The strategies for within-eye and between-eye clinical colocalization, as well as for generating between-eye ON axon density and axon size difference maps, are under development and will be the subject of future studies.
The study method has several limitations. First, the ON tissues used for this report were either perfusion-fixed or immersion-fixed immediately after sacrifice. While most experimental models can provide similar levels of tissue processing, the quality of axon preservation within human cadaver eyes may not be adequate for this technique without substantial adjustment of the algorithm. Second, the method was developed and validated using NHP ON images. For species with smaller axons, changes need to be made both to the acquisition system and to the software application in order to adequately capture images and accurately count optic nerve axons. Work on a rodent version of the APP will be started in the near future. Third, study calibration and validation studies were performed on two separate groups (n = 500 and n = 50, respectively) of images from a total of 17 normal and 13 experimental glaucoma eyes from 25 NHPs. The processing of these ON tissues occurred over a 10-year span and involved multiple technicians. While it is believed this is a reasonable representation of the variability inherent within the animals, eyes, and tissue processing, it is also believed that as the study method is implemented in other laboratories, similar calibration studies specific to each laboratory will be required. Finally, some degree of misalignment between the camera and stage axes will always occur and its reduction is difficult to attain particularly at high magnification. At present, the study's overlap calculation assumes no misalignment and it is done purely on the basis of the coordinates of each image. While this assumption does not introduce any error in the accuracy of each image density estimate, it can potentially affect the overall axon count depending on the amount of misalignment, size of the ON, and size of the axons in the overlap areas. Future experiments are planned to accurately determine and minimize the misalignment effects in the study's method.
It is important to note that the system and method presented in this manuscript can be easily expanded to other species. For rodents, for example, changes in the optics and small software adjustments to accommodate for the smaller axon size would be required. Also, similar studies as the ones presented herein would also be necessary to validate the accuracy of the APP. However, due to the smaller ON size, the time required to produce validation data (manual counts) would be greatly reduced and a large dataset could be easily assembled. The process to expand the software in this direction is already underway.
Finally, while compensation of the automated axon counts was required to most fairly compare the performance of the APP to that of the SAM in the validation study, it is not believed that the automated counts should be compensated in actual scientific application. Doing so would imply that the study manual counts are the gold standard and, thus, more accurate than the study's automated algorithm—something that is difficult, if not impossible, to determine. Therefore, the results reported in Figure 7 are raw (uncompensated) automated axon counts.
In summary, this study presented a method for fully automated 100% orbital ON axon counts in NHP ONs. Study calibration and validation data indicate that its counts are similar to SAM counts through a wide range of ON damage—from normal through to end-stage axon loss—in NHP eyes. While the imaging hardware necessary to implement this method is expensive, most laboratories already have parts that can be integrated to produce a solid imaging solution. Due to its low computational requirements, any recent computer (PC or Mac) should be capable of running the software, which is available upon request.
Appendix A
Algorithm.
The algorithm consists of the following steps:
Read the image and obtain its x, y coordinates. Because the APP is not designed to work with color images (e.g., fluorescently labeled images with multiple labels), only the green channel is currently used.
Create a binary image map. Every pixel in the image is categorized as either myelin (black) or other (white) using a fuzzy c-means classifier.1
Find all the white pixel clusters using ImageJParticleAnalyzer plugin. Every cluster is then filtered based on its size and circularity measure. Clusters that fall within myelin boundaries generally meet normal axon requirements and are kept. Other clusters representing vessels and voids in the ON cross section are rejected.
Write the results to a comma-delimited output file. Each line in this file represents a normal axon found in the image and includes the axon's centroid, area, and circularity value.
After all images have been counted, a separate utility gathers all output files from step 4 and produces a total axon count for the entire ON cross section and average axon size and density for every image. A scaled composite of the raw acquired images (composite image) is then created. An additional composite colored according to axon density is also produced (density composite image).
