Abstract
Genetic mapping of wheat, maize, and rice and other grass species with common DNA probes has revealed remarkable conservation of gene content and gene order over the 60 million years of radiation of Poaceae. The linear organization of genes in some nine different genomes differing in basic chromosome number from 5 to 12 and nuclear DNA amount from 400 to 6,000 Mb, can be described in terms of only 25 “rice linkage blocks.” The extent to which this intergenomic colinearity is confounded at the micro level by gene duplication and micro-rearrangements is still an open question. Nevertheless, it is clear that the elucidation of the organization of the economically important grasses with larger genomes, such as maize (2n = 10, 4,500 Mb DNA), will, to a greater or lesser extent, be predicted from sequence analysis of smaller genomes such as rice, with only 400 Mb, which in turn may be greatly aided by knowledge of the entire sequence of Arabidopsis, which may be available as soon as the turn of the century. Comparative genetics will provide the key to unlock the genomic secrets of crop plants with bigger genomes than Homo sapiens.
In the mid 1980s when restriction fragment length polymorphism (RFLP) technology was first applied to plants, the objectives of the early experiments—in tomato, Lycopersicon esculentum, by Steve Tanksley in New Mexico, in maize, Zea mays, by Tim Helentjaris in Utah, and ourselves in bread wheat, Triticum aestivum, in Cambridge—were no more ambitious than to produce a new generation of markers for use by breeders. In the race to build the first dense genetic maps, the early reports of synteny across genomes in 1988—between tomato and potato (1) and between the three diploid genomes of hexaploid wheat (2)— were interesting but not remarkable. Later the cross-genome comparisons became more compelling. These comparisons all employed hybridization-based mapping procedures, which, with variable stringency conditions, allowed the detection of similar but imperfectly matched DNA sequences. Large numbers of characterized DNA probes were still not available, and researchers of the day, therefore, used RFLP probes available in one species to create genetic maps in related genomes. Examples are the use of maize probes to map sorghum, Sorghum bicolor (3), and wheat probes to map rye, Secale cereale (4), and both studies revealed yet more colinearity. Nevertheless, it was not clear at the time that intergenomic synteny extends only to the genes themselves. Since the early 1990s alternative marker systems based on PCR have complemented RFLPs. However, cross-genome signals are only infrequently observed by using sequence-tagged sites or microsatellite primers because the sequences must match the template DNA precisely. In fact, had PCR been discovered 5 years earlier we may still have been ignorant about the conservation of gene order among plant species.
A Consensus Grass Map
Soon, wider comparisons between Tribes were reported. Ahn and Tanksley (5) showed the rice, Oryza sativa, and maize genomes to be closely related; Kurata et al. (6) showed rice and wheat to be colinear; Devos et al. (7) showed maize and wheat to have retained colinearity; Van Deynze et al. (8) extended the rice, maize, and wheat comparison to include an Avena atlantica × A. hirtular diploid oat map; Devos et al. (9) compared foxtail millet, Setaria italica, and rice; the complex polyploid sugarcane was mapped alongside maize and sorghum by Grivet et al. (10) and rice by Dufour et al. (11). More work, as yet unreported, will demonstrate that pearl millet, Pennisetum glaucum (K.M.D., T. S. Dugdale, M. Couchman, and M.D.G., unpublished data), finger millet, Eleusine corocana (M. Dida, M.D.G., and K.M.D., unpublished data), and rye grass, Lolium perenne (M. Hayward, I. P. King, H. Martin Thomas, J. King, I. Armstead, J. Forster, M. Humphries, and G. Morgan, unpublished data) also have genomes that are closely related to the other economic grass species.
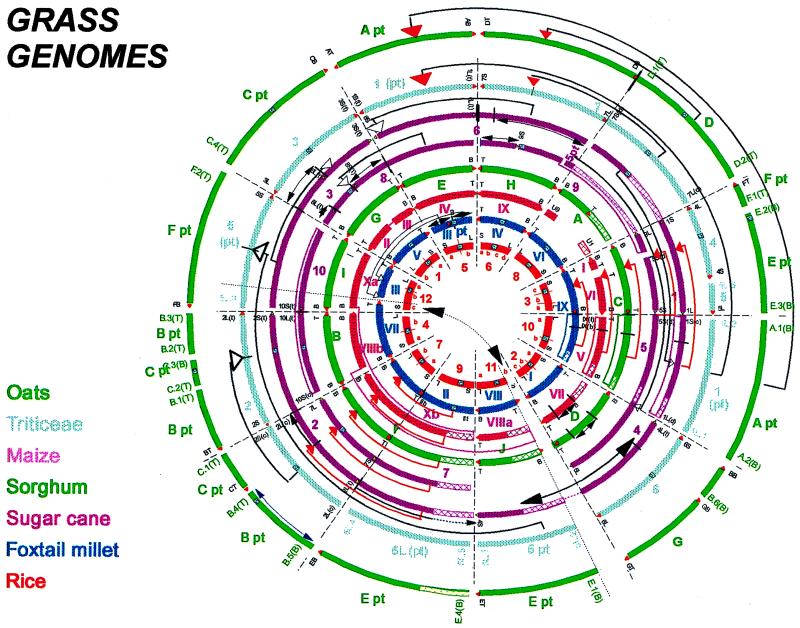
The first consensus grass map aligning the genomes of seven different grass species was shown by Moore et al. (12), and an extended and more detailed version is shown in Fig. 1. This map describes the different grass genomes in terms of “rice linkage blocks.” Fig. 1 shows, the several areas of uncertainty (shown by hatched regions) notwithstanding, that now nine different genomes—diploid oats, a wheat and barley consensus, the two genomes of maize, sorghum, the two genomes, S. spontaneum and S. officinarum, of sugarcane, foxtail millet, and rice—can be described by only 25 rice linkage blocks. Undoubtedly this estimate will grow as more detail is added. However, already the consensus can be used to rapidly construct maps of other grass species by using a set of anchor probes evenly spaced around the circles, and to predict the locations of key genes for adaptation from one crop species to another.
Figure 1.
A consensus grass comparative map. The comparative data have been drawn from many sources: Oats-wheat-maize-rice (8); wheat-rice (6, 30), and unpublished data; maize-rice (5, 10); maize-wheat (7); maize-sorghum-sugarcane (10, 11); and foxtail millet-rice (9). Arrows indicate inversions and transpositions necessary to describe present-day chromosomes. Locations of telomeres (▵) and centromeres (□) are shown where known. Hatched areas indicate chromosome regions for which very little comparative data exist. Chromosome nomenclatures and arm (short/long or top/bottom) designations are as described by O’Donoughue et al. (31) for oats, Pereira et al. (32) for sorghum, Dufour (11) for the sugarcane genomes, Wang et al. (33) in foxtail millet, and Singh et al. (34) in rice.
Major Gene Synteny
Although most of the mapped loci that anchor the consensus map are detected by RFLP probes of unknown function, some clear relationships between a few genes of economic and adaptive importance are already apparent. An underlying assumption to the consensus shown in Fig. 1 is that if, as presently estimated in Arabidopsis, cereal genomes carry about 25,000 genes, then one should be able to draw 25,000 radii around the circles to pass through homoeoloci in the different genomes.
These relationships can clearly be seen among the many isozyme loci that have been mapped in the different species. Other examples include waxy (Wx) genes in all of the species in rice linkage block 6a; the liguless (Lg) loci in barley, maize, and rice in RLB4b; genes controlling gibberellin insensitivity and plant height in wheat (Rht) and maize (d8, d9) in RLB3b; and red grain color in wheat (R) and rice (Rd) in RLB1b. In other cases, genes controlling major mutant phenotypes can be aligned with genetic factors with lesser effects, which have been measured as quantitative trait loci (QTLs) in other genomes. An example is the alignment of the major maize dwarfing loci, br1, an1, d3, and py1, with QTLs for plant height in sorghum (13). Similarly a major gene for shattering, a key component of domestication of crop plants, is aligned exactly with QTLs for shattering in rice and maize (14). Thus, many of the major gene mutants mapped in barley, maize, and rice may be used as pointers to homoeogenes with more subtle, exploitable effects in the same or other genomes.