Image Reader.
A custom BIF image reader was developed to support Bioquant's native BIF format. For TIFF and PNG images, the APP uses Java's ImageIO classes to read the input images and to produce the global composites and density maps.
Fuzzy C-Means Classifier.
The most important task of the APP is to automatically assign every pixel within the image to one of two categories: “myelin” or “other” (neural tissue, vessel, empty area, etc.). This “membership” association is difficult to obtain using a simple threshold technique because of image quality issues such as uneven stain uptake, exposure, and focus across the ON section, which reduce the simple threshold's performance. A more robust technique that is less affected by these factors and that can operate without direct user intervention is thus necessary. The FCM clustering method1 is well suited for this task. Using the pixel's grayscale value as input, the FCM assigns each pixel a probability of belonging to either the “myelin” (dark) or “other” (bright) category. Because the FCM operates on each image independently, its results are optimized for each individual image. This is particularly important in images from areas of moderate axon damage where there is less contrast between myelin and neural areas.
ImageJ's Particle Analyzer.
This plugin is a powerful utility that automatically counts and analyzes groups of adjacent pixels having the same binary value (particles). In this case, these particles represent normal axons, the area enclosed by healthy myelin. The plugin uses the binary map created in step 2 above to count, calculate the area, and fit an ellipse to the edge of all particles present in the image. Using the area and fitted ellipse information, the plugin then filters out particles that do not meet the supplied area and circularity criteria. Based on previous work,2 the minimum and maximum allowed particle size was set to 42 and 4000 square pixels, respectively. The circularity criteria was set to 0.5 to be able to retain particles that have an elongated shape in order to account for axons that appear elliptical in the ON cross sections due to their natural shape or because they are not perpendicular to the plane of the cross section.
Footnotes
Supported in part by National Institutes of Health Grant R01-EY011610; Merck Pharmaceuticals and Legacy Good Samaritan Foundation (CFB and GAC); and Sears Medical Trust and the Alcon Research Institute (CFB).
Disclosure: J. Reynaud, None; G. Cull, None; L. Wang, None; B. Fortune, None; S. Gardiner, None; C.F. Burgoyne, None; G.A. Cioffi, None
References
- 1. Marina N, Bull ND, Martin KR. A semiautomated targeted sampling method to assess optic nerve axonal loss in a rat model of glaucoma. Nat Protoc. 2010;5:1642–1651 [DOI] [PubMed] [Google Scholar]
- 2. Chauhan BC, Levatte TL, Garnier KL, et al. Semiquantitative optic nerve grading scheme for determining axonal loss in experimental optic neuropathy. Invest Ophthalmol Vis Sci. 2006;47:634–640 [DOI] [PubMed] [Google Scholar]
- 3. Cioffi GA, Wang L, Fortune B, et al. Chronic ischemia induces regional axonal damage in experimental primate optic neuropathy. Arch Ophthalmol. 2004;122:1517–1525 [DOI] [PubMed] [Google Scholar]
- 4. Cull G, Cioffi GA, Dong J, Homer L, Wang L. Estimating normal optic nerve axon numbers in non-human primate eyes. J Glaucoma. 2003;12:301–306 [DOI] [PubMed] [Google Scholar]