Plainly there is a need to continue the merging of the old “classical” maps with the newer “molecular” maps. The recent explosion of expressed sequence tag (EST) data in rice and maize and their localization on genetic or physical maps combined with the rapidly expanding gene sequence databases will make a powerful gene-mining tool.
Major Evolutionary Chromosomal Rearrangements
Inferences can also be made from the major genomic rearrangements that have taken place during evolution and were revealed through the comparative analysis. In fact, conservation of gene order is so much the rule that the differences in organization between genomes can be used for meaningful taxonomic analysis [see Kellogg (15) in this issue of the Proceedings].
With reference to Fig. 1, the insertion of rice linkage block 10 into 5 describes the present-day Triticeae group 1 chromosomes and oats chromosome A and is likely to be common to all Pooideae. Similarly, genomes of the Panicoideae species are all defined by the insertions of rice linkage blocks 9 into 7 and 10 into 3. All of these rearrangements are likely to date back 60–100 millions of years to the early radiation of the Poaceae. As to which arrangement, if any of those extant today, is the most primitive is still not clear. However, parsimony would argue that the fact that rice linkage block 10, freestanding in rice, is today found in two different chromosomal environments in the Pooideae and the Panicoideae indicates that the rice genome itself is the most primitive.
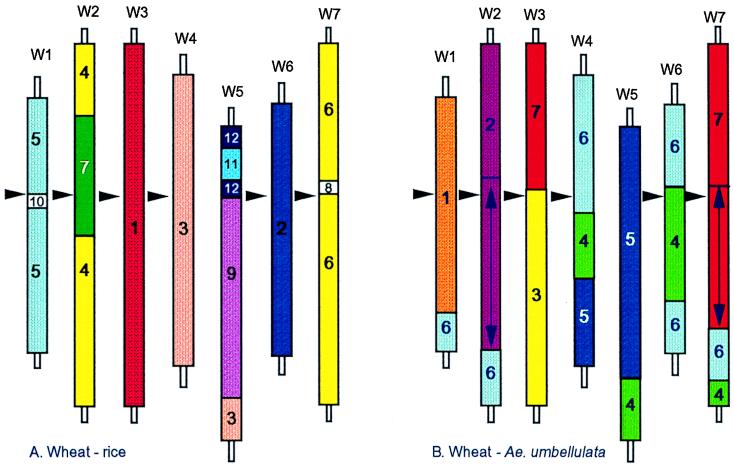
Comparison of the organization of the different grass genomes raises the interesting question of why any of these chromosome rearrangements have occurred and become fixed. Triticeae cereals, represented by the D genome of hexaploid bread wheat, and rice comparative maps (Fig. 2A) can, within the limits of the present analysis, be drawn with only 11 breaks in colinearity, despite the fact that their chromosome numbers are different (x = 7 and 12 respectively). Wheat and rye (4) or wheat and Aegilops umbellulata [Fig. 2B; after Zhang et al. (16)], on the other hand, all with the same basic chromosome reference (x = 7), show 11 and 12 breaks in colinearity, respectively, even though they have diverged after speciation less than eight million years ago. These results must be considered alongside comparisons of the D genome of bread wheat and the barley, Hordeum vulgare, genome, which diverged before wheat and rye, and shows almost complete colinearity. Plainly, in some instances, divergence of genome structure appears to be driven possibly to reinforce the process of speciation. Whatever the underlying mechanisms, the assumption that these events occur randomly and thus can be used as a measure of evolutionary time (17) appears to be unwarranted.
Figure 2.
Comparative maps of the wheat genome described in terms of the rice genome (A) and the Ae. umbellulata genome (B). (A) Data from Kurata et al. (6), Van Deynze et al. (30), and unpublished work. (B) Data from Zhang et al. (9).
Major Duplications
Much work is still required to develop the consensus map to be universally useful as an accurately predictive tool. Some regions (the hatched areas in Fig. 1) are, as yet, very poorly defined. Indeed, very few intragenomic duplications, besides the major duplication involving the short arms of rice chromosomes 11 and 12 (18), have been described. There are, however, likely to be many of these, judging by the frequency that single-copy RFLP probes in one species show two or more hybridizing DNA fragments in others. Their definition and the identification of odd nonhomoeologous copies of genes, such as sedoheptulose-1,7-bishosphatase (19) and acyl carrier proteins (20) in wheat, are vital to the interpretation of apparently noncolinear, intergenomic mapping information.
Colinearity at Micro Level
To date the comparative information derives only from the location of a limited number of genes and anonymous DNA sequences, often spaced at many millions of DNA base pairs apart, that have been cross-mapped in different genomes. This does not provide the answer to the key questions as to whether all of the genes are present in the same order, or whether individual species have some unique genes that have arisen to cope with the special and different environments to which each species is adapted. Moreover, the regions that appear colinear at the genetic map level could still be considerably rearranged at the micro level. The answers are vital to those teams that are now embarked on comparative map-based gene-isolation experiments, in which the relatively small genome and extensive genomic tools available in rice are being exploited to isolate genes of importance in grass-crop species with much larger genomes.
The information available to date is equivocal and probably will remain so until the genomic DNAs from several colinear regions in several different grasses have been sequenced. Bennetzen’s group (21) and Chen et al. (22) have obtained sequence of the region spanning the A1 and Sh2 loci in sorghum and rice, in which the region extends over 19 kb of genomic DNA. In both species the intervening region contains only the same single, unknown gene. Interestingly, the homoeology extends only to exons where it is nearly complete, whereas the introns and intergenic regions appear to be completely different and species-specific, confirming that colinearity is a function only of the genes themselves. Again on the positive side, Foote et al. (23), working in a region of wheat chromosome 5B carrying a gene, Ph1, which controls homoeologous chromosome pairing, find apparent colinearity of about 20 genes over a 3-Mb region. However, this experiment has also revealed genes or small linkage segments involved in duplications. A 500-kb segment on rice chromosome 9 is duplicated a few map units below the critical region on wheat chromosome 5B (23). Even more perplexing is the discovery, in the latter experiment, of a small region with synteny corresponding to rice chromosome 11 interpolated into a region with apparent total colinearity with rice 9.
Conclusions
The growing appreciation of the extent of conservation of genes and gene order is already significantly affecting the way in which we think about the genetics of the major cereals. As yet the linking information between different genomes is still too sparse to accurately pinpoint candidate homoeogenes except in the few cases where the similarities in phenotypes are obvious. However, this limitation will lift as more comparative data are added to the maps and the underpinning bioinformatics is developed. The time is fast approaching when the grasses, including all of the major cereals, can be considered as a single entity and all of the information available on gene structure, gene action, metabolism, physiology, and phenotype accumulated over the past century in the different species can be pooled. An immediate practical implication is that breeders need no longer be restricted to their own species in their search for exploitable variation. Homoeogenes and all of their alleles in all species will be available to the cereal breeder/genetic engineer of the early 21st century.
The extent to which synteny will be a useful tool throughout all crop species has yet to be determined; however, evidence for similar conserved genome relationships are already well developed in legumes (24, 25) and crucifers (26, 27), which include the main brassica crop species and the model plant arabidopsis. The possibility that usable synteny extends over the monocot–dicot divide, which may represent 130–200 million years of independent evolution (28, 29), is still open. Intriguing glimpses of conserved regions are, however, being noted from preliminary analysis of the Arabidopsis genomic sequences as it is produced (W. R. McCombie, L. Parnell, R. Wing, and R. Martienessen, unpublished data).