- 5. Jia L, Cepurna WO, Johnson EC, Morrison JC. Patterns of intraocular pressure elevation after aqueous humor outflow obstruction in rats. Invest Ophthalmol Vis Sci. 2000;41:1380–1385 [PubMed] [Google Scholar]
- 6. Morrison JC, Nylander KB, Lauer AK, Cepurna WO, Johnson E. Glaucoma drops control intraocular pressure and protect optic nerves in a rat model of glaucoma. Invest Ophthalmol Vis Sci. 1998;39:526–531 [PubMed] [Google Scholar]
- 7. Ogden TE, Miller RF. Studies of the optic nerve of the rhesus monkey: nerve fiber spectrum and physiological properties. Vision Res. 1966;6:485–506 [PubMed] [Google Scholar]
- 8 . Mikelberg FS , Yidegiligne HM, Schulzer M. Optic nerve axon count and axon diameter in patients with ocular hypertension and normal visual fields. Ophthalmology. 1995;102:342–348 [DOI] [PubMed] [Google Scholar]
- 9. Mikelberg FS, Drance SM, Schulzer M, Yidegiligne HM, Weis MM. The normal human optic nerve. Axon count and axon diameter distribution. Ophthalmology. 1989;96:1325–1328 [DOI] [PubMed] [Google Scholar]
- 10. Kupfer C, Chumbley L, Downer JC. Quantitative histology of optic nerve, optic tract and lateral geniculate nucleus of man. J Anat. 1967;101:393–401 [PMC free article] [PubMed] [Google Scholar]
- 11. Morrison JC, Cork LC, Dunkelberger GR, Brown A, Quigley HA. Aging changes of the rhesus monkey optic nerve. Invest Ophthalmol Vis Sci. 1990;31:1623–1627 [PubMed] [Google Scholar]
- 12. Brooks DE, Strubbe DT, Kubilis PS, MacKay EO, Samuelson DA, Gelatt KN. Histomorphometry of the optic nerves of normal dogs and dogs with hereditary glaucoma. Exp Eye Res. 1995;60:71–89 [DOI] [PubMed] [Google Scholar]
- 13. Harwerth RS, Wheat JL, Rangaswamy NV. Age-related losses of retinal ganglion cells and axons. Invest Ophthalmol Vis Sci. 2008;49:4437–4443 [DOI] [PubMed] [Google Scholar]
- 14. Harwerth RS, Vilupuru AS, Rangaswamy NV, Smith EL., III The relationship between nerve fiber layer and perimetry measurements. Invest Ophthalmol Vis Sci. 2007;48:763–773 [DOI] [PubMed] [Google Scholar]
- 15. Harwerth RS, Carter-Dawson L, Smith EL, III, Barnes G, Holt WF, Crawford ML. Neural losses correlated with visual losses in clinical perimetry. Invest Ophthalmol Vis Sci. 2004;45:3152–3160 [DOI] [PubMed] [Google Scholar]
- 16. Garway-Heath DF, Poinoosawmy D, Fitzke FW, Hitchings RA. Mapping the visual field to the optic disc in normal tension glaucoma eyes. Ophthalmology. 2000;107:1809–1815 [DOI] [PubMed] [Google Scholar]
- 17. Potts AM, Hodges D, Shelman CB, Fritz KJ, Levy NS, Mangnall Y. Morphology of the primate optic nerve. 3. Fiber characteristics of the foveal outflow. Invest Ophthalmol. 1972;11:1004–1016 [PubMed] [Google Scholar]
- 18. Potts AM, Hodges D, Shelman CB, Fritz KJ, Levy NS, Mangnall Y. Morphology of the primate optic nerve. II. Total fiber size distribution and fiber density distribution. Invest Ophthalmol. 1972;11:989–1003 [PubMed] [Google Scholar]
- 19. Buckingham BP, Inman DM, Lambert W, et al. Progressive ganglion cell degeneration precedes neuronal loss in a mouse model of glaucoma. J Neurosci. 2008;28:2735–2744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cepurna WO, Kayton RJ, Johnson EC, Morrison JC. Age related optic nerve axonal loss in adult Brown Norway rats. Exp Eye Res. 2005;80877–884 [DOI] [PubMed] [Google Scholar]