The impact of the discovery of conserved synteny is also most important in the consideration of genomics programs for the major cereals, wheat and maize. Both of these staple crops have genomes larger than the human genome; however, for rice, at only four times the size of the arabidopsis genome, major sequencing initiatives are already under discussion in China, Japan, Europe, and the U.S. It is very possible that the genomes of maize and wheat can both be defined completely by application of the arabidopsis and rice genomic sequences, complemented by long-range mapping and a little sequencing in the target crop.
ABBREVIATIONS
- RFLP
restriction fragment length polymorphism
- QTL
quantitative trait loci
References
- 1.Bonierbale M W, Plaisted R L, Tanksley S D. Genetics. 1988;120:1095–1103. doi: 10.1093/genetics/120.4.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chao S, Sharp P J, Gale M D. In: Proc. 7th Int. Wheat Genet. Symp. Miller T E, Koebner R M D, editors. Cambridge Laboratory, Cambridge: IPSR; 1988. pp. 493–498. [Google Scholar]
- 3.Melake-Berhan A, Hulbert S H, Butler L G, Bennetzen J L. Theor Appl Genet. 1993;86:598–604. doi: 10.1007/BF00838715. [DOI] [PubMed] [Google Scholar]
- 4.Devos K M, Atkinson M D, Chinoy C N, Harcourt R L, Koebner R M D, Liu C J, Masojc P, Xie D X, Gale M D. Theor Appl Genet. 1993;85:673–680. doi: 10.1007/BF00225004. [DOI] [PubMed] [Google Scholar]
- 5.Ahn S N, Tanksley S D. Proc Natl Acad Sci USA. 1993;90:7980–7984. doi: 10.1073/pnas.90.17.7980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kurata N, Moore G, Nagamura Y, Foote T, Yano M, Minobe Y, Gale M D. Bio/Technology. 1994;12:276–278. [Google Scholar]
- 7.Devos K M, Chao S, Li Q Y, Simonetti M C, Gale M D. Genetics. 1994;138:1287–1292. doi: 10.1093/genetics/138.4.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Van Deynze A E, Nelson J C, O’Donoughue L S, Ahn S N, Siripoonwiwat W, Harrington S E, Yglesias E S, Braga D P, McCouch S R, Sorrells M E. Mol Gen Genet. 1995;249:349–356. doi: 10.1007/BF00290536. [DOI] [PubMed] [Google Scholar]
- 9.Devos, K. M., Wang, Z. M., Beales, J., Sasaki, T. & Gale, M. D. (1998) Theor. Appl. Genet., in press.
- 10.Grivet L, D’Hont A, Dufour P, Hamon P, Roques D, Glaszmann J C. Heredity. 1994;73:500–508. [Google Scholar]
- 11.Dufour P. Ph.D. thesis. France: Université de Paris Sud; 1996. [Google Scholar]
- 12.Moore G, Devos K M, Wang Z M, Gale M D. Curr Biol. 1995;5:737–739. doi: 10.1016/s0960-9822(95)00148-5. [DOI] [PubMed] [Google Scholar]
- 13.Pereira M G, Lee M. Theor Appl Genet. 1995;90:380–388. doi: 10.1007/BF00221980. [DOI] [PubMed] [Google Scholar]
- 14.Paterson A H, Lin Y-R, Li Z, Schertz K F, Doebley J F, Pinson S R M, Liu S-C, Stansel J W, Irvine J E. Science. 1995;269:1714–1718. doi: 10.1126/science.269.5231.1714. [DOI] [PubMed] [Google Scholar]
- 15.Kellogg E A. Proc Natl Acad Sci USA. 1998;95:2005–2010. doi: 10.1073/pnas.95.5.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang, H., Jia, J., Gale, M. D. & Devos, K. M. (1998) Theor. Appl. Genet., in press.
- 17.Paterson A H, Lan T-H, Reischmann K P, Chang C, Lin Y-R, Liu S-C, Burow M D, Kowalski S P, Katsar C S, DelMonte T A, Feldmann K A, Schertz K F, Wendel J F. Nat Genet. 1996;14:380–382. doi: 10.1038/ng1296-380. [DOI] [PubMed] [Google Scholar]
- 18.Nagamura Y, Inoue T, Antonio B A, Shimano T, Kajiya H, Shomura A, Lin S Y, Kuboki Y, Harushima Y, Kurata N, Minobe Y, Yano M, Sasaki T. Breed Sci. 1995;45:373–376. [Google Scholar]
- 19.Devos K M, Atkinson M D, Chinoy C N, Lloyd J C, Raines C A, Dyer T A, Gale M D. Theor Appl Genet. 1992;85:133–135. doi: 10.1007/BF00222849. [DOI] [PubMed] [Google Scholar]
- 20.Devos K M, Chinoy C N, Atkinson M D, Hansen L, von Wettstein-Knowles P, Gale M D. Theor Appl Genet. 1991;82:3–5. doi: 10.1007/BF00231269. [DOI] [PubMed] [Google Scholar]
- 21.Bennetzen J L, SanMiguel P, Chen M, Tikhonov A, Francki M, Avramova Z. Proc Natl Acad Sci USA. 1998;95:1975–1978. doi: 10.1073/pnas.95.5.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen, M., SanMiguel, P., de Oliveira, A. C., Woo, S. S., Zhang, H., Wing, R. A. & Bennetzen, J. L. (1998) Proc. Nat. Acad. Sci. USA, in press. [DOI] [PMC free article] [PubMed]
- 23.Foote T, Roberts M, Kurata N, Sasaki T, Moore G. Genetics. 1997;147:801–807. doi: 10.1093/genetics/147.2.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weeden N F, Muehlbauer F J, Ladizinsky G. J Hered. 1992;83:123–129. [Google Scholar]
- 25.Menancio-Hautea D, Fatokun C A, Kumar L, Danesh D, Young N D. Theor Appl Genet. 1993;86:797–810. doi: 10.1007/BF00212605. [DOI] [PubMed] [Google Scholar]
- 26.Lagercrantz U, Lydiate D J. Genetics. 1996;144:1903–1910. doi: 10.1093/genetics/144.4.1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lagercrantz U, Putterill J, Coupland G, Lydiate D. Plant J. 1996;9:13–20. doi: 10.1046/j.1365-313x.1996.09010013.x. [DOI] [PubMed] [Google Scholar]
- 28.Wolfe K H, Gouy M, Yang Y-W, Sharp P M, Li W-H. Proc Natl Acad Sci USA. 1989;86:6201–6205. doi: 10.1073/pnas.86.16.6201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Crane P R, Friis E M, Pedersen K R. Nature (London) 1995;374:27–33. [Google Scholar]
- 30.Van Deynze A E, Nelson J C, Yglesias E S, Harrington S E, Braga D P, McCouch S R, Sorrells M E. Mol Gen Genet. 1995;248:744–754. doi: 10.1007/BF02191715. [DOI] [PubMed] [Google Scholar]
- 31.O’Donoughue L S, Wang Z, Röder M S, Kneen B, Leggett M, Sorrells M E, Tanksley S D. Genome. 1992;35:765–771. [Google Scholar]
- 32.Pereira M G, Lee M, Bramel-Cox P, Woodman W, Doebley J, Whitkus R. Genome. 1994;37:236–243. doi: 10.1139/g94-033. [DOI] [PubMed] [Google Scholar]
- 33.Wang, Z. M., Devos, K. M., Liu, C. J. & Gale, M. D. (1997) Theor. Appl. Genet., in press.
- 34.Singh K, Ishii T, Parco A, Huang N, Brar D S, Khush G S. Proc Natl Acad Sci USA. 1996;93:6163–6168. doi: 10.1073/pnas.93.12.6163. [DOI] [PMC free article] [PubMed] [Google Scholar]