- 21. Inman DM, Sappington RM, Horner PJ, Calkins DJ. Quantitative correlation of optic nerve pathology with ocular pressure and corneal thickness in the DBA/2 mouse model of glaucoma. Invest Ophthalmol Vis Sci. 2006;47:986–996 [DOI] [PubMed] [Google Scholar]
- 22. Chauhan BC, Pan J, Archibald ML, LeVatte TL, Kelly ME, Tremblay F. Effect of intraocular pressure on optic disc topography, electroretinography, and axonal loss in a chronic pressure-induced rat model of optic nerve damage. Invest Ophthalmol Vis Sci. 2002;43:2969–2976 [PubMed] [Google Scholar]
- 23. Yucel YH, Gupta N, Kalichman MW, et al. Relationship of optic disc topography to optic nerve fiber number in glaucoma. Arch Ophthalmol. 1998;116:493–497 [DOI] [PubMed] [Google Scholar]
- 24. Sanchez RM, Dunkelberger GR, Quigley HA. The number and diameter distribution of axons in the monkey optic nerve. Invest Ophthalmol Vis Sci. 1986;27:1342–1350 [PubMed] [Google Scholar]
- 25. Potts AM, Hodges D, Shelman CB, Fritz KJ, Levy NS, Mangnall Y. Morphology of the primate optic nerve. I. Method and total fiber count. Invest Ophthalmol. 1972;11:980–988 [PubMed] [Google Scholar]
- 26. Quigley HA , Addicks EM, Green WR. Optic nerve damage in human glaucoma. III. Quantitative correlation of nerve fiber loss and visual field defect in glaucoma, ischemic neuropathy, papilledema, and toxic neuropathy. Arch Ophthalmol. 1982;100:135–146 [DOI] [PubMed] [Google Scholar]
- 27. Yang H, Thompson H, Roberts MD, Sigal IA, Downs JC, Burgoyne CF. Deformation of the early glaucomatous monkey optic nerve head connective tissue after acute IOP elevation in 3-D histomorphometric reconstructions. Invest Ophthalmol Vis Sci. 2011;52:345–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Reynaud J, Cull G, Wang L, Burgoyne CF, Cioffi GAA. New hybrid algorithm for automated axon counting in normal optic nerves. Invest Ophthalmol Vis Sci. 2007;48:E-Abstract 3297 [Google Scholar]
- 29. Reynaud J, Cull G, Wang L, Burgoyne CF, Cioffi G. Bilateral distribution of axon density and size in optic nerves. Invest Ophthalmol Vis Sci. 2008;49:E-Abstract 3665 [Google Scholar]
- 30. Reynaud J, Cull G, Dyrud E, Burgoyne CF, Cioffi GA. An algorithm for counting axons in mild to severe damage optic nerve images. Invest Ophthalmol Vis Sci. 2010;51:E-Abstract 2143 [Google Scholar]
- 31. Reynaud J, Cull G, Wang L, et al. Automated detection of regional axonal damage in optic nerve cross-section images. Invest Ophthalmol Vis Sci. 2009;50:E-Abstract 5829 [Google Scholar]
- 32. Rasband WS. ImageJ. Bethesda, Maryland: U.S. National Institutes of Health,http://imagej.nih.gov/ij/1997–2011. [Google Scholar]
- 33. Deming W. Statistical Adjustment of Data. New York, NY:Wiley;1943. [Google Scholar]
- 34. Yang H, Downs JC, Bellezza AJ, Thompson H, Burgoyne CF. 3-D Histomorphometry of the normal and early glaucomatous monkey optic nerve head: prelaminar neural tissues and cupping. Invest Ophthalmol Vis Sci. 2007;48:5068–5084 [DOI] [PubMed] [Google Scholar]
References
- 1. Bezdek JC. Pattern Recognition with Fuzzy Objective Function Algorithms. Norwell, MA: Kluwer Academic Publishers; 1981. [Google Scholar]
- 2. Reynaud J, Cull G, Wang L, Burgoyne CF, Cioffi GA. A new hybrid algorithm for automated axon counting in normal optic nerves . Invest Ophthalmol Vis Sci. . 2007;. 48 : E-Abstract 3297 . [Google Scholar]